DYRK1A interacts with the tuberous sclerosis complex and promotes mTORC1 activity
Figures
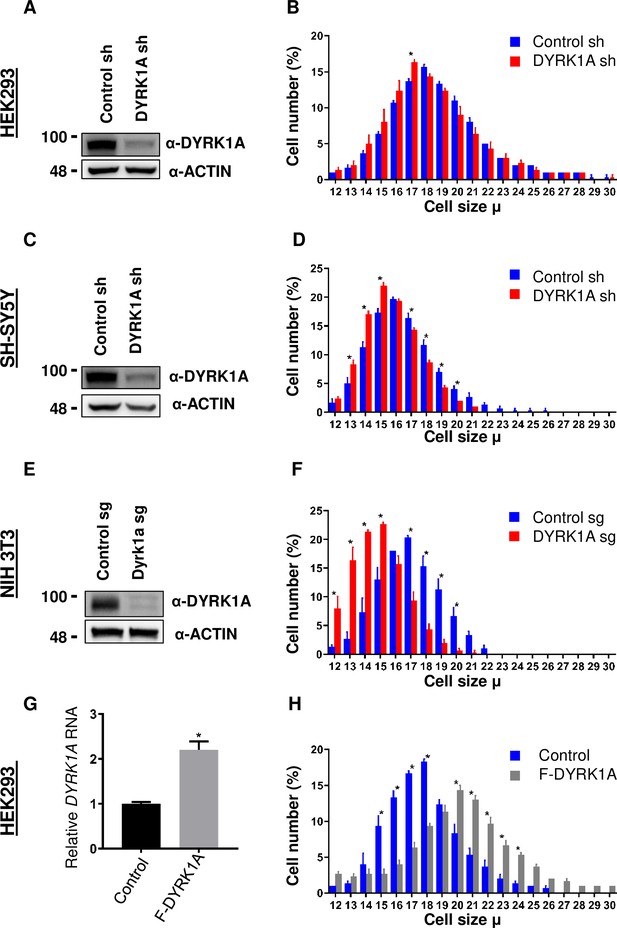
Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) regulates cell size.
shRNA-mediated knockdown of DYRK1A was performed in (A, B) HEK293 and (C, D) SH-SY5Y cells using lentivirus. Transduced cells were selected for four days before analysis. Western blot shows the efficiency of DYRK1A knockdown. (E, F) NIH3T3 cells were treated with Dyrk1a-targeting sgRNA expressing lentivirus and selected for four days before analysis. Western blot shows the efficiency of DYRK1A knockdown. (G, H) HEK293 cells expressing Flag-DYRK1A and the parental cells were treated with 40ng/ml Doxycycline for 48hr and analyzed for cell size. (G) Overexpression was analyzed by qRT-PCR. GAPDH mRNA was used to normalize RNA in q-RT-PCR samples. Data represent the mean ± SD (n=3 biological replicates).
-
Figure 1—source data 1
Uncropped blots for Figure 1A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw blots showing knockdown of DYRK1A for Figure 1A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data2-v1.zip
-
Figure 1—source data 3
Uncropped blots for Figure 1C.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data3-v1.zip
-
Figure 1—source data 4
Raw blots showing knockdown of DYRK1A for Figure 1C.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data4-v1.zip
-
Figure 1—source data 5
Uncropped blots for Figure 1E.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data5-v1.zip
-
Figure 1—source data 6
Raw blots showing knockdown of Dyrk1a for Figure 1E.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig1-data6-v1.zip
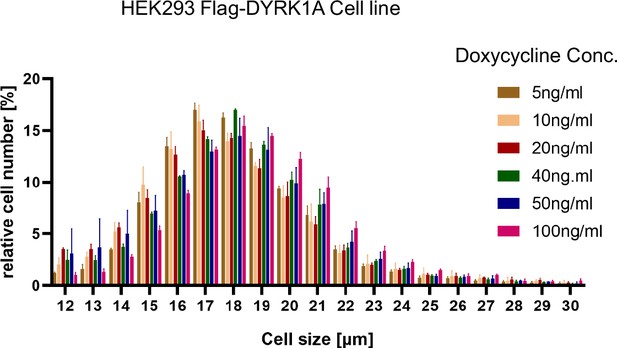
Analysis of cell size after induction of dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) expression with increasing dosage of Doxycycline.
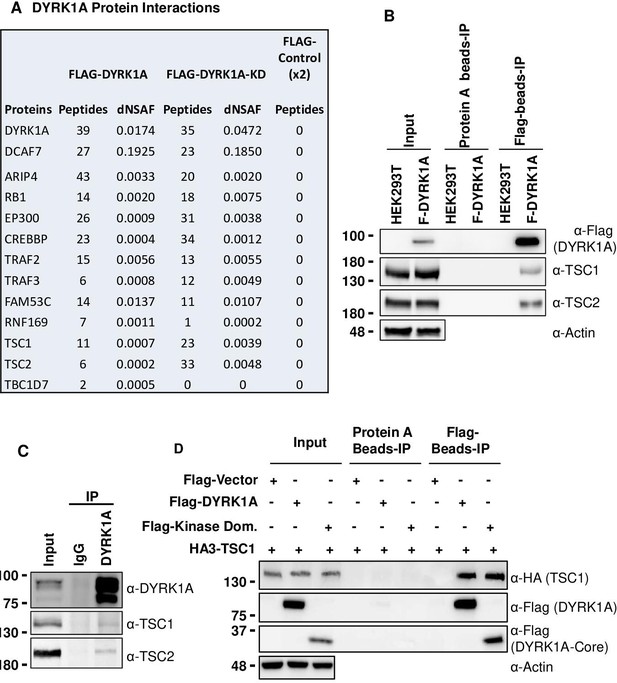
Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) interacts with the tuberous sclerosis complex (TSC).
(A) The tandem mass spectrometry (MS/MS) datasets previously acquired by MudPIT analyses of FLAG-DYRK1A affinity purifications and negative FLAG controls (Li et al., 2018) were searched against the most recent releases of the human protein sequence databases (built by collating and removing redundant entries from NCBI Homo sapiens RefSeq GCF_000001405.40_GRCh38.p14 and GCF_009914755.1_T2T-CHM13v2.0). Highly enriched proteins include known and novel DYRK1A-interacting partners and are reported with their peptide counts and distributed normalized spectral abundance factor (dNSAF) values, which reflect their relative abundance in the samples (Zhang et al., 2010). (B) Flag beads were used to pull down Flag-DYRK1A from whole cell extracts of HEK293 transfected with Flag-DYRK1A, and Protein A beads were used as a control. The blots were probed with TSC1 and TSC2 antibodies. Actin was used to normalize the lysate inputs. (C) Endogenous DYRK1A was immunoprecipitated with DYRK1A antibody from HEK293 cytoplasmic fraction generated using the Dignam protocol (Li et al., 2018) and probed with antibodies against endogenous DYRK1A, TSC1, and TSC2. Rabbit IgG was used as the IP control (D) Flag-DYRK1A and Flag-DYRK1A kinase domain constructs were affinity purified using Flag-beads from HEK293 cells co-transfected with HA3-TSC1 and probed with α-HA and α-Flag antibodies. Actin was used as the loading control.
-
Figure 2—source data 1
Uncropped blots for Figure 2B.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw data of Flag-DYRK1A affinity purification and Flag, TSC1 and TSC2 blot for Figure 2B.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data2-v1.zip
-
Figure 2—source data 3
Uncropped blots for Figure 2C.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data3-v1.zip
-
Figure 2—source data 4
Raw data of interaction between endogenous DYRK1A and TSC1 and TSC2 for Figure 2C.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data4-v1.zip
-
Figure 2—source data 5
Uncropped blots for Figure 2D.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data5-v1.zip
-
Figure 2—source data 6
Raw data of interaction between Kinase domain of Flag-DYRK1A and HA-TSC1 for Figure 2D.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-data6-v1.zip
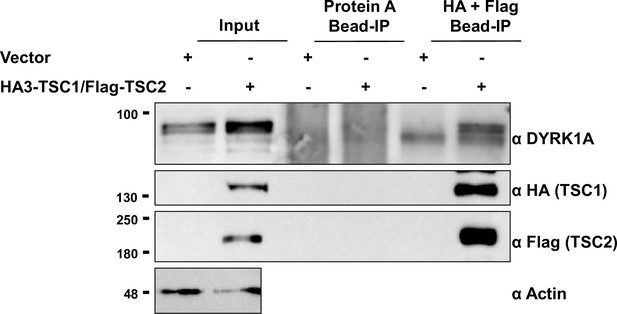
TSC1/TSC2 interact with dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A).
Flag and HA beads were used to pull down HA3-TSC1 and Flag-TSC2 from whole cell extracts of HEK293 transfected with HA3-TSC1 and Flag-TSC2. Blots were probed with Flag, HA, and DYRK1A antibodies. Actin was used as the loading control.
-
Figure 2—figure supplement 1—source data 1
Uncropped blots for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Raw data of interaction between overexpressed TSC1 and TSC2 with endogenous DYRK1A for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-figsupp1-data2-v1.zip
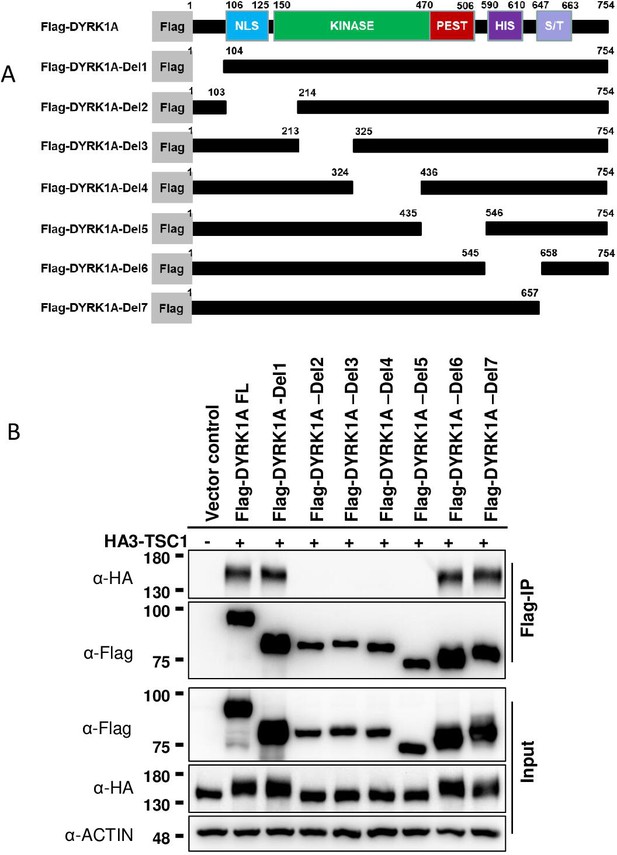
Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) kinase domain interacts with TSC1.
(A) Schematic of DYRK1A truncated constructs. All the truncated DYRK1A forms carry a Flag tag at the N-terminus. (B) Flag-DYRK1A and Flag-DYRK1A truncated constructs were affinity purified using Flag-beads from HEK293 cells co-transfected with HA3-TSC1 and probed with α-HA and α-Flag antibodies. All DYRK1A constructs, except with deletions in the kinase domain, immunoprecipitated HA3-TSC1. Note that Flag-DYRK1A kinase domain deletion constructs were expressed at lower levels than other constructs. Actin was used as lysate control.
-
Figure 2—figure supplement 2—source data 1
Uncropped blots for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
Raw data of interaction between truncated DYRK1A and full length-HA-TSC1 for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig2-figsupp2-data2-v1.zip
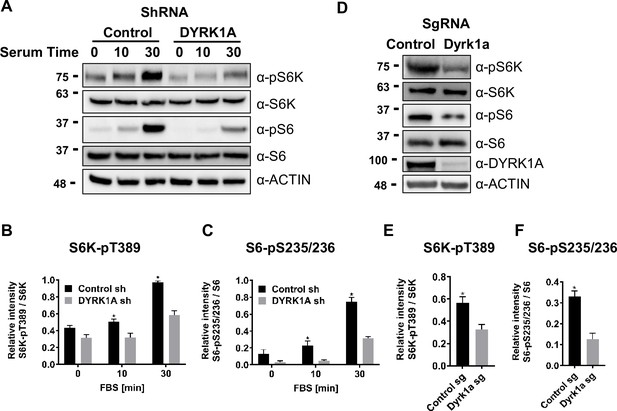
Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) promotes the activation of mTORC1 pathway in human and mouse cells.
(A) HEK293 cells treated with DYRK1A short hairpin RNA (shRNA) or control shRNA were serum starved for 12 hr before being activated with serum for the indicated times. Cells were then harvested, lysates, and probed with the indicated antibodies. Actin was used as the loading control. (B, C) Quantification of proteins in (A), levels of pS6K (T389), S6K, pS6 (pS235/236), and S6 were quantified using Image J software and the ratio of pS6K/S6K and pS6/S6 were plotted (n=3 biological replicates). (D) NIH3T3 cells were treated with sgRNA-targeting Dyrk1a or non-targeting control and selected for four days with Puromycin before harvesting. Lysates were probed with indicated antibodies. (E, F) Quantification of proteins in (D), levels of pS6K (T389), S6K, pS6 (pS235/236), and S6 were quantified (as described for B and C) and ratios were plotted (n=3 biological replicates). Student’s t-tests were done to compare samples. p-value = *p<0.05.
-
Figure 3—source data 1
Uncropped blots for Figure 3A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw data showing phosphorylation status of S6k and S6 after knockdown of DYRK1A for Figure 3A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig3-data2-v1.zip
-
Figure 3—source data 3
Uncropped blots for Figure 3D.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig3-data3-v1.zip
-
Figure 3—source data 4
Raw data showing phosphorylation status of S6K and S6 after CRISPR knockout of Dyrk1a in NIH3T3 cells for Figure 3D.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig3-data4-v1.zip
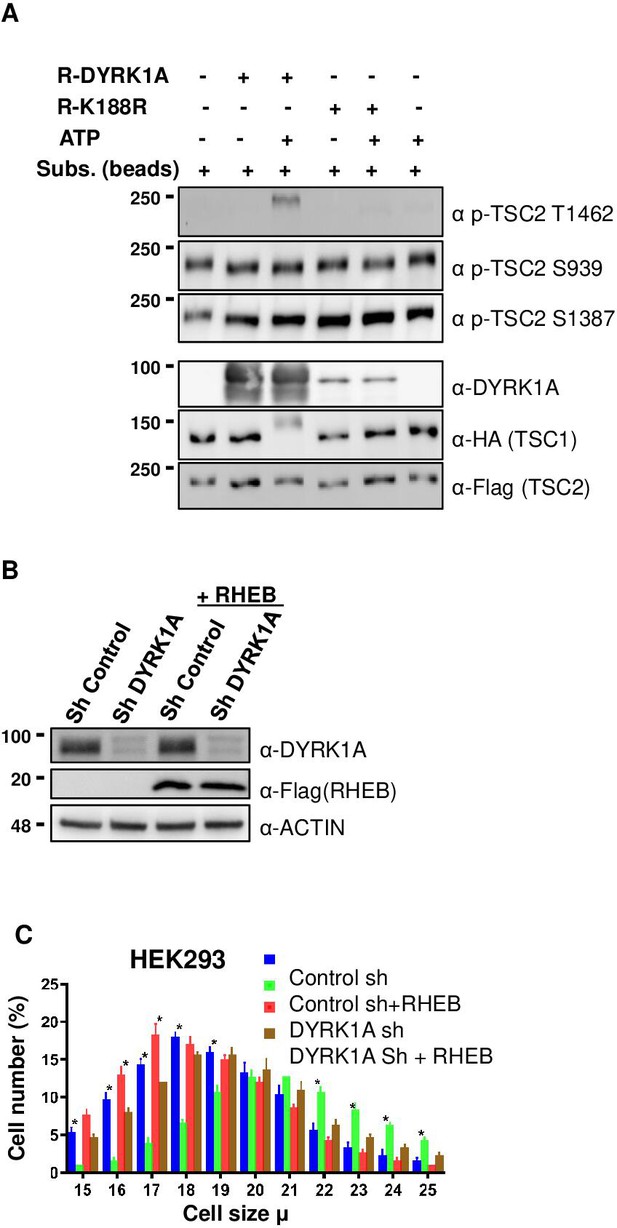
Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) phosphorylates TSC2 at T1462 in vitro, and Ras Homolog Enriched in Brain (RHEB) overexpression rescues mTORC1 activity in cells.
(A) An in-vitro kinase assay was performed using DYRK1A and kinase-dead DYRK1A (K188R) that were purified from bacteria. Flag-TSC2 and HA3-TSC1 were co-expressed in HEK293 cells and purified using a combination of (1:1) of HA and Flag beads. Beads were equilibrated with kinase assay buffer before the reactions were initiated on beads. After incubation for 30 min at 30°C, reactions were stopped by the addition of SDS loading buffer. Since bacterially purified DYRK1A is autophosphorylated, it exhibits a fuzzier signal, whereas kinase-dead DYRK1A is incapable of phosphorylation and appears as a sharp signal. (B, C) RHEB overexpression partially rescues the size of HEK293 cells. HEK293 cells were first transduced with short hairpin RNA (shRNA) lentivirus targeting DYRK1A or control and selected with 1 ug/ml Puromycin for three days, after which they were re-transduced with lentivirus expressing Flag-RHEB. The concentration of Puromycin was raised to 2 ug/ml for the next 48 hr in order to select for the second round of transduction. (B) Panel shows knockdown efficiency of DYRK1A and overexpression of RHEB. (C) Lower panel shows cell size analysis. Data represent the mean ± SD (n=3 biological replicates). Student’s t-test was done to compare samples. Significant difference in p-value = *p<0.05.
-
Figure 4—source data 1
Uncropped blots for Figure 4A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig4-data1-v1.zip
-
Figure 4—source data 2
Raw data showing in vitro kinase assay using recombinant DYRK1A and K188R for Figure 4A.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig4-data2-v1.zip
-
Figure 4—source data 3
Uncropped blots for Figure 4B.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig4-data3-v1.zip
-
Figure 4—source data 4
Raw data showing knockdown of DYRK1A and overexpression of Flag-RHEB for Figure 4B.
- https://cdn.elifesciences.org/articles/88318/elife-88318-fig4-data4-v1.zip
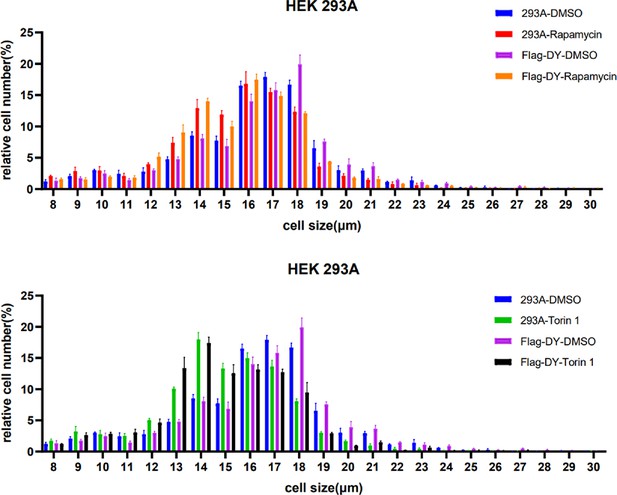
mTORC1 inhibitors block the increase in cell size mediated by dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A).
HEK293 cells expressing Flag-DYRK1A and the parental cells were treated with 40ng/ml Doxycycline. At 24hr mTOR inhibitors Torin1/Rapamycin were added and the cells were further incubated for 24hr. Data represent the mean ± SD (n=3 biological replicates).
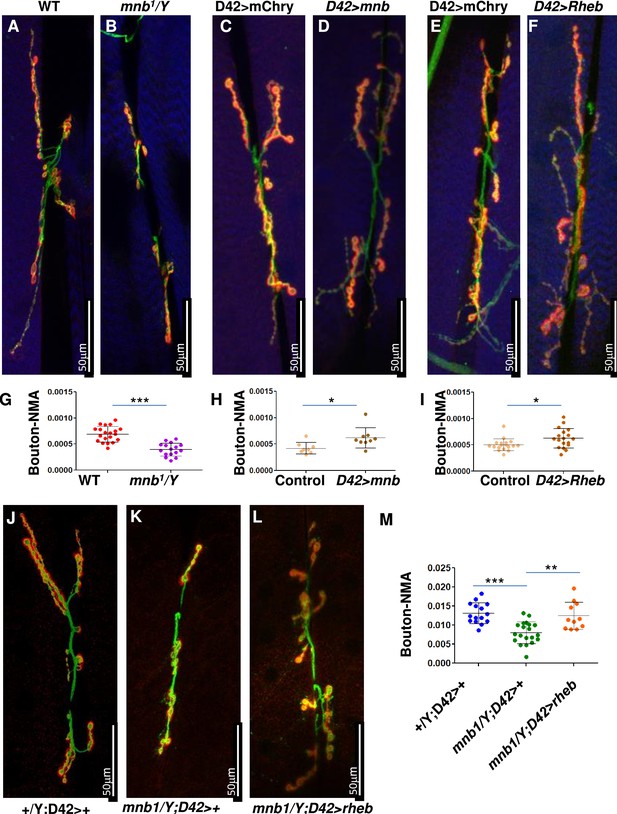
mnb mutant phenotype can be rescued by TOR activation in flies.
(A–F and J–L) Third instar larval neuromuscular junction (NMJ) (muscles 6/7) were stained using anti-HRP (Green) and anti-Dlg (Red). Muscles are stained with phalloidin (Blue, A–F). HRP (green) stains the entire neuron and Dlg (red) stains only boutons (Red + Green). (G–I, M) Quantification of bouton numbers, normalized to muscle area (Bouton-NMA). Error bars represent standard deviation. Statistical significance (p-values: ***<0.001; **<0.01; *<0.05) is calculated by unpaired student’s t-test. (A, B, G) mnb1 alleles show fewer boutons numbers as compared to wild-type (WT, Canton S) control (B). Data are quantified in G. (C, D, H) mnb overexpression (D42-Gal4>UAS mnb, D) increases bouton numbers as compared with mCherry overexpression (D42-Gal4>UAS-mCherry, Control, C). D42-Gal4 is a motor-neuron-specific driver. Data are quantified in H. (E, F, I) Rheb overexpression (D42-Gal4>UAS Rheb, F) increases bouton numbers as compared with mCherry overexpression (D42-Gal4>UAS-mCherry, Control, E). Data are quantified in I. (J–M) Rheb overexpression in mnb mutant (mnb1/Y D42-Gal4>UASRheb, L) suppressed bouton phenotype as compared to mnb mutant (mnb1/Y D42-Gal4/+, K). Wild-type is heterozygous D42-Gal4 (+/Y; D42-Gal4/+, J). Data is quantified in (M).
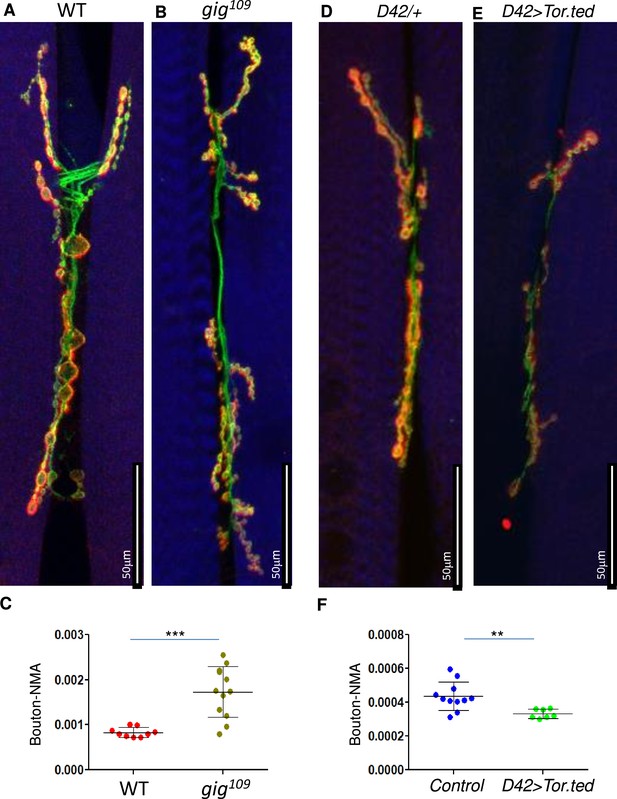
Neuromuscular junction (NMJ) phenotypes due to TOR gain or loss.
NMJ (muscles 6/7) are stained using anti-HRP (Green) and anti-Dlg (Red). Muscles are stained with phalloidin (Blue, A–F). HRP (green) stains the entire neuron and Dlg (red) stains only boutons (Red + Green). (G–I, M) Quantification of bouton numbers - normalized to muscle area (Bouton-NMA). Error bars represent standard deviation. Statistical significance (p-values: ***<0.001; **<0.01; *<0.05) is calculated by unpaired student’s t-test. (A–C) gig109 alleles show increased bouton numbers (B) as compared to wild-type (WT, Canton S) control (B). Data is quantified in C. (D–F) Expression of dominant negative TOR (D42-Gal4>UAS TOR.ted, E) decreases bouton numbers as compared to control (D42-Gal4/+, D). D42-Gal4 is a motor neuron-specific driver. Data is quantified in F.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Drosophila melanogaster) | Canton S | Bloomington Stock Center | ||
| Strain (D. melanogaster) | mnb[1] | Tejedor et al., 1995 | FBal0012364 | gift from Francisco J. Tejedor |
| Genetic reagent (D. melanogaster) | UAS-mnb | Shaikh et al., 2016 | FBtp0114512 | gift from Francisco J. Tejedor |
| Genetic reagent (D. melanogaster) | UAS-RHEB | Bloomington Stock Center | FBst0009688 | |
| Genetic reagent (D. melanogaster) | D42-Gal4 | Bloomington Stock Center; Gustafson and Boulianne, 1996 | FBti0002759 | |
| Genetic reagent (D. melanogaster) | P[(45)w[+mC]=UAS Tor.TED]II | Bloomington Stock Center; Shaikh et al., 2016 | FBti0026636 | |
| Genetic reagent (D. melanogaster) | UAS-mCherry | Bloomington Stock Center | FBti0147460 | |
| Chemical compound | 4% paraformaldehyde | Himedia | Cat# TCL119 | |
| Antibody | Mouse anti-DLG | DSHB; Shaikh et al., 2016 | CatID# 4F3 | IF (1:500) |
| Antibody | Rabbit anti-HRP conjugated with alexa488 | Jackson | CatID# 123-545-021 | IF (1:500) |
| Antibody | Goat anti-mouse conjugated with Alexa 555 | Invitrogen | CatID# A28180 | IF (1:500) |
| Other | Microsocope: Leica Stellaris 5 | Leica | PL APO 40 X/1.30 oil objective | |
| Other | Microsocope: Olympus FV3000 | Olympus | UPLFLN 40 X/1.30 oil objective | |
| Cell line (Homo sapiens) | HEK293 | ATCC | CRL-1573 | |
| Cell line (H. sapiens) | 293T | ATCC | CRL-3216 | |
| Cell line (Mus musculus) | NIH3T3 | ATCC | CRL-1658 | |
| Cell line (H. sapiens) | SH-SY5Y | ATCC | CRL-2266 | |
| Transfected construct (H. sapiens) | DYRK1A shRNA | ThermoFisher; Li et al., 2018 | Lentiviral construct to transduce and express the shRNA. | |
| Transfected construct (M. musculus) | Dyrk1a sgRNA | This paper | Lentiviral construct to transduce and mediate Dyrk1a knockout | |
| Antibody | anti-Actin (rabbit monoclonal) | Abclonal | Cat# AC026 | WB (1:5000) |
| Antibody | anti-HA (mouse monoclonal) | Abclonal | Cat# AE008 | WB (1:5000) |
| Antibody | anti-TSC1 (rabbit polyclonal) | Cell Signaling Technology | Cat# 4906 | WB (1:1000) |
| Antibody | anti-TSC2 (rabbit monoclonal) | Cell Signaling Technology | Cat# 4308 | WB (1:1000) |
| Antibody | anti- p70 S6 Kinase (rabbit monoclonal) | Cell Signaling Technology | Cat# 2708 | WB (1:1000) |
| Antibody | anti- Phospho-p70 S6 Kinase (Thr389) (rabbit polyclonal) | Cell Signaling Technology | Cat# 9205 | WB (1:1000) |
| Antibody | anti- S6 Ribosomal Protein (mouse monoclonal) | Cell Signaling Technology | Cat# 2317 | WB (1:1000) |
| Antibody | anti- Phospho-S6 Ribosomal Protein (Ser235/236) (rabbit monoclonal) | Cell Signaling Technology | Cat# 4856 | WB (1:1000) |
| Antibody | anti-Flag(mouse monoclonal) | MBL | Cat# M185 | WB (1:5000) |
| Antibody | anti-Phospho-TSC2-T1462(rabbit polyclonal) | Abclonal | Cat# AP0866 | WB (1:1000) |
| Antibody | anti-Phospho-TSC2-S1387 (rabbit polyclonal) | Abclonal | Cat# AP1117 | WB (1:1000) |
| Antibody | anti-Phospho- TSC2-S939 (rabbit polyclonal) | Cell Signaling Technology | Cat# 3615 | WB (1:1000) |
| Antibody | anti-DYRK1A | PMID:30137413 | WB (1:2000) | |
| Recombinant DNA reagent | pcDNA3-HA3-TSC1 (plasmid) | Addgene | RRID:Addgene_19911 | |
| Recombinant DNA reagent | pcDNA3 Flag TSC2 (plasmid) | Addgene | RRID:Addgene_14129 | |
| Recombinant DNA reagent | LentiCRISPR v2(plasmid) | Addgene | RRID:Addgene_52961 | |
| Sequence-based reagent | DYRK1A-RT-F | This paper | RT-qPCR primers | AAGCTCAGGTGGCTCATCGG |
| Sequence-based reagent | DYRK1A-RT-R | This paper | RT-qPCR primers | TCTCGCAGTCCATGGCCTG |
| Sequence-based reagent | GAPDH-RT-F | This paper | RT-qPCR primers | ACAACTTTGGTATCGTGGAAGG |
| Sequence-based reagent | GAPDH-RT-R | This paper | RT-qPCR primers | GCCATCACGCCACAGTTTC |
| Sequence-based reagent | Control-sgRNA | This paper | SgRNA target sequences for mouse cells | CGAGGTATTCGGCTCCGCG |
| Sequence-based reagent | Dyrk1a-sgRNA | This paper | SgRNA target sequences for mouse cells | CGCTTTTATCGGTCTCCAG |
| Commercial assay or kit | BCA Protein Quantification Kit | Meilunbio | Cat# MA0082 | |
| Commercial assay or kit | HiScript II 1st Strand cDNA Synthesis Kit | Vazyme | Cat# R212 | |
| Chemical compound, drug | Puromycin | Solarbio | Cat# P8230 | |
| Chemical compound, drug | Doxycycline | MCE | Cat# HY-N0565 | |
| Software, algorithm | GraphPad | Prism v.7.00 | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | ImageJ v.1.53 | RRID:SCR_003070 | |
| Other | Anti-Flag Beads | Smart-Lifesciences | Cat# SA042005 | |
| Other | r Protein A/G MagPoly Beads | Smart-Lifesciences | Cat# SM015001 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88318/elife-88318-mdarchecklist1-v1.docx
-
Supplementary file 1
Table S1: SgRNA target sequences for mouse cells.
- https://cdn.elifesciences.org/articles/88318/elife-88318-supp1-v1.docx





