The seminal odorant binding protein Obp56g is required for mating plug formation and male fertility in Drosophila melanogaster
Figures
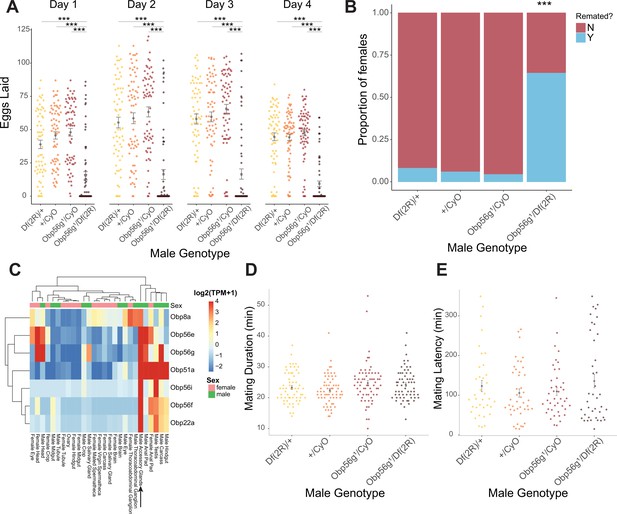
Seminal Obp gene expression and fecundity/remating defects in females mated to Obp56g1 null males.
(A) Egg counts from CS females mated to Df(2 R)/+, CyO/+Obp56 g1/CyO, or Obp56g1/Df(2 R) males from 1 to 4 days after mating. Significance indicated from pairwise comparisons of male genotypes within days using emmeans on a Poisson linear mixed effects model. Error bars represent mean +/-SEM. (B) Proportion of females who did or did not remate with a standard CS male on the fourth day after mating within a one-hour timeframe. Significance indicated from tests of equality of proportions. (C) Median centered log2 normalized TPM values for the seven seminal Obp genes in adult tissues from FlyAtlas2.0 bulk RNAseq data. Arrow points to male accessory gland sample. (D) Mating duration of CS females with indicated males (all pairwise comparisons using emmeans p>0.05). (E) Mating latency of CS females with indicated males (all pairwise comparisons using emmeans p>0.05). For A,B, D, and E, n=61–65. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant.
-
Figure 1—source data 1
Remating counts and percentages for data shown in Figure 1B.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig1-data1-v2.xlsx
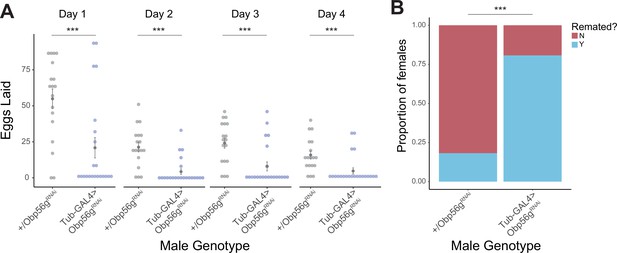
Whole body knockdown of Obp56g using Tubulin-GAL4 results in loss of post-mating response phenotypes in females.
(A) Counts of eggs from mated CS females over 4 days. Females mated to Tubulin-GAL4 >Obp56gRNAi males lay significantly fewer eggs than females mated to control males (p<0.001, n=20–24). Significance indicated from pairwise comparisons of male genotype within day using emmeans on a Poisson linear mixed effects model. (B) CS females mated to Tubulin-GAL4 >Obp56gRNAi males are significantly more likely to remate 4 days post-mating relative to control males (p<0.001, n=26–33). Significance indicated from Fisher’s exact test. Error bars represent mean +/-SEM. Significance level: * p<0.05, ** p<0.01, *** p<0.001.
-
Figure 1—figure supplement 1—source data 1
Remating counts and percentages for data shown in Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig1-figsupp1-data1-v2.xlsx
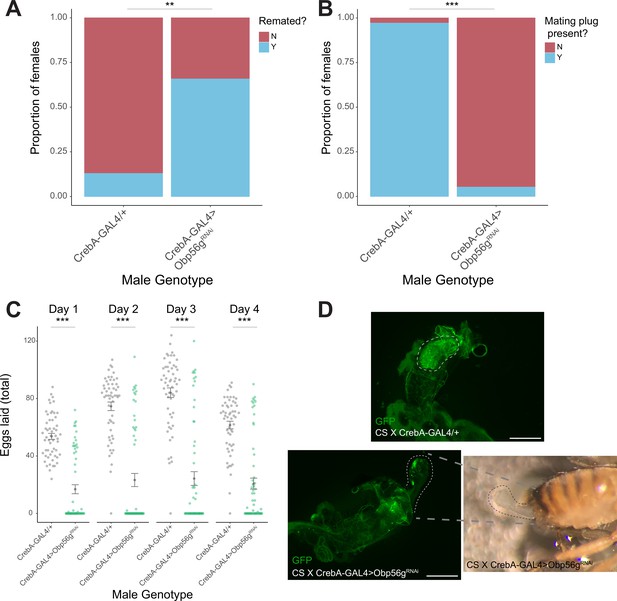
Male reproductive tract knockdown of Obp56g with CrebA-GAL4 is required for the post-mating response and mating plug formation.
(A) CS females mated to CrebA-GAL4 >Obp56gRNAi males are significantly more likely to remate 4 days post-mating relative to control males (p=0.001, n=55–60). (B) A significantly reduced proportion of females mated to CrebA-GAL4 >Obp56gRNAi males have fully formed mating plugs in their bursa immediately after the end of mating relative to females mated to control males (p<0.001, n=35–40). (C) Counts of eggs from mated CS females over 4 days. CS females mated to CrebA-GAL4 >Obp56gRNAi lay significantly fewer numbers of eggs relative to CS females mated to control males (p<0.001, n=55–60). (D) Fluorescent image of bursa from CS female mated to control (top) or knockdown (bottom) males. Dotted line shows position of WT mating plug (top) or ejaculate loss/uncoagulated mating plug from the bursa observed in females mated to CrebA-GAL4 >Obp56gRNAi males. Scale bar = 130 µm. Error bars represent mean +/-SEM. Significance level: * p<0.05, ** p<0.01, *** p<0.001.
-
Figure 1—figure supplement 2—source data 1
Counts and percentages for data shown in Figure 1—figure supplement 2A and B.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig1-figsupp2-data1-v2.xlsx
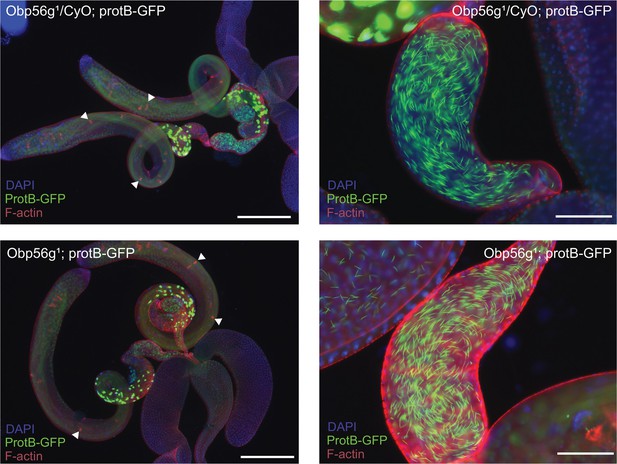
Spermatogenesis appears normal in Obp56g1;ProtB-eGFP males relative to Obp56g1/CyO;ProtB-eGFP control males.
Left: testes, accessory glands, and ejaculatory ducts dissected from 3- to 5-day-old males. Tissue morphology and major stages of sperm development are present in both genotypes (white arrows point to progressing individualization cones). Right: seminal vesicles from males of each genotype have mature and individualized sperm present.
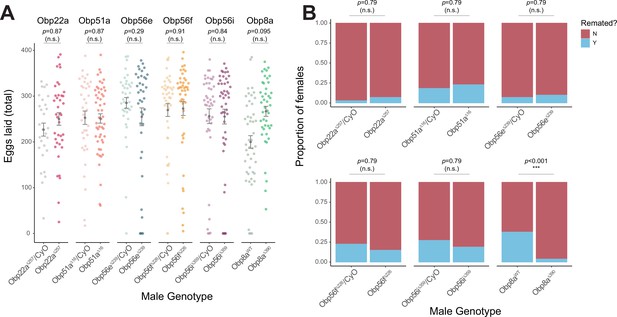
CRISPR/Cas9-generated mutants of Obp22a, Obp51a, Obp56e, Obp56f, Obp56i, and Obp8a have no or marginal effects on female fecundity and remating rates.
(A) Egg counts from CS females mated to homozygous null or heterozygous control males (except for Obp8a, the control of which is from an unedited sibling line) from 1 to 4 days after mating. Significance indicated from Poisson linear models with Benjamini-Hochberg corrections for multiple comparisons. Error bars represent mean +/-SEM. (B) Proportion of females who did or did not remate with a standard CS male on the fourth day after mating within a one-hour timeframe. Significance indicated from Fisher’s exact tests with Bejamini-Hochberg correction. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant. For A and B, n=28–51.
-
Figure 2—source data 1
Remating counts and percentages for data shown in Figure 2B.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig2-data1-v2.xlsx
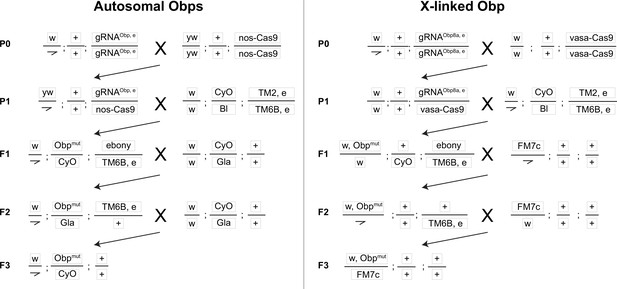
Crossing scheme to generate CRISPR mutants in autosomal (Obp22a, Obp51a, Obp56e, Obp56f, Obp56i) and X-linked (Obp8a) Obp genes used in this study, with text boxes representing chromosomes X/Y, 2, and 3 (dot chromosome not shown).
The kinked line represents the Y chromosome. Obp and ebony CRISPR editing takes place in the germline of individuals in the P1 generation. ebony editing can happen on either the gRNA or Cas9 chromosomes (written out in the F1 generation as “ebony” for simplicity), which are removed from the genetic background before assaying males for reproductive phenotypes.
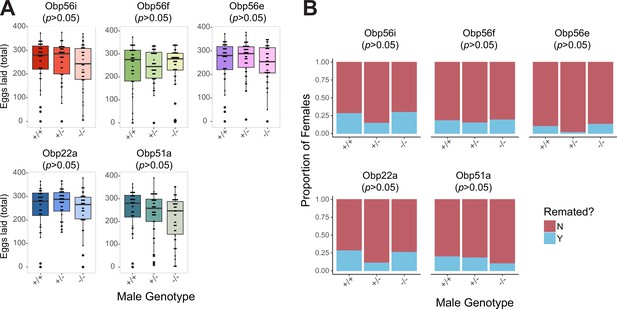
No effect of heterozygosity in PMR phenotypes relative to homozygous WT or homozygous CRISPR mutant males.
(A) Egg counts from CS females mated to +/+, +/-, -/- CRISPR mutant males from 1 to 4 days after mating. Significance indicated from pairwise comparisons of male genotypes within days using emmeans on a Poisson linear mixed effects model (p value adjusted for multiple comparisons, all p>0.05). (B) Proportion of female who did or did not remate with a standard CS male on the fourth day after mating. Significance indicated from pairwise Fisher’s exact tests (p value adjusted for multiple comparisons, all p>0.05). n for A and B=37–42. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant.
-
Figure 2—figure supplement 2—source data 1
Counts and percentages for data shown in Figure 2—figure supplement 2B.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig2-figsupp2-data1-v2.xlsx
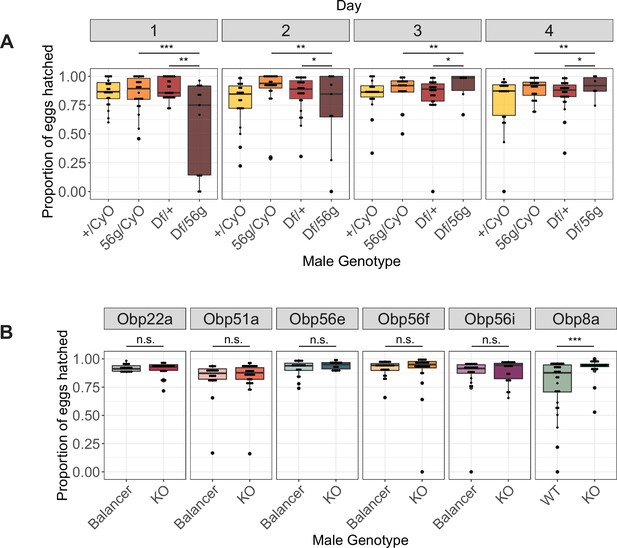
Box plots of hatchability estimates from CS females mated to Obp56g or CRISPR mutant males.
(A) Proportion of eggs hatched over 4 days from females mated to Df(2 R)/+, CyO/+Obp56 g1/CyO, or Obp56g1/Df(2 R) males. Significance indicated from pairwise comparisons of male genotypes across days using emmeans on a binomial mixed effects model. (B) Proportion of eggs hatched over 4 days from females mated to CRISPR mutant males. Significance indicated from binomial linear models with Benjamini-Hochberg corrections for multiple comparisons. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant.
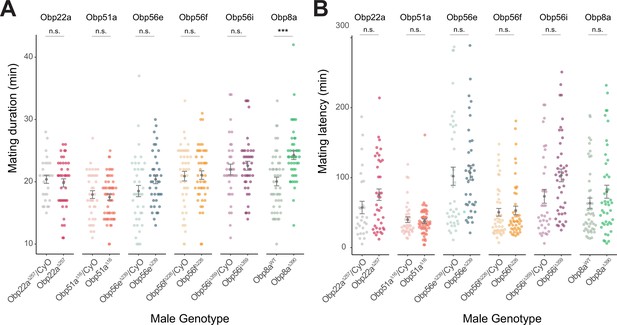
Mating latency and duration measurements from CRISPR-generated Obp mutants with CS females.
(A & B) Obp8aΔ390 mutant flies mate for longer duration than Obp8aWT control flies (p<0.001)though no other statistically significant differences were observed between mating duration of mutant or control males for other genotypes Significance indicated for male genotype term from linear mixed effects models. n for each genotype ranged from 28 to 51. Error bars represent mean +/-SEM. Significance level: * p<0.05, ** p<0.01, *** p<0.001, n.s. not significant.
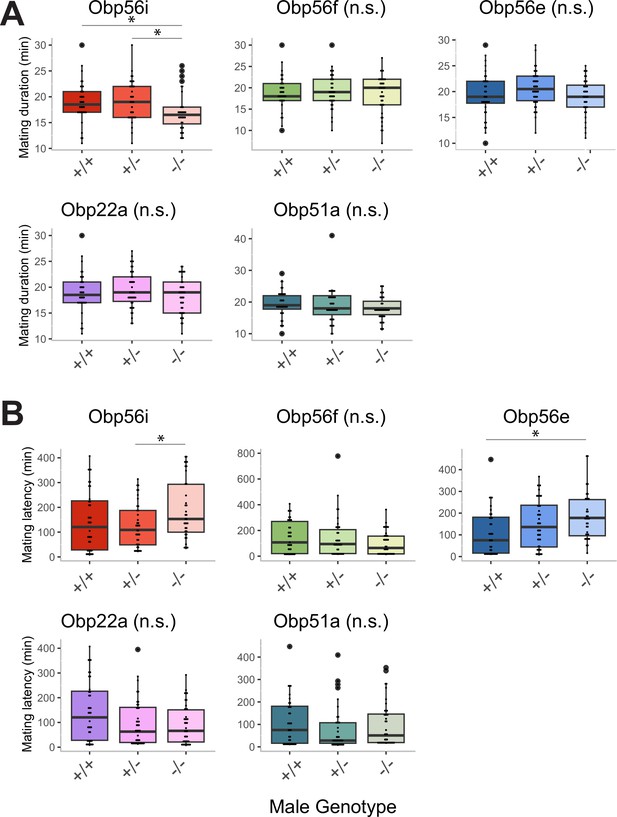
Mating duration (A) and latency (B) measurements from homozygous wildtype (+/+), heterozygous mutant (+/-) and homozygous CRISPR mutant (-/-) males mated to CS females.
Significance indicated from pairwise comparisons between male genotype using emmeans in a linear mixed effects model (p-value adjusted for multiple comparisons). n for A and B=37–42. Significance level: * p<0.05, ** p<0.01, *** p<0.001, n.s. not significant.
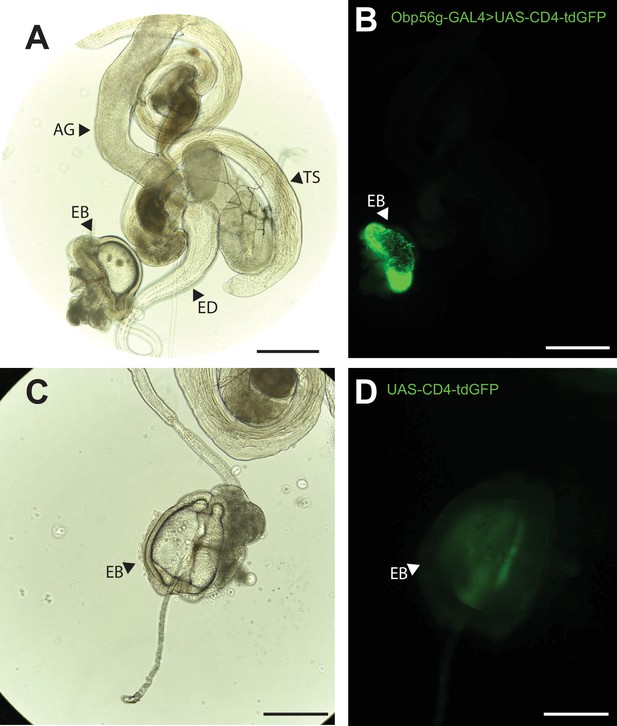
Obp56g is expressed in the Drosophila male ejaculatory bulb of the reproductive tract.
(A) Brightfield and (B) GFP fluorescent microscopy image of a reproductive tract dissected from a Obp56g-GAL4>UAS-CD4-tdGFP male, where the following tissues are labeled: AG, accessory gland. TS, testes. ED, ejaculatory duct. EB, ejaculatory bulb. (C) Brightfield and (D) GFP fluorescent microscopy images from UAS-CD4-tdGFP control males, showing only the EB portion of the tract. Scale bars in A&B=130 um, C&D=70 µm.
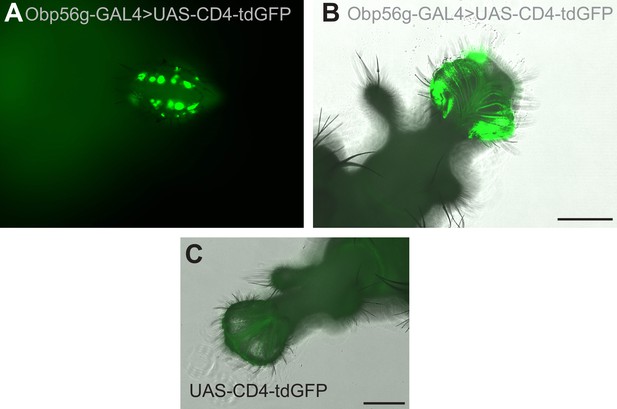
Expression of Obp56g-GAL4 in the gustatory bristles of the labellum.
(A) GFP expression from Obp56g-GAL>UAS-CD4-tdGFP males in the head. This sample is not placed under a coverslip. (B) GFP expression in the same genotype, with the proboscis dissected off and gently pressed under a coverslip. (C) GFP expression in UAS-CD4-tdGFP control male labellum. Scale bar = 130 µm.
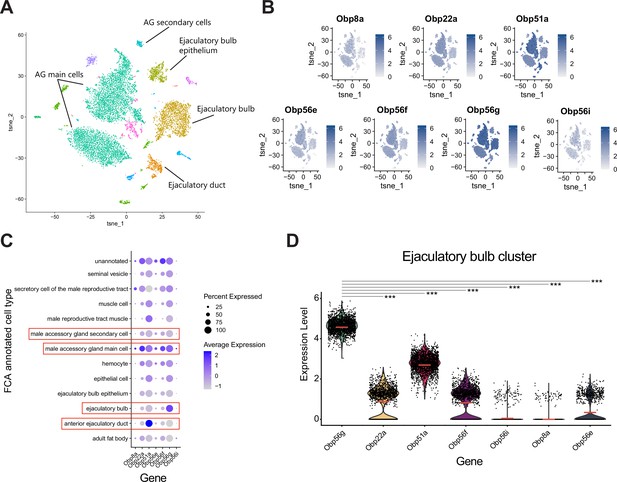
Obp56g is the most highly expressed seminal Obp in the ejaculatory bulb.
(A) Seurat tSNE dimensionality reduction plot of single nucleus RNAseq expression data from the male reproductive tract (without testes) and their major cell type annotations according to Li et al., 2022. (B) Feature plots from Seurat showing expression of the seminal Obp genes across single nuclei from A. (C) Seurat dot plot of scaled average gene expression across annotated cell types for seminal Obps. Dot size indicates the percentage of cells within a cluster that express each Obp gene. Red boxes indicate major tissues of the male reproductive tract that secrete SFPs (accessory gland main and secondary cells, ejaculatory bulb, and ejaculatory duct). (D) Violin plots of normalized expression level for each seminal Obp gene within the ejaculatory bulb cluster. Red line indicates the mean. Significance indicated from Wilcoxon rank sum tests of Obp56g vs. each other gene.
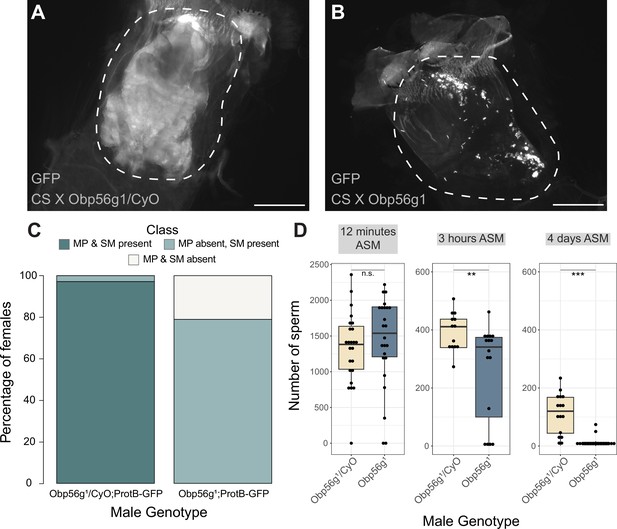
Females mated to Obp56g1 null males have defects in mating plug formation and sperm storage after mating.
(A) Fluorescent GFP microscopy image of the bursa of a CS female mated to a Obp56g1/CyO;ProtB-eGFP control male, with the mating plug surrounded by a dotted white line. Females were frozen in liquid nitrogen immediately after the end of mating. The mating plug is autofluorescent. (B) Fluorescent GFP microscopy image of the bursa of a CS female mated to a Obp56g1;ProtB-eGFP mutant male, where a similar region in the bursa as (A) is shown in the dotted white line. (C) Proportion of females mated to Obp56g1/CyO;ProtB-eGFP control or Obp56g1;ProtB-eGFP mutant males who had mating plugs or sperm masses present or absent immediately after the end of mating (n=35–38). MP, mating plug. SM, sperm mass. (D) Box plots of sperm counts in the storage organs of CS females mated to control (Obp56g1/CyO;ProtB-eGFP) or mutant (Obp56g1;ProtB-eGFP) males at 12 min, 3 hr, or 4 days (ASM, after the start of mating). n=13–24 for each group. Significance indicated from Student’s t-tests. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant. Scale bar = 130 µm.
-
Figure 4—source data 1
Counts and proportions for data shown in Figure 4C.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig4-data1-v2.xlsx
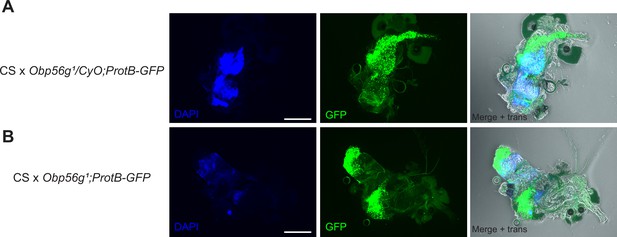
Obp56g1 mutant males do not have gross issues with sperm transfer during mating at the 12 min ASM time point.
(A) Reproductive tract of a representative CS female mated to Obp56g1/CyO;ProtB-eGFP control males, showing DAPI (mating plug), GFP (sperm heads), and merge +transillumination microscopy images and 9/9 females mated to these males had mating plugs, and 9/9 had sperm masses present in their bursas. (B) Reproductive tract of a representative CS female mated to Obp56g1;ProtB-eGFP mutant males. 0/10 females had mating plugs, though 10/10 had sperm masses present in their bursas.
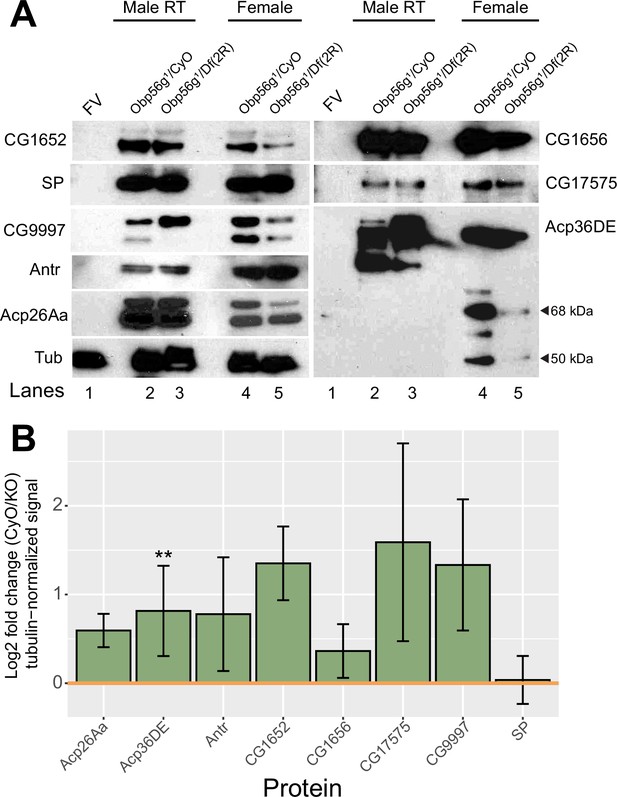
Western blot of major SFPs from CS females mated to Obp56g null and control males at 35 minutes ASM.
(A) Females mated to Obp56g1 null males have reduced amounts of Acp36DE in their bursas at 35 min ASM. Representative Western blots for SFPs in (lane 1) unmated female reproductive tracts (FV=female virgin), (lanes 2-3) male reproductive tracts, or (lanes 4-5) mated female reproductive tracts from CS females mated to either Obp56g1/CyO control or Obp56g1/Df(2 R) males at 35 min ASM. All flies are 3–5 days old. Tubulin is shown as a loading control. Cleavage products of Acp36DE (68 kDa and 50 kDa) are shown with black arrows. (B) Quantification of three replicate blots shown in A of log2 fold change of CyO/KO signal. Yellow line at zero indicates equal signal intensity for each genotype. Significance indicated from male genotype term in a linear model. Significance levels: *p<0.05, **p<0.01, ***p<0.001, n.s. not significant.
-
Figure 4—figure supplement 2—source data 1
Raw film images and uncropped, labeled western blots for data shown in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig4-figsupp2-data1-v2.zip
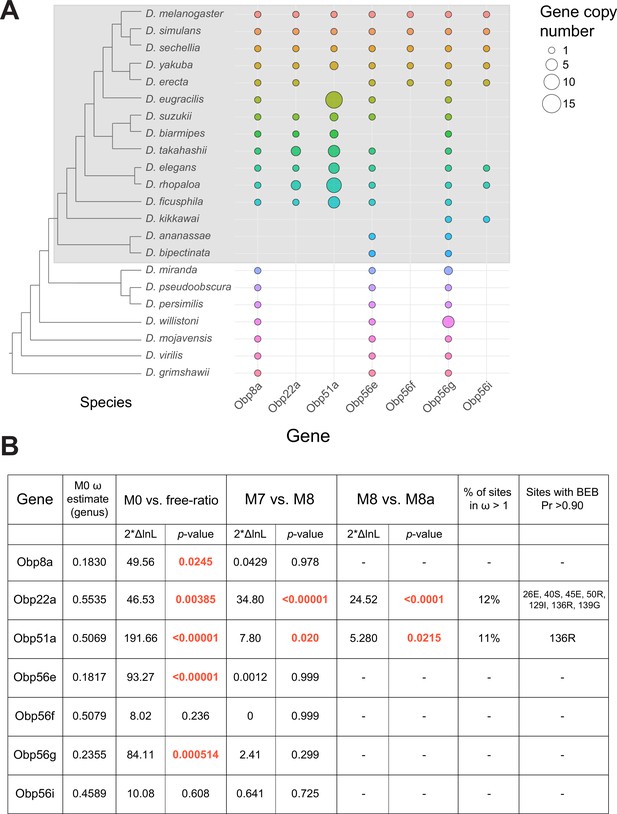
Dynamic changes in copy number, presence/absence, and evolutionary divergence rates of seminal Obp genes across the Drosophila genus.
(A) Inferred copy number of seminal Obp genes across Drosophila. Species without a dot represent an inferred loss based on syntenic analysis. Increased size of the dot represents increased gene copy number. Phylogeny on the left from McGeary and Findlay, 2020. Grey box surrounds species of the melanogaster group. (B) PAML results for the seminal Obp genes from analysis spanning the Drosophila genus (M0 ω estimate, M0 vs. free ratio test) or spanning the melanogaster group (M7 vs. M8, M8 vs. M8a tests). Bold and red text indicates statistically significant comparisons. Amino acid residues with >0.90 probability of being under positive selection are indicated, with the number/letter indicative of the D. melanogaster position within the alignment.
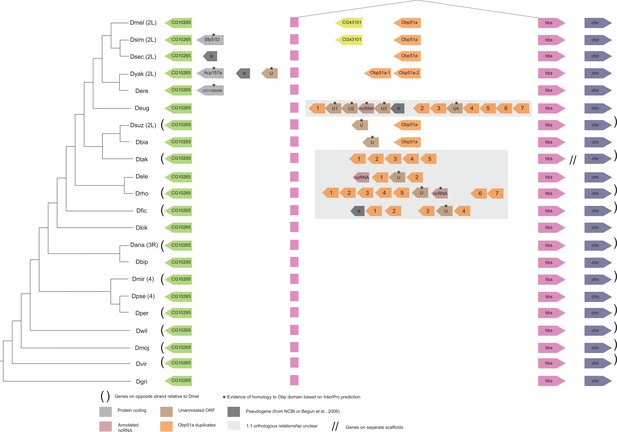
Synteny plot for Obp51a, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
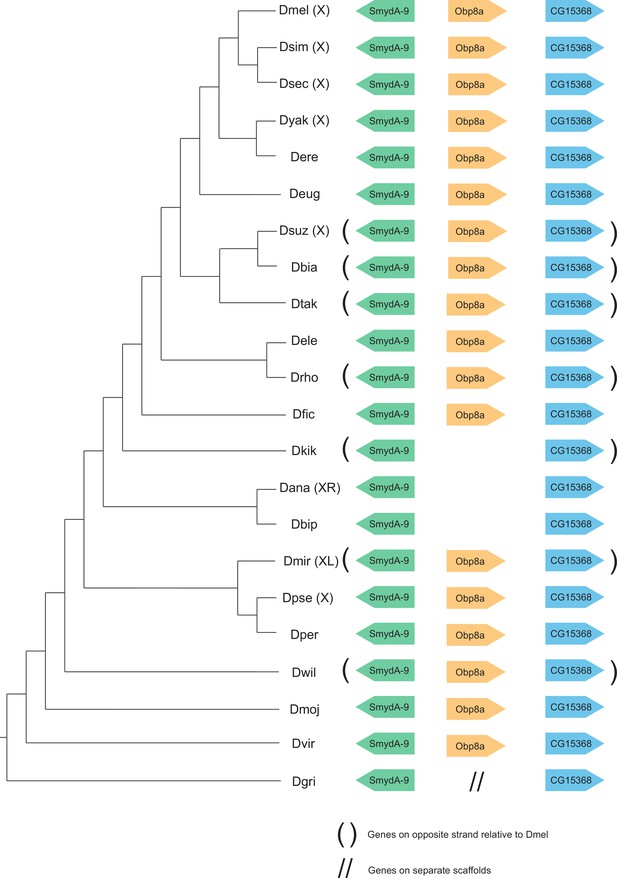
Synteny plot for Obp8a, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
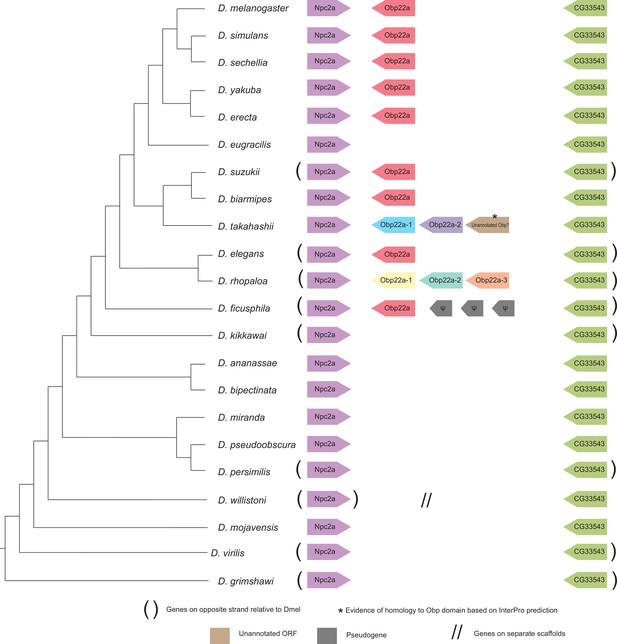
Synteny plot for Obp22a, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
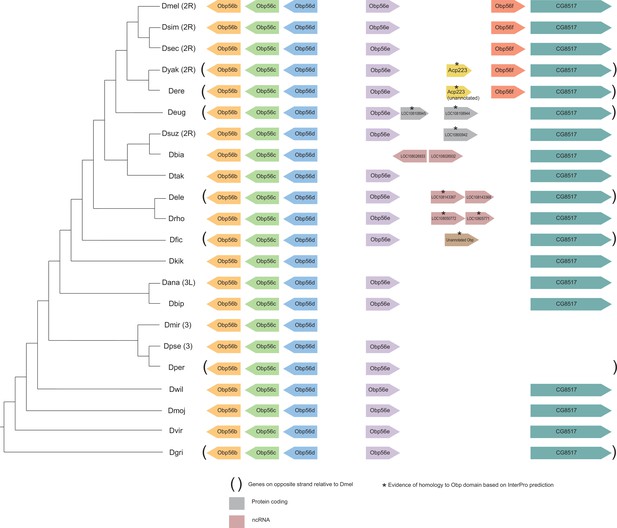
Synteny plot for Obp56e and Obp56f, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
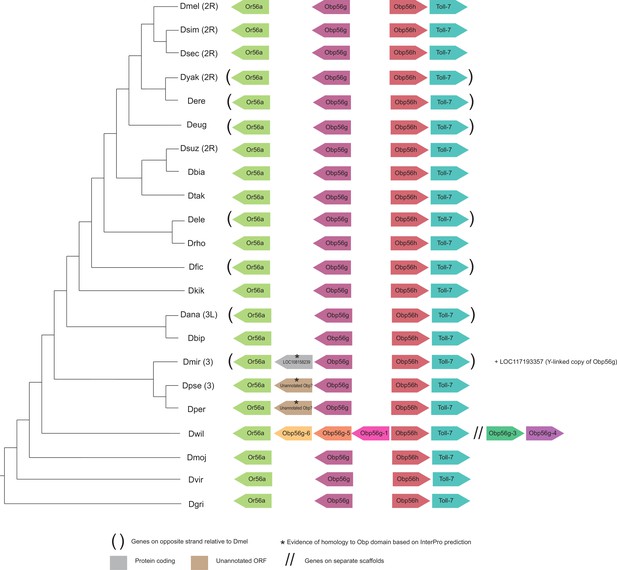
Synteny plot for Obp56g, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
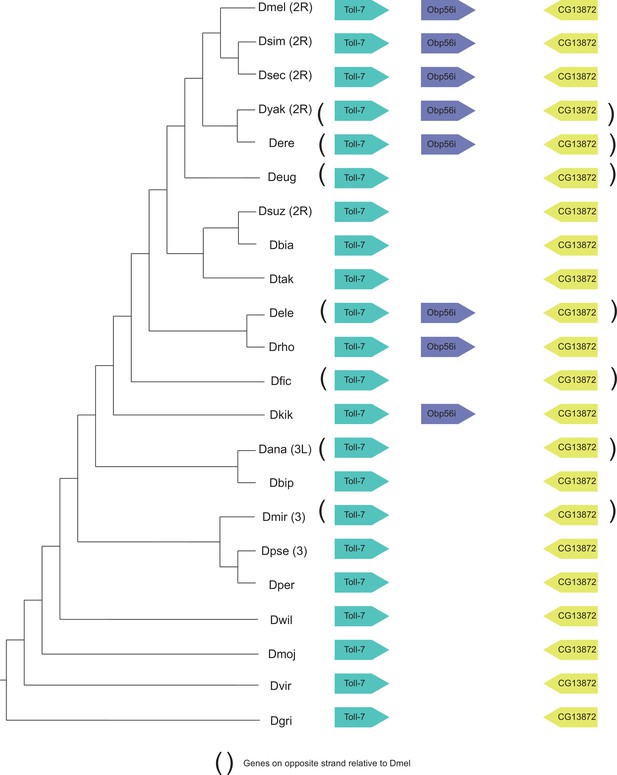
Synteny plot for Obp56i, phylogeny on the left from McGeary and Findlay, 2020.
Surrounding gene names represent gene names in D. melanogaster.
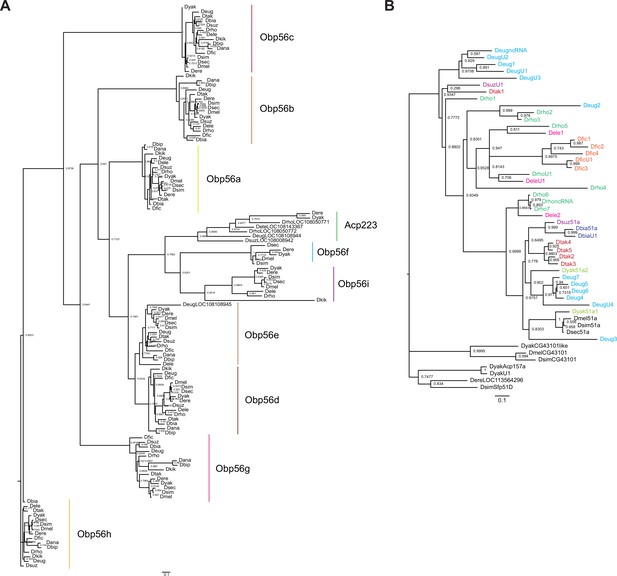
RAXML-NG maximum likelihood inferred trees for genes in the (A) Obp56 cluster across melanogaster group species, or (B) Obp51a cluster, where genes are colored as in Figure 5—figure supplement 1.
Node values are bootstrap support estimates based on 1,000 replicates. CG43101 is a gene located next to Obp51a in D. melanogaster, which has 6 cysteines in a pattern reminiscent of the Obp ‘domain’ but is not a predicted Obp based on InterProScan searches.
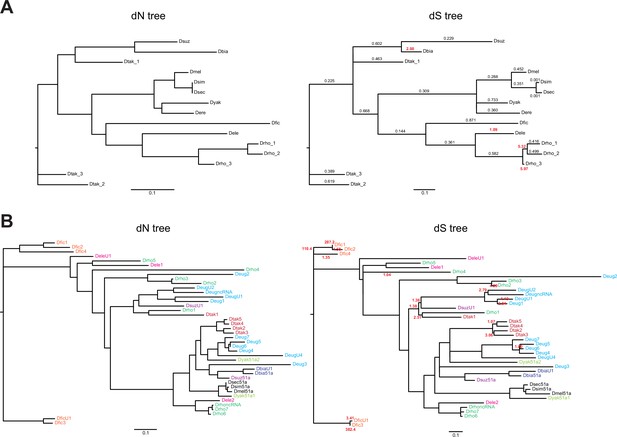
Gene trees for Obp22a and Obp51a.
(A) Obp22a and (B) Obp51a maximum likelihood inferred gene trees, where branch lengths are proportional to either estimates of dN (left) or dS (right) from PAML. Values indicated on the dS tree represent ML-inferred estimates of ω from PAML’s free ratio model, where the value is bold and red if ω>1. Values on the Obp51a dS tree are only shown if ω>1 for clarity. Genes in (B) are color-coded by species if more than one paralog is present in that species’ genomes.
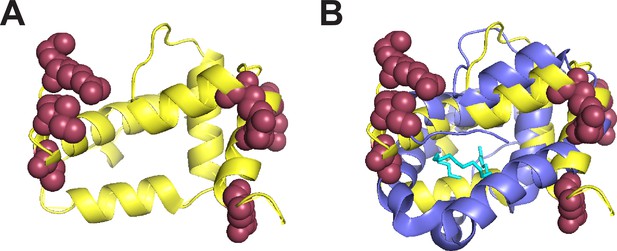
Positively selected sites in Obp22a cluster on the outward-facing region of the protein.
(A) Alphafold predicted protein structure (yellow) of Obp22a with the positively selected sites (Pr >0.90 BEB from model M8 of PAML) shown in maroon (Jumper et al., 2021). (B) The same structure as in A with a superimposed alignment of the crystal structure of Obp76a (LUSH) from Laughlin et al., 2008, (purple). The cyan molecule represents cVA and the inferred region of the binding pocket.
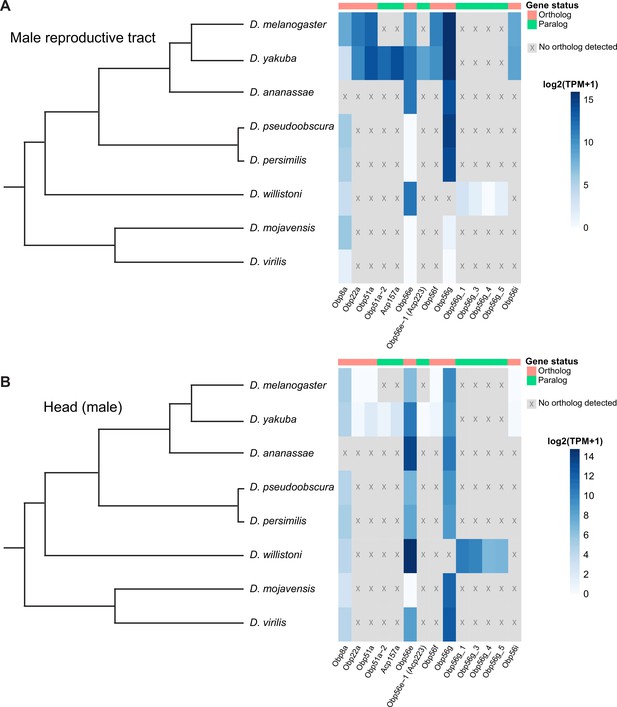
Seminal Obp genes show changes in expression pattern across species from bulk RNAseq data published in Yang et al., 2018.
(A) log2 normalized TPM expression values (averaged across four biological replicates) of seminal Obp genes and their associated orthologs and paralogs in male reproductive tissue (including accessory glands, ejaculatory duct, ejaculatory bulb, and terminal genitalia for all species except D. melanogaster, which includes all tissues aside from the genitalia) of different Drosophila species. Grey indicates that no ortholog could be detected in that species. (B) log2 normalized TPM expression values of seminal Obp gene orthologs and paralogs in male head tissue.
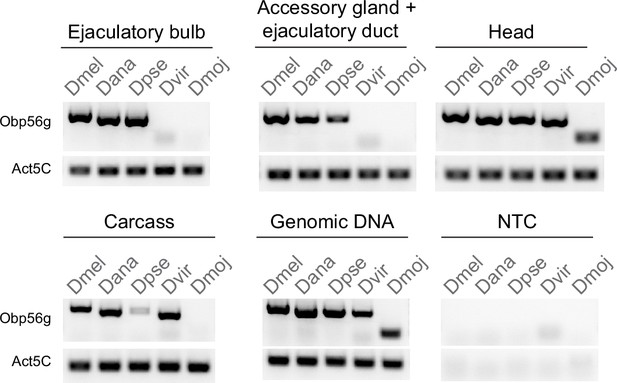
Semi-quantitative RT-PCR data from dissected tissues (head, accessory gland +ejaculatory duct, ejaculatory bulb, and carcass) from D. melanogaster (Dmel), D. ananassae (Dana), D. pseudoobscura (Dpse), D. virilis (Dvir), and D. mojavensis (Dmoj) males after 35 cycles of PCR. NTC = no template control.
-
Figure 6—figure supplement 1—source data 1
Raw and uncropped, labeled gel images for data shown in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/86409/elife-86409-fig6-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Obp56g | FlyBase | FLYB: FBgn0034474 | |
| Gene (D. melanogaster) | Obp56i | FlyBase | FLYB: FBgn0043532 | |
| Gene (D. melanogaster) | Obp56f | FlyBase | FLYB: FBgn0043533 | |
| Gene (D. melanogaster) | Obp56e | FlyBase | FLYB: FBgn0034471 | |
| Gene (D. melanogaster) | Obp22a | FlyBase | FLYB: FBgn0043539 | |
| Gene (D. melanogaster) | Obp51a | FlyBase | FLYB: FBgn0043530 | |
| Gene (D. melanogaster) | Obp8a | FlyBase | FLYB: FBgn0030103 | |
| Genetic reagent (D. melanogaster) | Obp56g1 | Bloomington Drosophila Stock Center | BDSC:55079; FBst0055079; RRID:BDSC_55079 | FlyBase genotype: w*; TI{GAL4}Obp56g1 |
| Genetic reagent (D. melanogaster) | UAS-CD4-tdGFP | Bloomington Drosophila Stock Center | BDSC:35836; FBst0035836; RRID:BDSC_35836 | FlyBase genotype: w1118; PBac{UAS-CD4-tdGFP}VK00033 |
| Genetic reagent (D. melanogaster) | Lhm; PBac{Ubnls-eGFP, ProtB-eGFP}(3) | Gift from John Belote and Scott Pitnik | ||
| Genetic reagent (D. melanogaster) | w;Df(2 R)/CyO | Bloomington Drosophila Stock Center | BDSC:25678; FBst0025678; RRID:BDSC_25678 | FlyBase genotype: w1118; Df(2 R)BSC594/CyO |
| Genetic reagent (D. melanogaster) | w;CyO/Bl;TM2/TM6B | Bloomington Drosophila Stock Center | BDSC:3704 (formerly, stock no longer available) | Genotype: w[1118]/Dp(1;Y)y[+]; CyO/Bl[1]; TM2/TM6B, Tb[1] |
| Genetic reagent (D. melanogaster) | nos-Cas9attP2 | BestGene/Shu Kondo & Ryu Ueda | Genotype: y1 w1118; attP2{nos-Cas9}/TM6C,Sb Tb | |
| Genetic reagent (D. melanogaster) | w;vasa-Cas9 | Bloomington Drosophila Stock Center | BDSC:51324; FBst0051324; RRID:BDSC_51324 | FlyBase genotype: w1118; PBac{vas-Cas9}VK00027 |
| Genetic reagent (D. melanogaster) | Obp56gRNAi | Vienna Drosophila Resource Center | VDRC:23206 (GD); FBst0454878 | FlyBase genotype: w1118; P{GD13268}v23206/TM3 |
| Genetic reagent (D. melanogaster) | CrebA-GAL4 | Bloomington Drosophila Stock Center | BDSC: 49409; FBst0049409; RRID:BDSC_49409 | FlyBase genotype: w1118; P{GMR64E07-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | Tubulin-GAL4 | Findlay et al., 2014; 10.1371/journal.pgen.1004108 | ||
| Genetic reagent (D. melanogaster) | FM7c | Gift from Susan Younger | Genotype: C(1)DX, y[1] w[1] f[1]/FM7c, Kr-GAL4[DC1], UAS-GFP[DC5], sn[+];;; | |
| Genetic reagent (D. melanogaster) | Phi-C31 integrase attP9A | Bloomington Drosophila Stock Center / Rainbow Transgenics | BDSC: 35569; FBst0035569; RRID:BDSC_35569 | FlyBase genotype: y1 w* P{nanos-phiC31\int.NLS}X; PBac{y+-attP-9A}VK00027 |
| Genetic reagent (Drosophila ananassae) | Wildtype (Cebu, Philippines) | National Drosophila Species Stock Center | SKU: 14024–0371.37 | |
| Genetic reagent (Drosophila pseudoobscura) | Genome line | National Drosophila Species Stock Center | SKU: 14011–0121.94 | |
| Genetic reagent (Drosophila mojavensis) | Wildtype (Chocolate Mountains) | National Drosophila Species Stock Center | SKU: 15081–1352.00 | |
| Genetic reagent (Drosophila virilis) | Genome line | National Drosophila Species Stock Center | SKU: 15010–1051.87 | |
| Software, algorithm | CRISPR Optimal Target Finder (flyCRISPR) | Gratz et al., 2014; http://doi.org/10.1534/genetics.113.160713 | ||
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp8a, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp8a and ebony | |
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp56e, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp56e and ebony | |
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp56f, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp56f and ebony | |
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp56i, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp56i and ebony | |
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp22a, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp22a and ebony | |
| Genetic reagent (D. melanogaster) | w;;gRNA(Obp51a, ebony) | This paper | Transgenic stock carrying gRNAs targeting Obp51a and ebony | |
| Genetic reagent (D. melanogaster) | Obp8aΔ390 | This paper | CRISPR deletion of 390 bp in exon 2 of Obp8a | |
| Genetic reagent (D. melanogaster) | Obp22aΔ257 | This paper | CRISPR deletion of 257 bp in exon 2 of Obp22a | |
| Genetic reagent (D. melanogaster) | Obp51aΔ16 | This paper | CRISPR deletion of 16 bp in exon 1 of Obp51a | |
| Genetic reagent (D. melanogaster) | Obp56eΔ239 | This paper | CRISPR deletion of 239 bp in exon 2 of Obp56e | |
| Genetic reagent (D. melanogaster) | Obp56fΔ226 | This paper | CRISPR deletion of 226 bp in exon 2 of Obp56f | |
| Genetic reagent (D. melanogaster) | Obp56iΔ359 | This paper | CRISPR deletion of 359 bp in exon 2 of Obp56i | |
| Antibody | Anti-SP (rabbit polyclonal) | Wolfner lab | 1:1000 | |
| Antibody | Anti-CG1656 (rabbit polyclonal) | Wolfner lab | 1:500 | |
| Antibody | Anti-CG1652 (rabbit polyclonal) | Wolfner lab | 1:250 | |
| Antibody | Anti-Acp36DE (rabbit polyclonal) | Wolfner lab | 1:12,000 | |
| Antibody | Anti-Acp26Aa (rabbit polyclonal) | Wolfner lab | 1:5000 | |
| Antibody | Anti-CG9997 (rabbit polyclonal) | Wolfner lab | 1:750 | |
| Antibody | Anti-Antr (rabbit polyclonal) | Wolfner lab | 1:750 | |
| Antibody | Anti-CG17575 (rabbit polyclonal) | Wolfner lab | 1:500 | |
| Antibody | Anti-tubulin (mouse monoclonal) | Sigma-Aldrich | T5168 | 1:4000 |
| Software, algorithm | RAxML-NG | Kozlov et al., 2019; http://doi.org/10.1093/bioinformatics/btz305 | ||
| Software, algorithm | MEGA-11 | Tamura et al., 2021; http://doi.org/10.1093/molbev/msab120 | ||
| Software, algorithm | PAML (v4.9) | Yang, 2007; https://doi.org/10.1093/molbev/msm088 | ||
| Software, algorithm | R (4.2.1) | https://www.R-project.org/ |
Additional files
-
Supplementary file 1
gRNA sequences from flyCRISPR’s Optimal Target Finder tool for each Obp gene.
- https://cdn.elifesciences.org/articles/86409/elife-86409-supp1-v2.docx
-
Supplementary file 2
Primer sequences for cloning gRNAs from Supplementary file 1 into pAC-U63-tgRNA-Rev using pMGC as a PCR template (from Poe et al., 2019).
- https://cdn.elifesciences.org/articles/86409/elife-86409-supp2-v2.docx
-
Supplementary file 3
Primer sequences used in this study.
- https://cdn.elifesciences.org/articles/86409/elife-86409-supp3-v2.docx
-
Supplementary file 4
CRISPR mutant allele summary for each Obp gene.
- https://cdn.elifesciences.org/articles/86409/elife-86409-supp4-v2.docx
-
Supplementary file 5
Proportion of CS females mated to CRISPR mutant males with morphologically normal mating plugs assessed immediately after the end of mating.
- https://cdn.elifesciences.org/articles/86409/elife-86409-supp5-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86409/elife-86409-mdarchecklist1-v2.pdf





