CHD-associated enhancers shape human cardiomyocyte lineage commitment
Figures
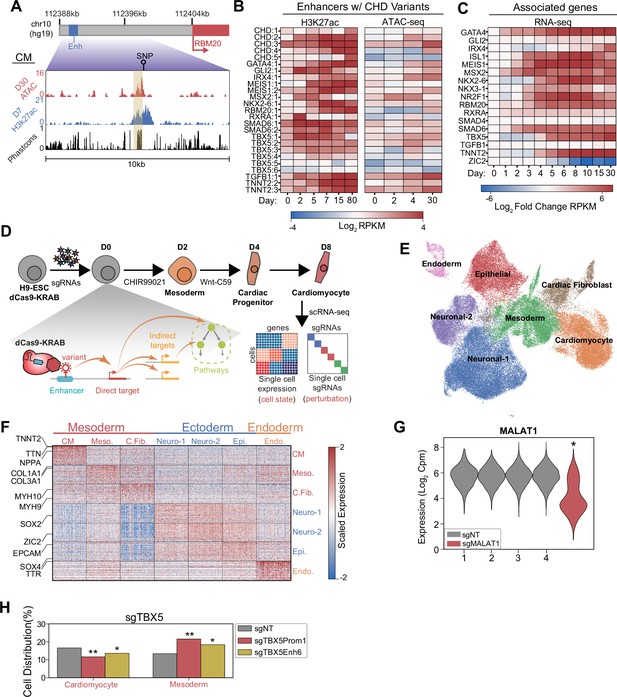
Single-cell screens of congenital heart defect (CHD)-associated enhancers during cardiomyocyte (CM) differentiation.
(A) Genome browser snapshot emphasizing features of a targeted enhancer about 15 kb upstream of the RBM20 gene. Yellow highlights enhancer region. (B) H3K27ac and open chromatin (ATAC-Seq) enrichment for targeted enhancers across multiple time points of CM differentiation (Liu et al., 2017; Tompkins et al., 2016; Zhang et al., 2019). (C) Expression of putative target genes across multiple stages of CM differentiation. Expression defined as fold change over the day 0 expression of a target gene (Tompkins et al., 2016). (D) Schematic of single-cell CRISPRi screen. H9-dCas9-KRAB cells are infected with a lentiviral sgRNA library and differentiated over 8 days into CMs followed by scRNA-seq. Individual cells are linked to sgRNA perturbations and changes in transcriptional cell state. (E) UMAP visualization of H9-derived cells after 8 days of differentiation. Seven Louvain clusters indicated. (F) Expression of the top 100 cluster defining genes for each Louvain cluster cell type. (G) MALAT1 expression in control (non-targeting) and sgMALAT1 cells (*p<0.05 by Mann-Whitney U). (H) Distribution of cells receiving sgNT, sgTBX5 PROM1, sgTBX5 ENH6 that differentiate into CM and mesoderm states (*p<0.05 and **p<0.001 by hypergeometric test).
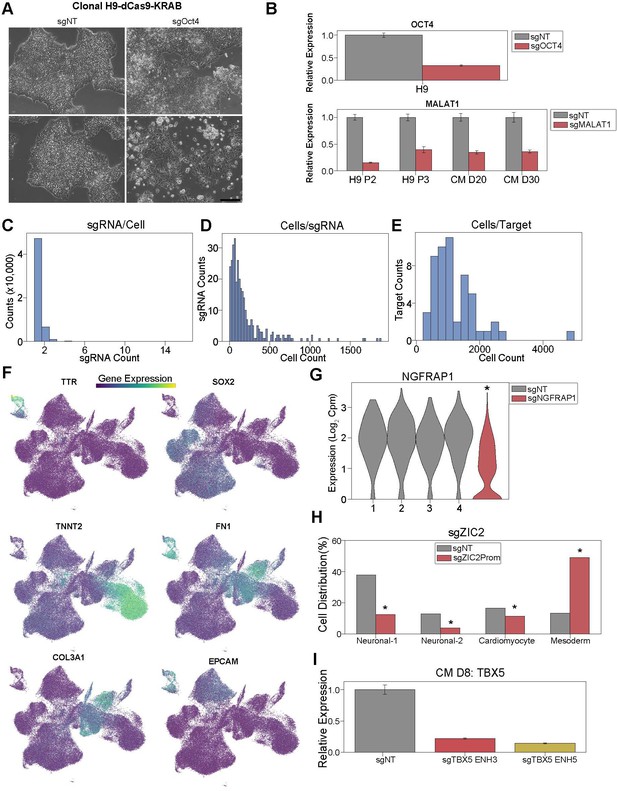
Single-cell CRISPRi screen validation and statistics.
(A) Brightfield images of H9-dCas9-KRAB cells receiving sgRNAs targeting OCT4 or NT control (Scale bar = 200μm). (B) (Top) OCT4 expression in cells receiving sgRNAs targeting OCT4 or NT control. For both, qPCR quantification normalized to reference gene (RPLP0) and then compared with control cells. (Bottom) MALAT1 expression in cells receiving sgRNAs targeting MALAT1 or NT control (n = 3 technical replicates). (C) Distribution of sgRNA counts in cells with sgRNAs detected. (D) Distribution of cell counts for each sgRNA. (E) Distribution of cell counts for each targeted region. (F) Feature plots of marker genes for neuronal (+SOX2), cardiomyocyte (+TNNT2), mesoderm (+FN1), epithelial (+EPCAM), cardiac fibroblast (+COL3A1), and endoderm cells (+TTR). (G) NGFRAP1 expression in cells receiving sgRNA targeting NGFRAP1 or NT controls (*p<0.05 by Mann-Whitney U). (H) Distribution of cells receiving sgRNAs targeting ZIC2 promoter or NT controls after differentiation (*p<0.05 by hypergeometric test). (I) TBX5 expression in cells receiving sgRNAs targeting TBX5 enhancer 3, enhancer 5, or non-targeting NT control (n = 3 technical replicates).
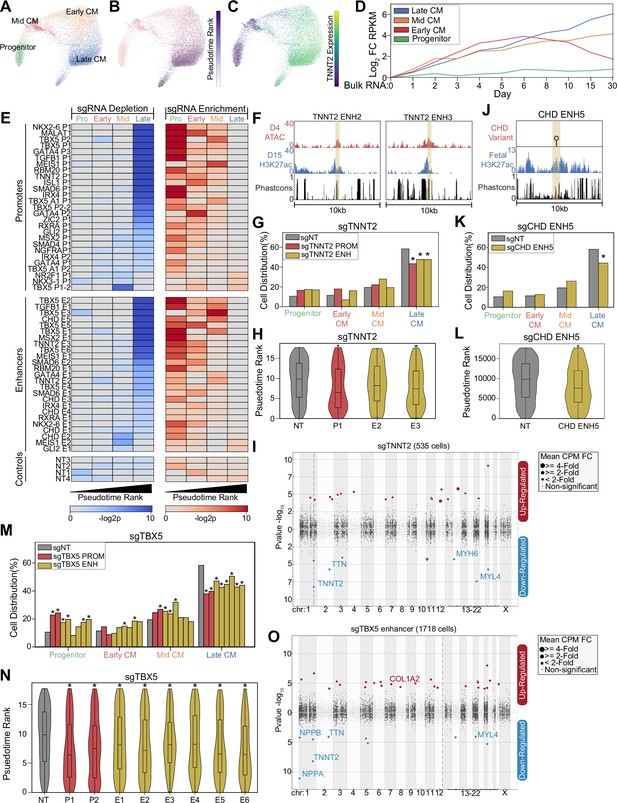
CRISPRi of congenital heart defect (CHD)-associated enhancers delays cardiomyocyte (CM) differentiation.
(A) PHATE visualization of CM cells with four Louvain clusters. (B) Feature plot of pseudotime ranking of cells across PHATE trajectory. (C) Feature plot of TNNT2 expression. We defined the top 100 genes for each of the four CM subtypes. Shown is the expression of these gene sets in bulk RNA-seq experiments of CM differentiation (Tompkins et al., 2016). Expression defined as fold change over day 0. (E) Enrichment (right) and depletion (left) of cells with distinct perturbations across CM subpopulations (p-values: hypergeometric test). Top: Targeted promoters; middle: targeted enhancers; bottom: non-targeting sgRNAs. (F) Genome browser tracks of chromatin and sequence conservation at two putative TNNT2 enhancers (ENH2 and ENH3). Yellow region denotes enhancer boundaries. (G) Distribution of states for cells receiving sgRNAs targeting TNNT2 promoter and enhancers (*p<0.05 by hypergeometric test). (H) Distribution of pseudotime rank for cells receiving sgRNAs targeting TNNT2 promoter and enhancers (*p<0.05 by Mann-Whitney U) (nNT = 983 cells, nP1 = 145 cells, nE2 = 86 cells, nE3 = 311 cells). (I) Differentially expressed genes in CM cells receiving sgRNAs targeting TNNT2 promoter or enhancers. In this Manhattan plot, the horizontal axis indicates genomic coordinates, with the dotted line indicating the targeted TNNT2 promoter. The vertical axis indicates differential expression (p-value), with positive values representing increased expression and negative values representing decreased expression. (J) Genome browser track of fetal human heart H3K27ac for CHD ENH5. Yellow region denotes enhancer boundaries. (K) Distribution of states for cells receiving sgRNAs targeting CHD ENH5 (*p<0.05 by hypergeometric test). (L) Distribution of pseudotime rank for cells receiving sgRNAs targeting CHD ENH5 (*p<0.05 by Mann-Whitney U) (nNT = 983 cells, nE5 = 281 cells). (M) Distribution of states for cells receiving sgRNAs targeting TBX5 promoters and enhancers (*p<0.05 by hypergeometric test). (N) Distribution of pseudotime rank for cells receiving sgRNAs targeting TBX5 promoter and enhancers (*p<0.05 by Mann-Whitney U) (nNT = 983 cells, nP1 = 179 cells, nP2 = 271 cells, nE1 = 348 cells, nE2 = 466 cells, nE3 = 437 cells, nE4 = 166 cells, nE5 = 205 cells, nE6 = 127 cells). (O) Differentially expressed genes in CM cells receiving sgRNAs targeting a TBX5 enhancer (as described in 2I).
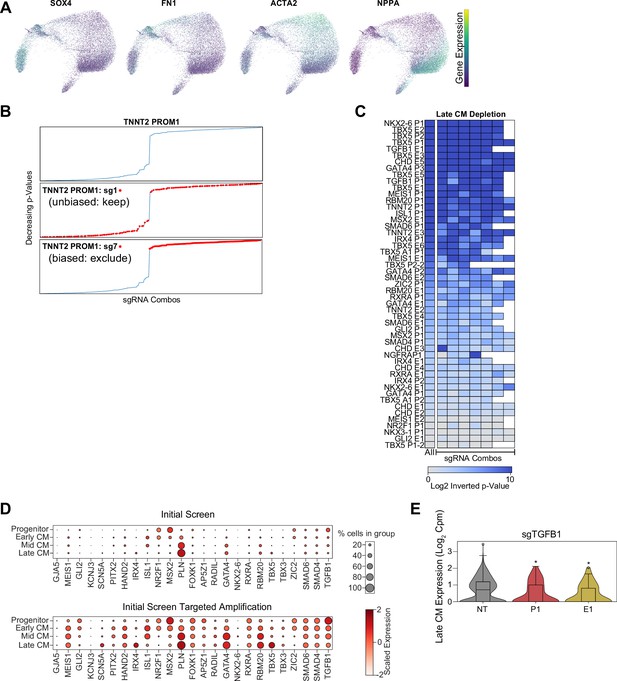
Single-cell CRISPRi screen cardiomyocyte (CM) subpopulation marker gene expression.
(A) Feature plots of marker genes for SOX4+ progenitors, FN1+ early-stage CMs, ACTA2+ mid-stage CMs, and NPPA+ atrial-like late-stage CMs. (B) Example of sgRNA filtering approach. (Top) p-Values (hypergeometric test) for late-CM depletion for all combinations of eight sgRNA targeting TNNT2 PROM1. (Middle) As above, but sgRNA combinations with TNNT2 PROM1 sgRNA 1 in red. This sgRNA does not bias p-value in either direction and is therefore kept for downstream analysis. (Bottom) As above, but sgRNA combinations with TNNT2 PROM1 sgRNA 7 in red. This sgRNA does bias p-values directionally, and is therefore excluded from downstream analysis. (C) To verify that multiple sgRNAs support the same CM differentiation phenotype, we performed n-1 analysis by removing each sgRNA in turn. In this heatmap, the left column represents cells with all sgRNAs targeting an enhancer or promoter. The right columns indicate p-values after removal of cells with individual sgRNAs (p-values: hypergeometric test). (D) (Top) Dotplot shows expression of cardiac genes across CM subpopulations. (Bottom) Expression of cardiac genes across CM subpopulations after targeted transcript amplification. (E) Expression of TGFB1 in late-CM cells receiving sgRNAs targeting TGFB1 promoter, enhancer, or non-genome targeting (NT) controls (*p<0.05 by Mann-Whitney U) (nNT = 967 cells, nP1 = 175 cells, nE1 = 111 cells).
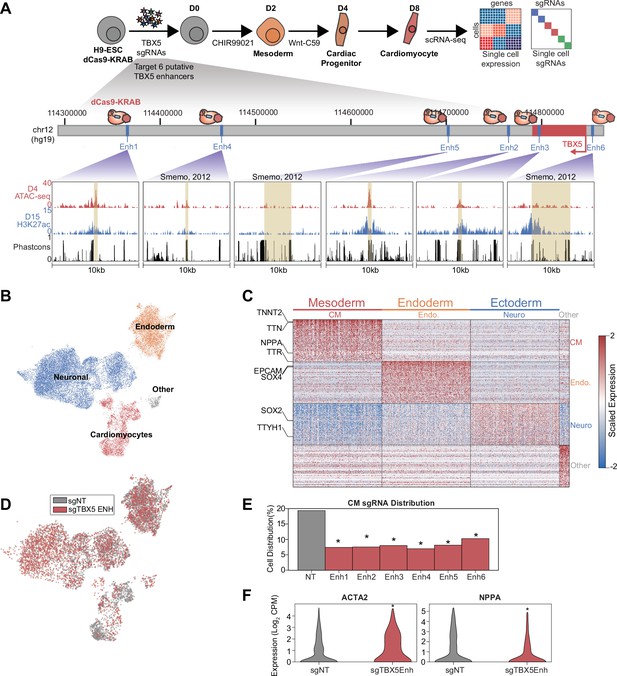
A focused validation screen demonstrates that TBX5 enhancers modulate cardiomyocyte (CM) cell fate.
(A) (Top) Schematic of validation CRISPRi screen. H9-dCas9-KRAB cells are infected with lentiviral sgRNA library targeting TBX5 enhancers and differentiated over 8 days into CMs followed by scRNA-seq. (Bottom) Genome browser tracks of chromatin status and sequence conservation for TBX5 enhancers. Yellow regions denote enhancers. (B) UMAP visualization of H9-derived cells after 8 days of differentiation. Four Louvain clusters indicated. (C) Expression of the top 100 cluster-defining genes for each Louvain cluster. (D) Feature plots of cells receiving sgRNAs targeting TBX5 enhancers (red) or non-targeting (NT) control (gray). (E) Distribution of cells receiving sgNT and sgTBX5 enhancers that differentiate into CMs (*p<0.05 by hypergeometric test). (F) Expression of ACTA2 (left) and NPPA (right) in sgTBX5 enhancer and control sgNT CMs (*p<0.05 by Mann-Whitney U).
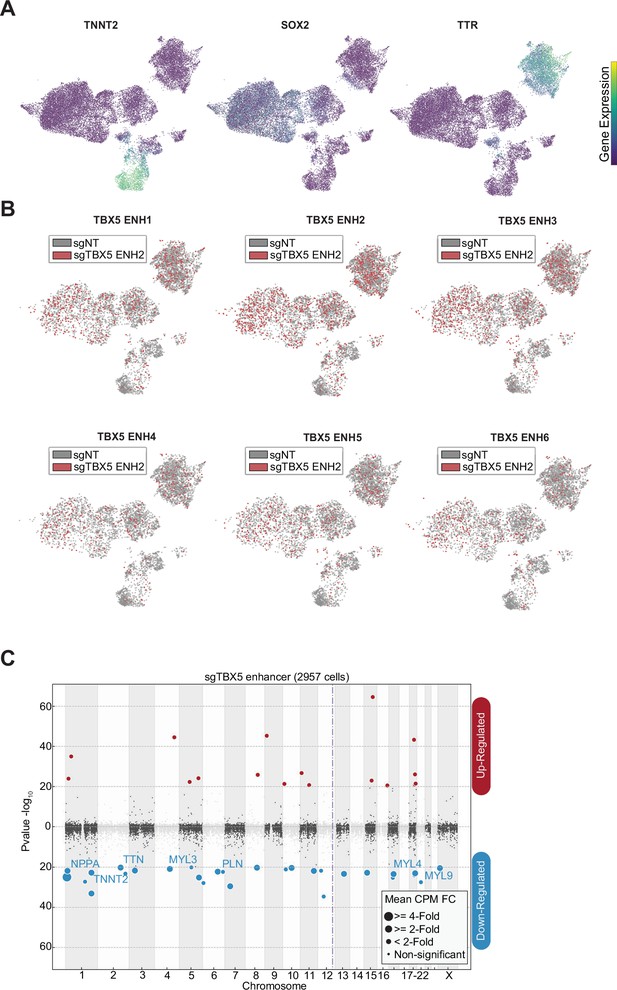
Focused screen marker expression and sgRNA distribution.
(A) Feature plots of marker genes for neuronal (+SOX2), cardiomyocytes (+TNNT2), endoderm (+TTR). (B) Feature plots of cells receiving sgRNAs targeting TBX5 enhancers (red) or non-genome targeting (NT) control (gray). (C) Differentially expressed genes in cells receiving sgRNAs targeting TBX5 enhancers. Please see description of Manhattan plot in Figure 2I.
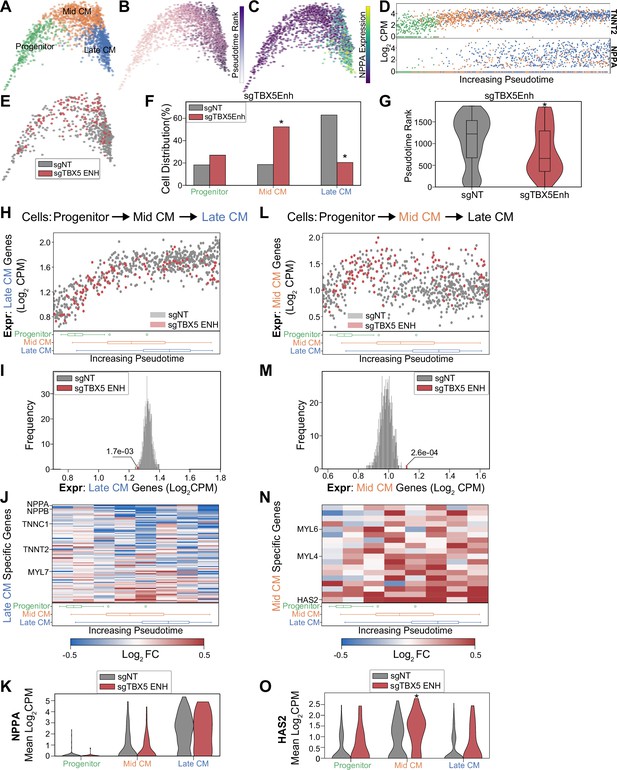
TBX5 enhancer repression alters cardiomyocyte (CM) molecular signatures.
PHATE visualization of CMs in the focused screen with three Louvain clusters. (B) Feature plot of pseudotime ranking of cells across PHATE trajectory. (C) Feature plot of NPPA expression. (D) Single-cell expression of TNNT2 (top) and NPPA (bottom) across pseudotime ordered CMs. (E) Feature plots of cells receiving sgRNAs targeting TBX5 enhancers or non-targeting non-genome targeting (NT) control. (F) Distribution of states for cells receiving sgRNAs targeting TBX5 enhancers or non-targeting NT control (*p<0.05 by hypergeometric test). (G) Distribution of pseudotime rank for cells receiving sgRNAs targeting TBX5 enhancers and NCs (*p<0.05 by Mann-Whitney U) (nNT = 558 cells, nTBX5 Enh = 122 cells). (H) (Top) We defined late-CM genes as those more expressed in late-CM cells. Shown is the average expression of late-CM genes in cells across pseudotime. Cells receiving sgRNAs targeting TBX5 enhancers (red) or non-targeting NT control (gray). (Bottom) Boxplots of CM subpopulation pseudotime ranks across pseudotime. (I) Average expression of late-CM genes across cells receiving sgRNAs targeting TBX5 enhancers (red) and 1000 random samplings of non-targeting control (sgNT) late-CM cells (gray) (*p<0.05 by Z-test). (J) (Top) For late-CM genes, shown is the relative expression in cells receiving sgRNAs targeting TBX5 enhancers compared with non-targeting NT control, across pseudotime. (Bottom) Boxplots of CM subpopulation pseudotime ranks across pseudotime. (K) NPPA expression in cells receiving sgRNAs targeting TBX5 enhancers and NC. (L) (Top) We defined mid-CM genes as those more expressed in mid-CM cells. Shown is the average expression of mid-CM genes in cells across pseudotime. Cells receiving sgRNAs targeting TBX5 enhancers (red) or non-targeting NT control (gray). (Bottom) Boxplots of CM subpopulation pseudotime ranks across pseudotime. (M) Averaged expression of mid-CM genes across cells receiving sgRNAs targeting TBX5 enhancers (red) and 1000 random samplings of non-targeting control (sgNT) late-CM cells (gray) (*p<0.05 by Z-test). (N) (Top) For mid-CM genes, shown is the relative expression in cells receiving sgRNAs targeting TBX5 enhancers compared with non-targeting NT control, across pseudotime. (Bottom) Boxplots of CM subpopulation pseudotime ranks across pseudotime. (O) HAS2 expression in cells receiving sgRNAs targeting TBX5 enhancers and non-targeting NT control.
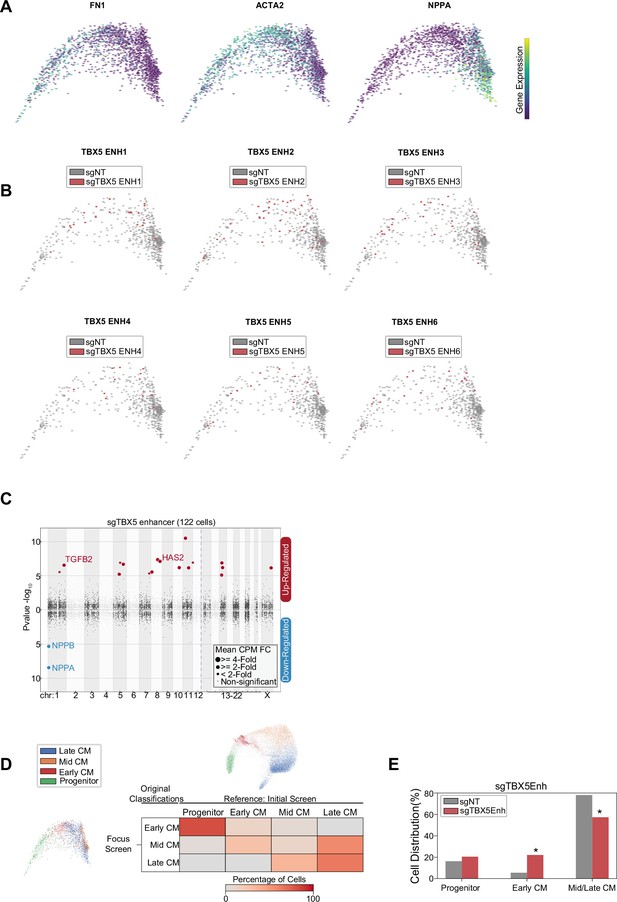
Focused screen cardiomyocyte (CM) subpopulation marker expression and sgRNA distribution.
(A) Feature plots of marker genes for CM progenitors (FN1+), mid-CM (ACTA2+), and late-CM (NPPA+). (B) Feature plots of cells receiving sgRNAs targeting TBX5 enhancers (red) or non-genome targeting (NT) control (gray). (C) Differentially expressed genes in CM cells receiving sgRNAs targeting TBX5 enhancers. Please see description of Manhattan plot in Figure 2I. (D) Reclustering of focused screen CM cells through cell label transfer using initial screen as reference. (E) Distribution of cells receiving sgRNAs targeting TBX5 enhancers across label transferred CM subpopulations (*p<0.05 by hypergeometric test).
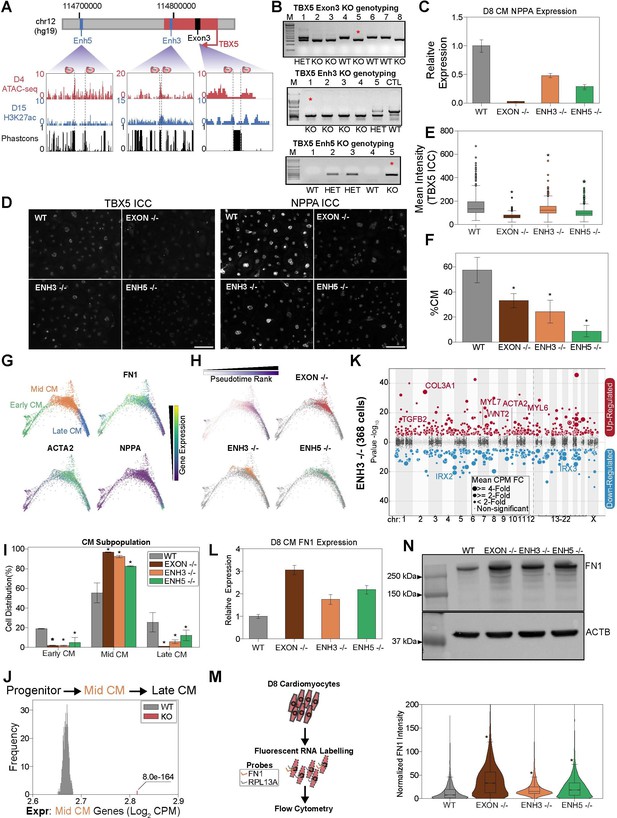
TBX5 enhancer knockouts recapitulate CRISPRi phenotypes.
(A) (Top) TBX5 enhancer knockout strategy. (Bottom) Genome browser snapshots of chromatin status and sequence conservation. Dotted lines denote sgRNA target sites. (B) Genotyping PCR to verify TBX5 knockouts of exon 3 (top), enhancer 3 (middle), and enhancer 5 (bottom). Red asterisk indicates clones used in downstream analysis. (C) NPPA transcript expression in exon and enhancer knockout cells after 8 days of cardiomyocyte (CM) differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with WT cells (n = 3 technical replicates and 2 biological replicates). (D) ICC for TBX5 (left) and NPPA (right) in WT and TBX5 exon 3, enhancer 3, and enhancer 5 knockout cells (Scale bar = 100μm). (E) Quantification of TBX5 ICC (mean intensity) across TBX5 genotyping (*p<0.05 by Mann-Whitney U) (nWT = 2003 cells, nEXON -/- = 780 cells, nENH3 -/- = 1357 cells, nENH5 -/- = 1592 cells). (F) Distribution of WT, TBX5 enhancer KO, or exon KO cells that differentiate into CMs relative to endoderm population (*p<0.05 by hypergeometric test) (nWT = 2 biological replicates, nEXON -/- = 2 biological replicates, nENH3 -/- = 2 biological replicates, nENH5 -/- = 2 biological replicates). (G) (Top left) PHATE visualization of CMs with three Louvain clusters. (Other quadrants) Feature plots of FN1, ACTA2, and NPPA expression. (H) (Top left) Feature plot of pseudotime ranking of CM cells across PHATE trajectory. (Other quadrants) Distribution of TBX5 exon KO and enhancer KO cells across CM trajectory. WT: gray. (I) Distribution of WT, TBX5 exon KO, and enhancer KO cells across three CM subpopulations (*p<0.05 by hypergeometric test) (nWT = 2 biological replicates, nEXON -/- = 2 biological replicates, nENH3 -/- = 2 biological replicates, nENH5 -/- = 2 biological replicates). (J) Averaged expression of mid-CM genes across TBX5 exon and enhancer KO cells (red) and 1000 random samplings of WT mid-CM cells (gray) (*p<0.05 by Z-test). (K) Differentially expressed genes in enhancer 3 KO cells in CM states. Please see description of Manhattan plot in Figure 2I. (L) FN1 transcript expression in exon and enhancer knockout cells after 8 days of CM differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with WT cells (n = 3 technical replicates). (M) (Left) Overview of FlowFISH experiment. (Right) Flow cytometry of FN1 RNA FISH intensity normalized by control RPL13A intensity in TBX5 WT and KO lines (*p<0.05 by Mann-Whitney U) (nWT = 4312 cells, nEXON -/- = 4034 cells, nENH3 -/- = 11166 cells, nENH5 -/- = 12286 cells). (N) (Top) FN1 and ACTB (bottom) protein expression in WT, TBX5 exon, and enhancer KO cells after 8 days of CM differentiation.
-
Figure 5—source data 1
Original genotyping gels for panel B.
Enhancer 3 KO genotyping gel, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data1-v2.zip
-
Figure 5—source data 2
Original genotyping gels for panel B.
Enhancer 3 KO genotyping gel, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data2-v2.zip
-
Figure 5—source data 3
Original genotyping gels for panel B.
Enhancer 5 KO genotyping gel, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data3-v2.zip
-
Figure 5—source data 4
Original genotyping gels for panel B.
Enhancer 5 KO genotyping gel, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data4-v2.zip
-
Figure 5—source data 5
Original genotyping gels for panel B.
TBX5 exon 3 KO genotyping gel, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data5-v2.zip
-
Figure 5—source data 6
Original genotyping gels for panel B.
TBX5 exon 3 KO genotyping gel, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data6-v2.zip
-
Figure 5—source data 7
Original western blots for panel N. Western blot of actin control, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data7-v2.zip
-
Figure 5—source data 8
Original western blots for panel N. Western blot of actin control, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data8-v2.zip
-
Figure 5—source data 9
Original western blots for panel N. Western blot of FN1, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data9-v2.zip
-
Figure 5—source data 10
Original western blots for panel N. Western blot of FN1, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig5-data10-v2.zip
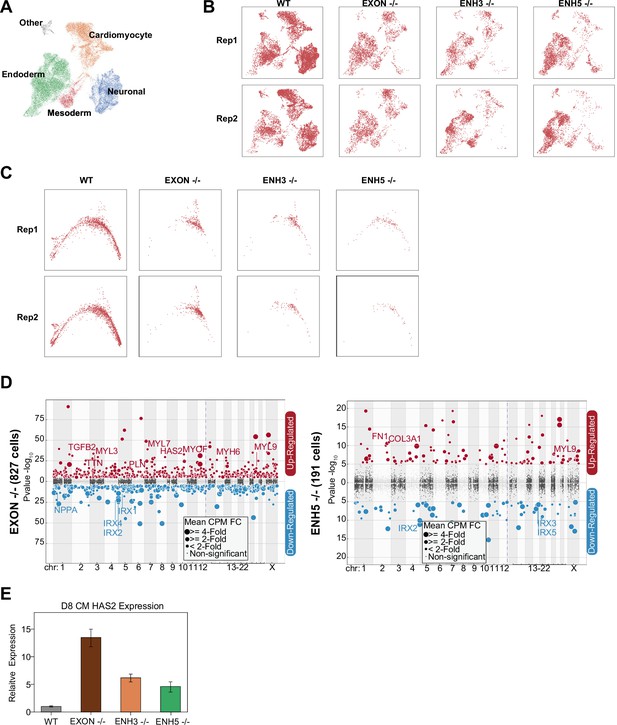
TBX5 enhancer knockout cell distribution.
(A) UMAP visualization of H9-derived cells after 8 days of differentiation. Five Louvain clusters indicated. (B) UMAP distribution of TBX5 exon KO and enhancer KO cells after cardiomyocyte (CM) differentiation. Shown are two biological replicates per genotype. (C) PHATE distribution of TBX5 exon and enhancer KO cells after CM differentiation. Shown are two biological replicates per genotype. (D) Differentially expressed genes in (left) TBX5 exon KO CM cells and (right) TBX5 enhancer 5 KO CM cells. (E) HAS2 transcript expression in exon and enhancer knockout cells after 8 days of CM differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with WT cells (n = 3 technical replicates).
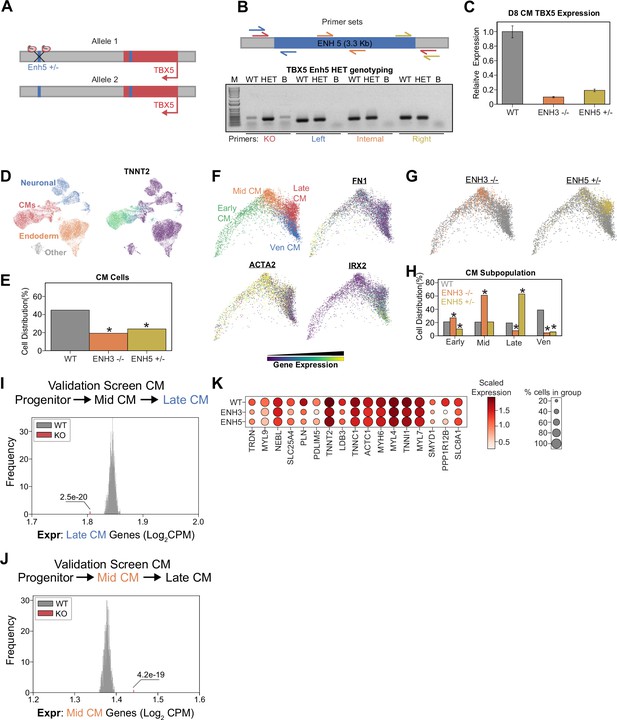
Heterozygous TBX5 enhancer 5 knockout (KO) displays reduced phenotypes.
(A) TBX5 enhancer 5 heterozygous KO strategy. (B) (Top) Position of primers used to validate TBX5 enhancer 5 heterozygous clone. Red: KO enhancer-spanning primers; blue: left junction primers; orange: enhancer internal primers; and yellow: right junction primers. (Bottom) Genotyping PCR to verify TBX5 enhancer 5 heterozygous deletion. Like the WT, the heterozygous clone retains left, internal, and right fragments, consistent with retaining a copy of enhancer 5. Unlike the WT, the heterozygous clone yields a small fragment when amplified with the KO enhancer-spanning primers. PCR conditions were not optimized for amplification of the WT large 3-kb+ fragment. WT: wild-type; HET: TBX5 enhancer 5 heterozygous clone; B: blank. (C) TBX5 transcript expression in enhancer KO cells after 8 days of cardiomyocyte (CM) differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with control cells (n = 3 technical replicates). (D) (Left) UMAP visualization of H9-derived cells after 8 days of differentiation. Four Louvain clusters indicated. (Right) Feature plot of TNNT2 expression. (E) Distribution of WT and TBX5 enhancer KO cells after differentiation (*p<0.05 by hypergeometric test). (F) (Top left) PHATE visualization of CM trajectory cells with four distinct Louvain clusters. (Other quadrants) Feature plot of FN1, ACTA2, and IRX2 expression. (G) Distribution of TBX5 enhancer KO cells across CM trajectory. WT: gray. (H) Cell distribution of TBX5 enhancer KO cells across four CM subpopulations (*p<0.05 by hypergeometric test). (I) Averaged expression of late-CM genes across TBX5 enhancer KO cells (red) and 1000 random samplings of WT late-CM cells (gray) (*p<0.05 by Z-test). (J) Averaged expression of mid-CM genes across TBX5 enhancer KO cells (red) and 1000 random samplings of WT late-CM cells (gray) (*p<0.05 by Z-test). (K) Dotplot shows the expression of cardiac genes in WT and KO cells belonging to the late-CM cluster.
-
Figure 6—source data 1
Original genotyping gels for panel B. Enhancer 5 HET genotyping, with labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig6-data1-v2.zip
-
Figure 6—source data 2
Original genotyping gels for panel B. Enhancer 5 HET genotyping, without labels.
- https://cdn.elifesciences.org/articles/86206/elife-86206-fig6-data2-v2.zip
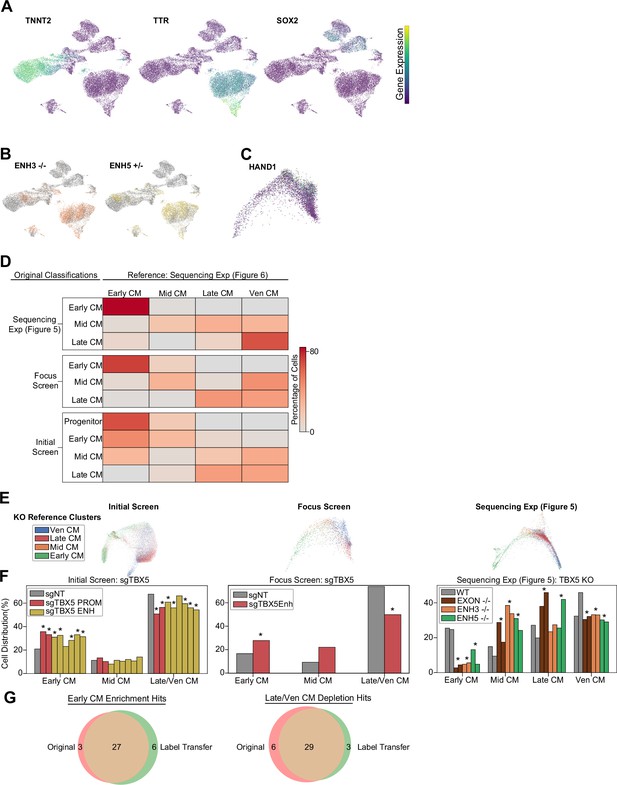
TBX5 heterozygous enhancer knockout validation.
(A) Feature plots of marker genes for neuronal (+SOX2), cardiomyocytes (CMs) (+TNNT2), endoderm (+TTR). (B) Feature plots of TBX5 enhancer 3 (orange), enhancer 5 (yellow), and WT (gray) CMs. (C) Feature plots of HAND1 expression. (D) Reclustering of CM cells from previous datasets through cell label transfer using sequencing exp (Figure 6) as reference. (E) PHATE distribution of CM cells clustered using four CM label transfer subpopulations. (F) Distribution of TBX5 enhancer perturbation cells across label transferred CM subpopulations (*p<0.05 by hypergeometric test). (G) Overlap of early-CM enriched (left) and late/ven-CM depleted (right) hits from original and label transfer clustering of initial screen CMs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Puromycin | Cayman Chemical | Cat#13884 | |
| Chemical compound, drug | Blasticidin | RPI | Cat#3513-03-9 | |
| Chemical compound, drug | Thiazovivin | Sigma-Aldrich | Cat#SML1045 | |
| Chemical compound, drug | TrypLE Select | Thermo Fisher | Cat#12563 | |
| Chemical compound, drug | CHIR99021 | Tocris | Cat#4423 | |
| Chemical compound, drug | Wnt-C59 | Cayman | Cat#16644 | |
| Chemical compound, drug | Accutase | Sigma-Aldrich | Cat#SCR005 | |
| Chemical compound, drug | Insulin | Gibco | Cat#12585014 | |
| Chemical compound, drug | B-27 | Thermo Fisher | Cat#17504044 | |
| Chemical compound, drug | Molecular Probes Fura-2 | Thermo Fisher | Cat#F1221 | |
| Chemical compound, drug | Pluronic F-127 | Thermo Fisher | Cat#P6867 | |
| Antibody | Anti-human-FN1 (Rabbit polyclonal) | Thermo Fisher | Cat#PA5-29578 | WB(1:1000) |
| Antibody | Anti-rabbit IgG | Cell Signaling Technology | CST #7074 | WB(1:400) |
| Chemical compound, drug | KAPA HiFi HS | KAPA | Cat#KK2502 | |
| Commercial assay, kit | PrimeFlow RNA Assay Kit | Thermo Fisher | Cat#88-18005-210 | |
| Chemical compound, drug | NEBNext High-Fidelity | New England Biolabs | Cat#M0541L | |
| Chemical compound, drug | Gibson Assembly Master Mix | New England Biolabs | Cat#E2611L | |
| Chemical compound, drug | mTeSR Plus | Stemcell Technologies | Cat#100–0276 | |
| Chemical compound, drug | Matrigel | Corning | Cat#354277 | |
| Commercial assay, kit | P3 Primary Cell 4D-Nucleofector X Kit | Lonza | Cat#V4XP-3024 | |
| Commercial assay, kit | 10× genomics Chromium Single Cell 3′ Kit V3.1 | 10× Genomics | Cat#PN-1000147 | |
| Commercial assay, kit | 10× CellPlex | 10× Genomics | Cat#PN-1000261 | |
| Commercial assay, kit | 10× Target Hybridization Kit | 10× Genomics | Cat#PN-1000248 | |
| Chemical compound, drug | RPMI 1640 | Thermo Fisher | Cat#11875093 | |
| Chemical compound, drug | KnockOut Serum | Thermo Fisher | Cat#10828028 | |
| Chemical compound, drug | HHBSS | Corning | Cat#21-023-CM | |
| Strain, strain background (Endura) | Endura ElectroCompetent Cells | Lucigen | Cat#60242–2 | Electrocompetent Cells |
| Strain, strain background (Escherichia coli) | Stellar Competent Cells | Clontech | Cat#636766 | |
| Other | Single-cell RNA-seq Data | This paper | GEO: GSE190475 | Sequencing data located at GEO |
| Cell line (Homo sapiens) | 293T cells | ATCC | CRL-3216 | Mycoplasma free; ATCC STR authenticated as CRL-3216 |
| Cell line (Homo sapiens) | H9 cells | WiCell | WA09 | Mycoplasma free; ATCC STR authenticated as WAe009-A-18 |
| Recombinant DNA reagent | Plasmid: pMD2.G | Addgene | RRID:Addgene_12259 | |
| Recombinant DNA reagent | Plasmid: psPAX2 | Addgene | RRID:Addgene_12260 | |
| Recombinant DNA reagent | Plasmid: lenti-dCas9-KRAB-Blast | Addgene | RRID:Addgene_89567 | |
| Recombinant DNA reagent | Plasmid: CROPseq-Guide-puro | Addgene | RRID:Addgene_86708 | |
| Sequence-based reagent | sgRNA oligos | This paper | sgRNA oligos | Supplementary file 1 |
| Sequence-based reagent | qPCR primers | This paper | qPCR primers | Supplementary file 2 |
| Software, algorithm | Star | PMID:23104886 | RRID:SCR_004463 | |
| Software, algorithm | Picard | Broad Institute | RRID:SCR_006525 | |
| Software, algorithm | FlowCal | PMID:27110723 | RRID:SCR_018140 | |
| Software, algorithm | FeatureCounts | DOI:10.1093/bioinformatics/btt656 | RRID:SCR_012919 | |
| Software, algorithm | 10× Genomics Cellranger | 10× Genomics | RRID:SCR_023221 | |
| Software, algorithm | Scanpy | PMID:29409532 | RRID:SCR_018139 | |
| Software, algorithm | IGV | Broad Institute | RRID:SCR_011793 | |
| Other | Illumina NextSeq 500 instrument | Illumina | Next-generation sequencer | |
| Other | Illumina NextSeq 2000 instrument | Illumina | Next-generation sequencer | |
| Other | Illumina NovaSeq 6000 instrument | Illumina | Next-generation sequencer | |
| Other | Agilent 2200 TapeStation instrument | Agilent | Automated electrophoresis instrument | |
| Other | Qubit Fluorometric Quantitation instrument | Thermo Fisher | Flurometer | |
| Other | EVOS FL Auto Imaging System | Thermo Fisher | Microscope |
Additional files
-
Supplementary file 1
List of target enhancers and sequences used in study.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp1-v2.xlsx
-
Supplementary file 2
Sequencing statistics.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp2-v2.xlsx
-
Supplementary file 3
Top 100 Louvain cluster defining genes for each dataset.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp3-v2.xlsx
-
Supplementary file 4
Differentially expressed genes through hypergeometric test.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp4-v2.xlsx
-
Supplementary file 5
Focus screen mid-CM and late-CM defining genes.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp5-v2.xlsx
-
Supplementary file 6
Differentially expressed genes between TBX5 enhancer 5 heterozygous KO cells and WT cells, in the late-CM state.
- https://cdn.elifesciences.org/articles/86206/elife-86206-supp6-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86206/elife-86206-mdarchecklist1-v2.docx





