The carboxyl-terminal sequence of PUMA binds to both anti-apoptotic proteins and membranes
Figures
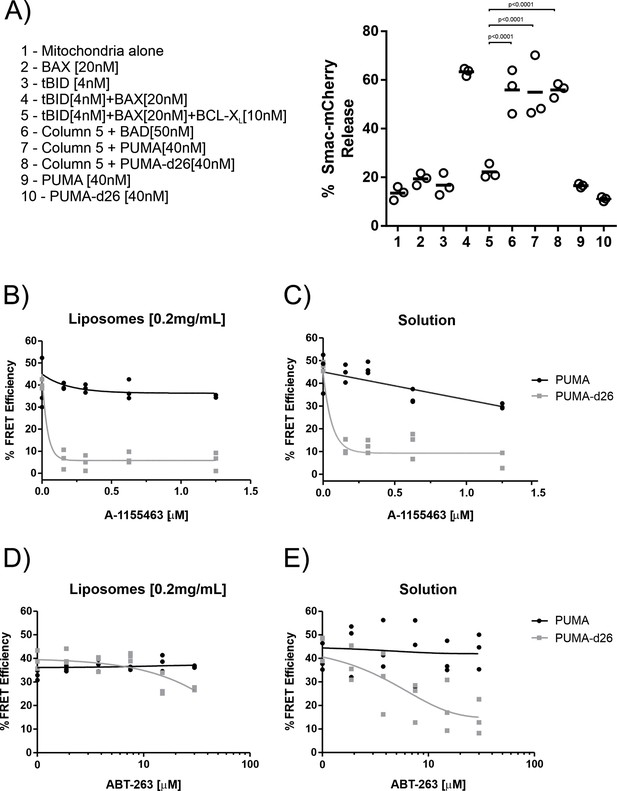
The PUMA CTS contributes to BH3-mimetic resistance independent of membranes in vitro.
(A) Left panel, legend indicating the combinations of recombinant proteins incubated with purified mitochondria encapsulating SMAC-mCherry. Right panel, release of SMAC-mCherry from mitochondria by the combination of recombinant proteins indicated by the legend numbers below. Each point represents a single technical replicate. The horizontal bar indicates the average of all three technical replicates per group. A one-way ANOVA and Dunnett’s multiple comparisons test resulted in the indicated p-values. (B–E) Alexa*568 labelled PUMA [5 nM] (black line) or PUMA-d26 [5 nM] (grey line) were incubated with Alexa*647 labelled BCL-XL [40 nM] in the presence of the indicated concentration of the BH3-mimetics (B and C) A-1155463 or (D and E) ABT-263. Each graph includes datapoints from three independent replicates. Total data were fit to a one phase exponential decay (grey and black lines as indicated). (B,D) Incubations of contained mitochondrial-like liposomes (0.2 mg/mL). (C,E) Solution indicates incubations that did not contain liposomes.
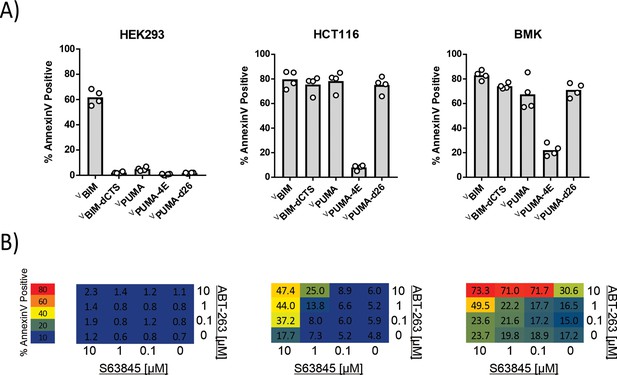
PUMA functions as a sensitizer in cells and kills BMK and HCT116 cells but not HEK293 cells.
(A) Transient transfection of HEK293, HCT116 and BMK cell lines to express exogenous BH3-only protein: BIM, PUMA, BIMdCTS, PUMAdCTS as well as PUMA-4E in which the 4 key hydrophobic residues in the BH3 motif of PUMA were changed to glutamic acid to disrupt the pro-apoptotic function of PUMA (I137E, L141E, M144E, and L148E). Cell death was scored using Annexin V staining to quantify cells that were both Venus and Annexin V positive out of the total Venus-positive cell population. (B) Two-way titration of two BH3-mimetics S63845 (MCL-1 inhibitor) and ABT-263 (BCL-XL, BCL-2, and BCL-W inhibitor) confirmed that HEK293 cells cannot be killed by inhibiting anti-apoptotic proteins (are unprimed) whereas BMK and HCT116 cells are primed for apoptosis and were killed by both BH3-mimetics. Annexin V positivity was scored as in (A) for cells that were incubated overnight with the indicated concentrations of S63845 and ABT-263. Results are displayed in a heatmap colored as indicated in the scale at the left, the values within each square indicate the average % Annexin V positive (dead cells) from three independent biological replicates.
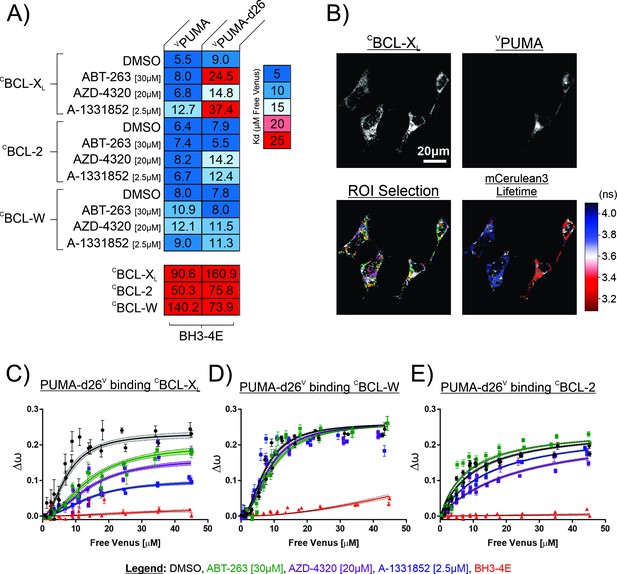
The PUMA CTS contributes to resistance to BH3-mimetics in live cells.
Quantitative Fast FLIM-FRET (qF3) was used to measure binding of PUMA and PUMA-d26 to the anti-apoptotic proteins: CBCL-XL, CBCL-W, and CBCL-2 in live BMK-dko cells. (A) Calculated apparent dissociation constants (Kd’s) for VPUMA and VPUMA-d26 binding to the indicated anti-apoptotic proteins are presented in a heatmap according to the scale at the right, with the calculated values specified inside the heatmap cells. VPUMA-d26 was displaced from CBCL-XL by all of the mimetics, as indicated by the increased apparent Kd in response to addition of BH3-mimetic. The protein pairs are indicated to the left and at the top. Final drug concentrations added to cells are indicated at the left. DMSO is the solvent control. BH3-4E below the panels indicates mutation of the BH3 protein listed at the top. Data is averages of three independent biological replicates. (B) Representative qF3 micrographs of BMK-dko cells stably expressing CBCL-XL, and transiently transfected with the plasmid to express VPUMA. Regions of interest identified automatically (ROI Selection) were assigned arbitrary colors to permit visualization. The FLIM image indicates the subcellular localization of VPUMA- CBCL-XL protein complexes (red, decreased mCerulean3 fluorescence lifetime) compared to unbound CBCL-XL (blue). (C–E) The effect of BH3 mimetics on the binding of PUMA-d26V to the anti-apoptotic proteins indicated above the graphs as measured by qF3. Binding data (Δω,from phasor plots)at different concentrations of unbound PUMA-d26V (means, symbols; error bars, SE) was fit to a Hill equation. Data points are averages from independent experiments. Line was fit to the data points from all three independent experiments. Lines with shaded areas indicate 90% confidence interval for the best fit. The results demonstrate displacement in (C) from BCL-XL, but not in (D) from BCL-W and to an intermediate extent in (E) from BCL-2 when incubated with the drugs indicated by the legend. DMSO is the solvent control and BH3-4E indicates the non-binding PUMA-d26V mutant used to control for collisions.
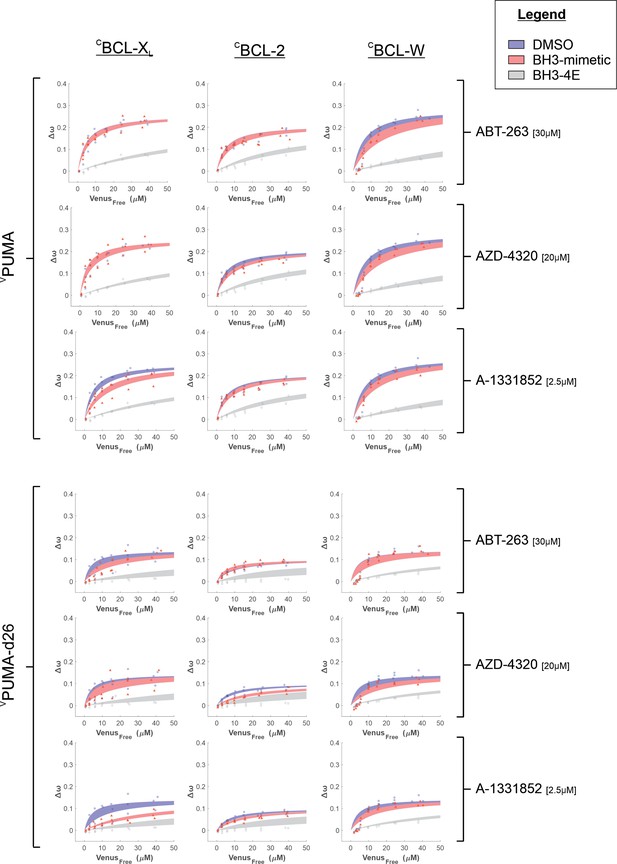
VPUMA binding to anti-apoptotic proteins is resistant to BH3-mimetics displacement in live cells due to the last 26 amino acids in the C-terminus of PUMA.
Binding curves generated from qF3 data demonstrate that both VPUMA and VPUMA-d26 bind to CBCL-XL, CBCL-2, and CBCL-W (blue lines) in a BH3-dependent manner as the corresponding BH3-4E mutant has dramatically reduced Δω values best fit with a straight-line indicating collisions rather than binding (grey lines). Addition of the specific BCL-XL inhibitor, A-1331852, only marginally affected Δω for VPUMA binding with CBCL-XL (left column, third panel down, compare blue and red lines) but reduced Δω for VPUMA-d26 (left column, sixth panel down) to a straight-line indicating addition of A-1331852 reduced binding of VPUMA-d26 with CBCL-XL to primarily collisions. For BCL-W at low concentrations of free VPUMA and VPUMA-d26 the data do not fit well to a Hill equation with a Hill slope of 1 suggesting binding of PUMA to BCL-W may be more complicated than a simple binary interaction. Data points are averages from independent experiments. The line was fit to the data points from the 3 independent experiments. Shaded areas indicate the 95% confidence interval for the best fit of the Hill equation to the data. In all Figures, where the lines overlapped completely only the red shaded area is visible as typically the 95% confidence interval is at least as great as for assays without the BH3 mimetic.
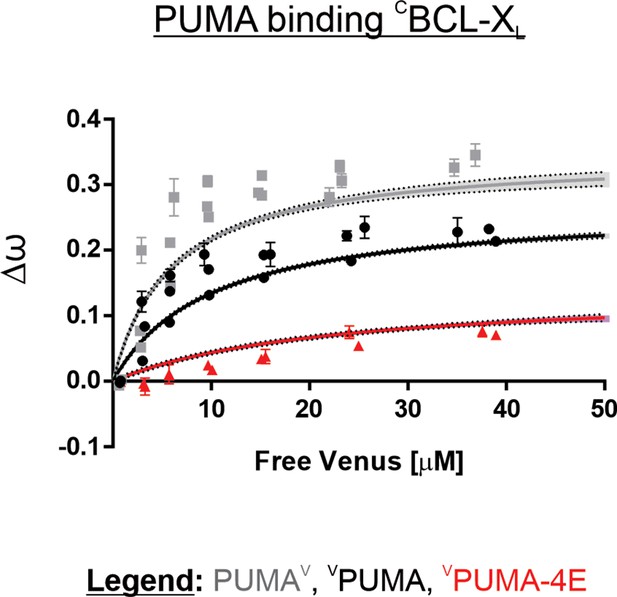
Venus fused to the C-terminus of PUMA (PUMAV, grey line) resulted in higher Δω for binding to CBCL-XL than when Venus was fused to the N-terminal of PUMA (VPUMA, black line).
Increased Δω translates to a larger dynamic range in assaying protein binding. Data points are averages from independent experiments. Line was fit to the data points from the 3 independent experiments. Shaded areas indicate the 95% confidence interval for the best fit of the Hill equation with a Hill slope of 1 to the data.
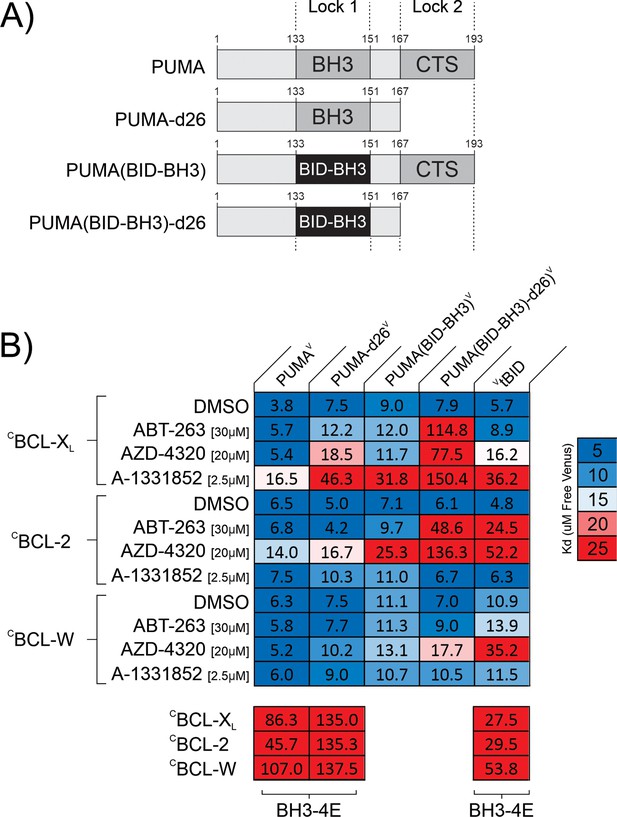
PUMA resists BH3-mimetic displacement by double-bolt locking to anti-apoptotic proteins.
(A) Linear depictions of the PUMAV mutants analyzed. Lock 1 indicates the BH3-motif, Lock 2 indicates the CTS. (B) Mutation of either the PUMA CTS or the BH3 region is sufficient to relieve resistance to a subset of the drugs resulting in apparent Kd values similar to those obtained for the BH3 mimetic sensitive control VtBID. Mutation of both sequences in PUMA results in a protein (PUMA(BID-BH3)-d26V) that can be displaced by the BH3 mimetic AZD-4320 from the three anti-apoptotic proteins, CBCL-XL, CBCL-2, and CBCL-W. Heatmaps displaying Kd values calculated from fitting qF3 binding curves and represented by color (scale to the right) to highlight changes in these binding affinities (binding (blue) to non-binding (red)) within the heatmap cells. The interacting protein pairs are indicated to the left and at the top. Final drug concentrations added are indicated at the left. DMSO is the solvent control. BH3-4E below the panels indicates mutation of the BH3 protein listed at the top. Binding by the BH3-mimetic sensitive control VtBID to the anti-apoptotic proteins was inhibited by the cognate BH3-mimetics, as expected. In the assay conditions used, inhibition of VtBID indicated that BCL-XL was inhibited primarily by AZD-4320 and A-1331852; BCL-2 was inhibited by ABT-263 and AZD-4320; CBCL-W was inhibited by only AZD-4320. Data is averages from three independent experiments.
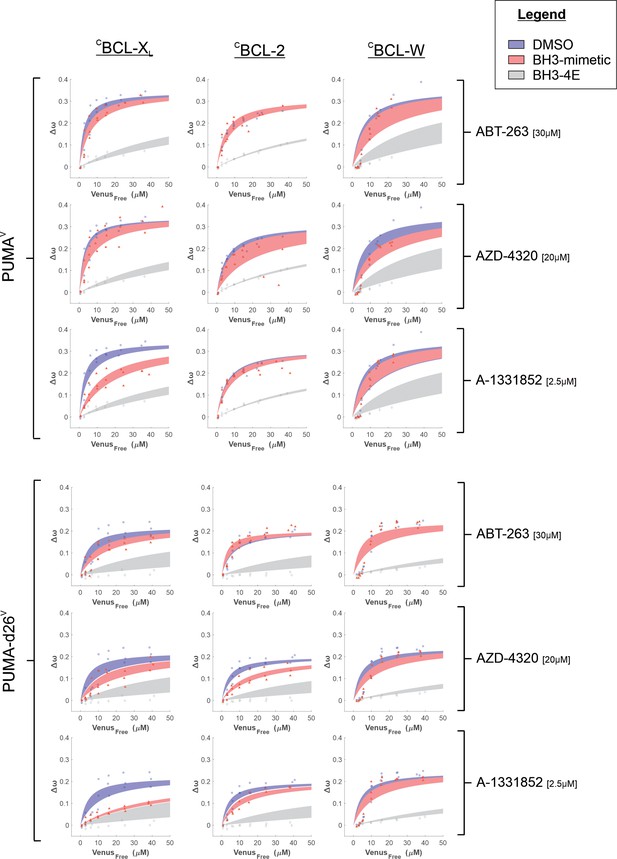
PUMAV binding to anti-apoptotic proteins is resistant to displacement by BH3-mimetics in live cells due to the last 26 amino acids in the C-terminus of PUMA.
Binding curves generated from qF3 data demonstrate that both VPUMA and VPUMA-d26 bind to CBCL-XL, CBCL-2, and CBCL-W (blue lines) in a BH3-dependent manner as binding of the corresponding BH3-4E mutant was dramatically reduced resulting in lower Δω values that are best fit with a straight-line, indicating collisions rather than binding (grey lines). VPUMA binding with CBCL-XL was slightly reduced by the specific BCL-XL inhibitor, A-1331852, (left column, third panel down, compare blue and red lines). Reduced binding of VPUMA-d26 (Δω, left column, sixth panel down) resulted in data best fit to a straight-line indicating only collisions were detected. Binding between VPUMA-d26 and CBCL-2 was disrupted by AZD-4320, while no BH3-mimetic disrupted the interaction between VPUMA-d26 and CBCL-W. Moreover, at low concentrations of free VPUMA and VPUMA-d26 the data do not fit well to a Hill equation suggesting binding of PUMA to BCL-W may be more complicated than simple 1:1 binding described by a Hill equation. Data points are averages from independent experiments. Line was fit to the data points from the three independent experiments. Shaded areas indicate the 95% confidence interval for the best fit of the data to a Hill equation.
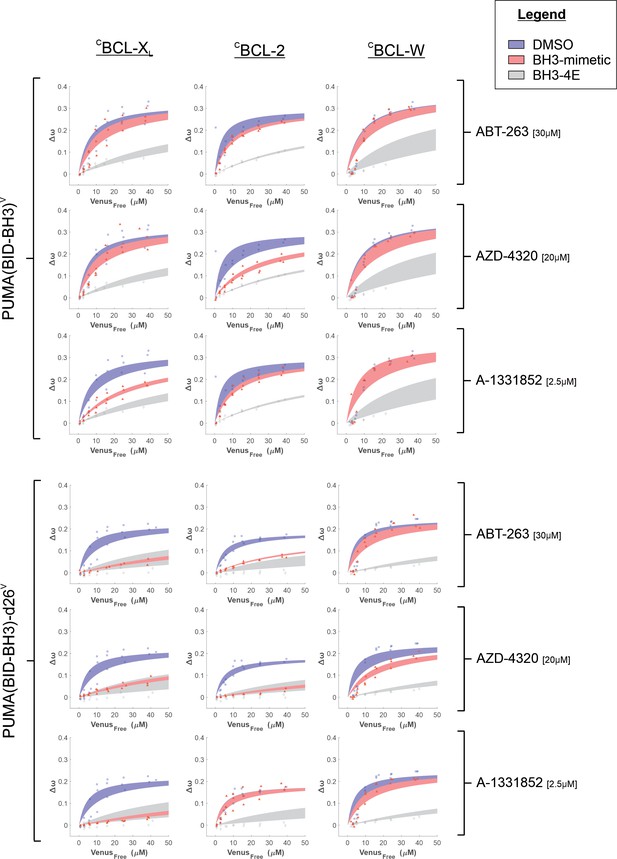
Binding curves generated from qF3 data demonstrate that both VPUMA(BID-BH3) and VPUMA(BID-BH3)-d26 bind to CBCL-XL, CBCL-2, and CBCL-W (blue lines).
Addition of the specific BCL-XL inhibitor, A-1331852, reduced Δω for both VPUMA(BID-BH3) and VPUMA(BID-BH3)-d26 binding to CBCL-XL (left column, third and sixth panel down, compare blue and red lines). Addition of ABT-263 and AZD-4320 displaced VPUMA(BID-BH3)-d26 from CBCL-XL and CBCL-2 (left and middle columns, fourth and fifth panels down). Only AZD-4320 reduced binding of VPUMA(BID-BH3)-d26 to CBCL-W (right column, fifth panel down). Moreover, for BCL-W and low concentrations of free VPUMA(BID-BH3) and VPUMA(BID-BH3)-d26 the data do not fit well to a Hill equation suggesting binding of PUMA to BCL-W may be more complicated. Data points are averages from independent experiments. Line was fit to the data points from the three independent experiments. Shaded areas indicate the 95% confidence interval for the best fit of the Hill equation to the data.
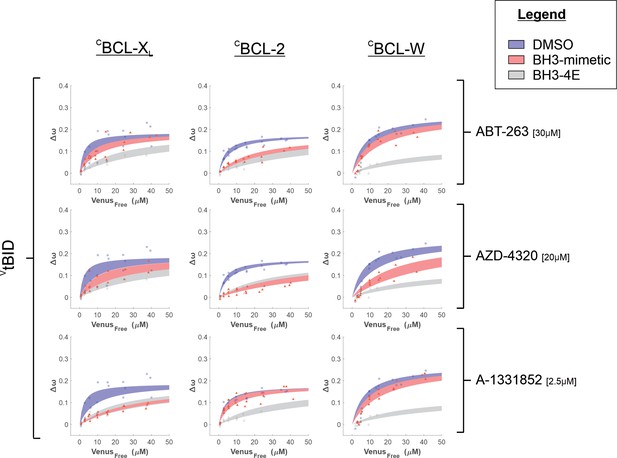
Binding curves generated from qF3 data demonstrate that VtBID binds to CBCL-XL, CBCL-2, and CBCL-W (blue lines) in a BH3-depdendent manner (VtBID-BH3-4E, grey lines).
The addition of A-1331852 and AZD-4320 reduced the affinity of VtBID binding to CBCL-XL (Left column, second and third panels down, compare blue and red lines) while AZD-4320 and ABT-263 reduced the binding affinity for VtBID binding with CBCL-2 (middle column, first and second panels down). VtBID binding with CBCL-W was disrupted by AZD-4320 (right column, second panel down). Data points are averages from independent experiments. Line was fit to the data points from the three independent experiments. Shaded areas indicate the 95% confidence interval for the best fit of the Hill equation with a slope of 1 to the data.
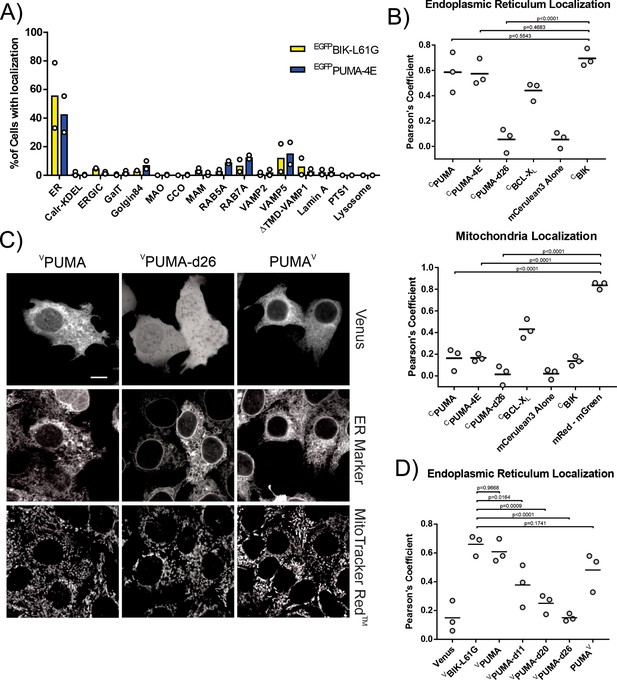
The CTS of PUMA localizes primarily to the endoplasmic reticulum.
(A) Lentiviral infected NMuMG cells expressing EGFP fused BH3-proteins with mutated BH3-motifs to prevent cell death, were imaged using confocal fluorescence microscopy. Images of individual cells were classified as one of the previously defined landmarks as described in Schormann et al., 2020. Most cells expressing EGFPPUMA-4E (blue) or the ER localized control EGFPBIK-L61G (yellow) were classified as most closely resembling ER landmarks with a small fraction of cells expressing EGFPPUMA-4E resembling localization in a variety of transport vesicles (RAB5A, RAB7A) or secretory pathway vesicles and plasma membrane (VAMP5). The landmarks tested included ER (3 resident endoplasmic reticulum membrane markers), Calr-KDEL (recycling between ER and Golgi), ERGIC (ER-Golgi intermediate compartment), GalT (trans-Golgi), Golgin84 (cis-Golgi), MAO (outer mitochondrial membrane), CCO (inner mitochondrial membrane), MAM (mitochondrial associated ER membrane), Rab5A, Rab7A, VAMP2 (transport and secretory vesicles), VAMP5 (Secretory pathway to the plasma membrane), ΔTMD-VAMP1 (cytoplasm), Lamin A (nuclear envelope), PTS1 (peroxisomes), LAMP1 (Lysosomes) (Schormann et al., 2020). (B) Quantification of fluorescence colocalization in BMK-dko cells indicates PUMA primarily localizes in a CTS-dependent manner to the ER (upper panel) and not Mitochondria (lower panel). Pearson’s correlation coefficients from three independent experiments are reported for the indicated proteins with the ER marker BODIPY FL thapsigargin (upper panel) or the mitochondrial marker Mitotracker Red (mRed, lower panel). The horizontal bars indicate the medians and mGreen indicates the stain Mitotracker Green. CBIK is an ER marker composed of mCerulean3 fused to the ER localized protein BIK. Data from three independent experiments are shown with horizontal bars indicating the medians. Each data point represents the average from a minimum of 50 cells. (C) Fusion of Venus to the C-terminus of PUMA (PUMAV) does not prevent localization at the ER, suggesting that the PUMA CTS is not a conventional TA that spans the bilayer. Top row: Micrographs of the Venus fluorescence from cells expressing VPUMA, VPUMA-d26, and PUMAV by transient transfection, as indicated above. Middle row: Micrographs of the ER marker. Bottom row: MitoTracker Red staining for the same cells. White scale bar is 5 μm. (D) Quantification of the extent to which the distribution of the various mutant proteins (indicated below) correlated with the distribution of the ER marker (cBIK) in BMK-dko cells. Data from three independent experiments are shown with horizontal bars indicating the medians. Each data point represents the average from a minimum of 50 cells.
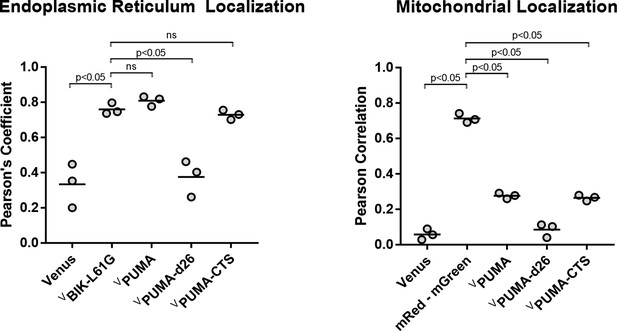
Quantification of fluorescence colocalization in BMK-dko cells indicates that fusion of just the PUMA CTS to the carboxyl-terminus of Venus (VPUMA-CTS) primarily localizes the fluorescent protein to the ER (left panel) with minimal localization to the Mitochondria, similar to the localization of VPUMA (right panel).
Pearson’s correlation coefficients from three independent experiments are reported for the proteins indicated at the bottom with the ER marker CBIK (left panel) or the mitochondrial marker Mitotracker Red (mRed, right panel). The horizontal bars indicate the means.
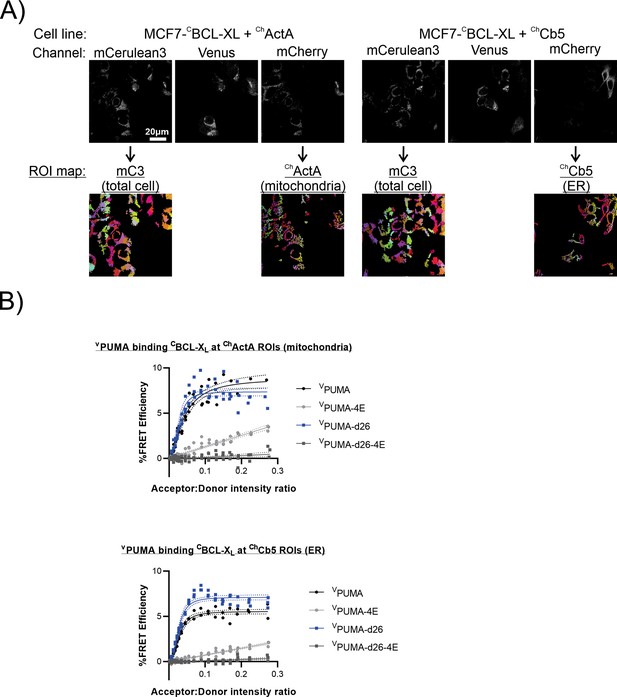
PUMA binds BCL-XL at ROIs corresponding to mitochondria and ER in live cells.
(A) Images of mCerulean3 and mCherry in MCF-7 cells expressing donor CBCL-XL and either ChActA or ChCb5(indicated above), with corresponding segmentation maps selected based on each channel below. ROIs obtained by segmentation of the total cell CBCL-XL (mCerulean3 signal) and ROIs identified from organelle marker-based segmentation maps are shown below the corresponding images. (B) FLIM–FRET binding curves for VPUMA binding to CBCL-XL were generated from ROIs identified from images of MCF-7 cells by organelle marker-based segmentation as indicated above each panel (n=3).
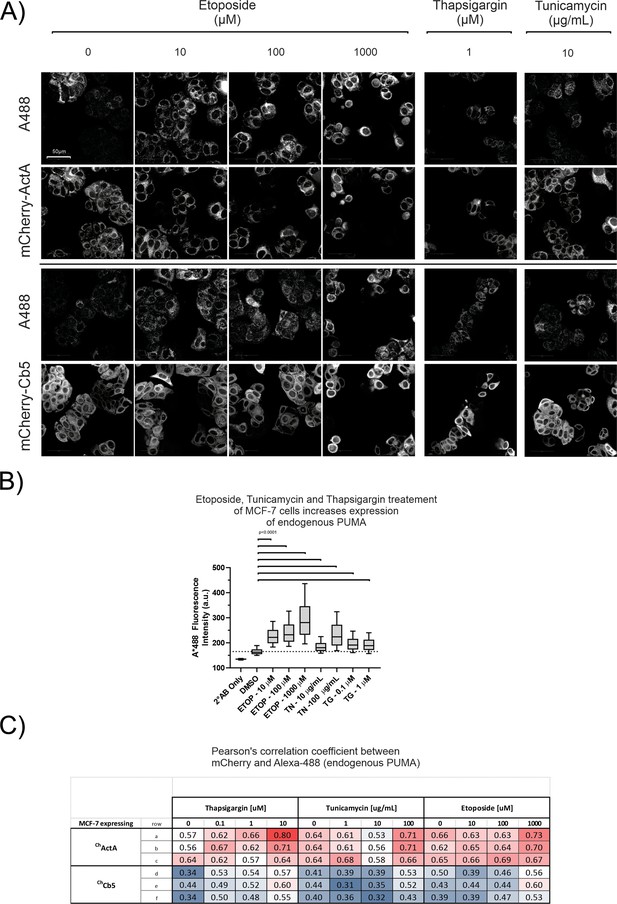
Genotoxic and ER stress inducing drugs cause upregulation of endogenous PUMA protein which localized with mitochondrial and ER markers in MCF-7 cells.
(A) Fluorescence images of MCF-7 cells expressing either mCherry-ActA (ChActA) (Top panels) or mCherry-Cb5 (ChCb5) (Bottom panels) were fixed and immunostained for PUMA (A488). Cells were treated with the drugs and concentrations indicated at the top of the panels for 24 hr before fixation and immunostaining. DMSO treated control cells were labeled as 0 µM drug. (B) Drug treatment increased the level of endogenous PUMA as shown by immunostaining in MCF-7 cells. Fluorescence intensity of Alexa-488 conjugated to the secondary antibody that recognizes the anti-PUMA (human) primary antibody were reported for individual cells and plotted as a box and whisker plot. ETOP, Etoposide; TN, Tunicamycin; TG, Thapsigargin. (C) Table containing Pearson correlation coefficient values from confocal micrographs for co-localization of endogenous PUMA with either ChActA for mitochondria or ChCb5 for ER in MCF-7 cells treated with the indicated concentrations of Thapsigargin or Tunicamycin or Etoposide. Three technical replicates (a, b, c and d, e, f) are represented as rows in the table. The values are color-coded to show the modest drug treatment induced increase in Pearson correlation coefficient from ≈0.4 to ≈0.5 for the co-localization of PUMA to the ER marker and from ≈0.6 to ≈0.7 for the co-localization of PUMA to mitochondria marker. The increase is largest at the highest concentration of the drugs compared to the baseline at 0 µM or 0 µg/mL.
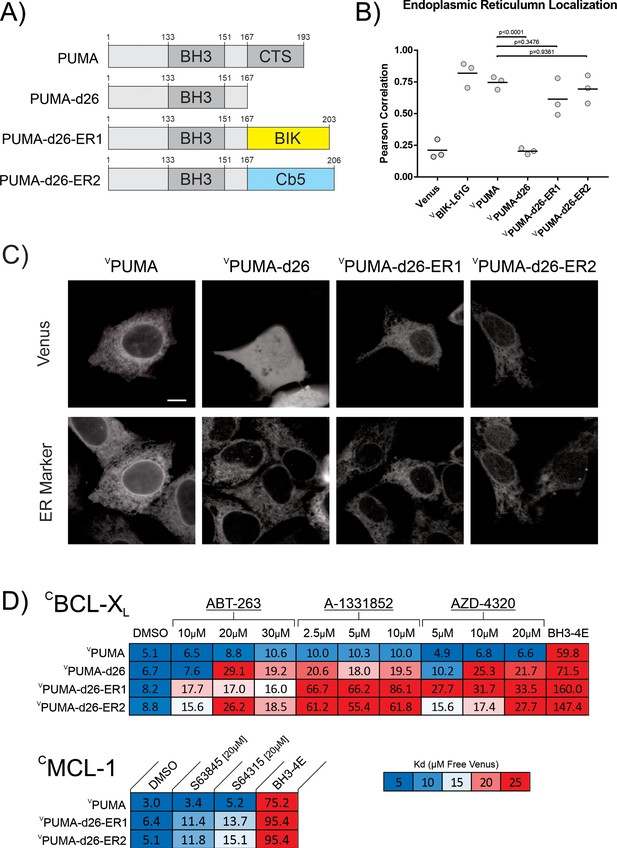
Restoring ER localization to PUMA-d26 does not confer resistance to BH3-mimetic displacement.
(A) Cartoon depiction of fusion proteins created. (B,C) Fusion of the canonical tail-anchors from BIK and CB5 to VPUMA-d26 restored ER localization when expressed in BMK-dko cells. (B) Pearson’s correlation with the ER marker protein CBIK in BMK-dko cells. Data points are averages from independent experiments. A one-way ANOVA and Dunnett’s multiple comparisons test were used to calculate the indicated p-values. (C) Micrographs illustrating subcellular localization by confocal microscopy of the indicated Venus fusion proteins co-expressed with the ER marker protein CBIK. The scale bar indicates 5 μm. (D) Heatmaps generated from qF3 data display calculated apparent Kd’s for binding of the indicated mutants to CBCL-XL and CMCL-1 in live BMK-dko cells. Restoring ER localization to PUMA-d26 (VPUMA-d26-ER1 and VPUMA-d26-ER2) did not restore resistance to BH3-mimetic displacement as indicated by increased dissociation constants in the presence of BH3-mimetic.
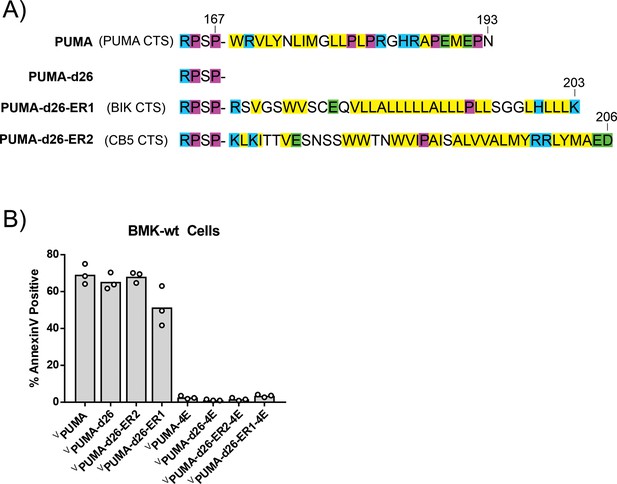
Exogenous expression of VPUMA and VPUMA-ER localized tail-anchor chimera mutants induce apoptosis in BMK cells.
(A) Single letter amino-acid code depiction of the PUMA CTS and the canonical tail-anchors (BIK CTS and CB5 CTS) fused to PUMA-d26. Color depicts amino acid chemical properties (yellow = hydrophobic, purple = helix breaking residue, blue = positively charged, green = negatively charged). One construct (VPUMA-d26-ER1) has the BIK CTS sequence (amino acids 126–160, uniprot: Q13323) and the other (VPUMA-d26-ER2) the CB5 CTS sequence (amino acids 97–132 uniprot: P00167). (B) Fusion proteins VPUMA-d26-ER1 and VPUMA-d26-ER2 are functional as expression kills BMK-wt cells in a BH3-dependent manner as shown by lack of cell death due to expression of proteins containing the 4E mutation. Cell death was measured by Annexin V staining of BMK-wt cells scored by automated confocal microscopy.
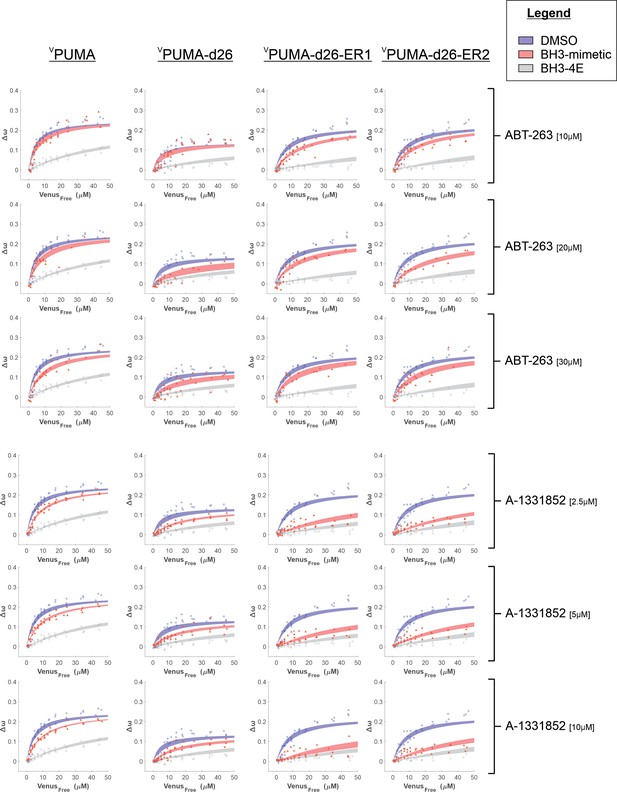
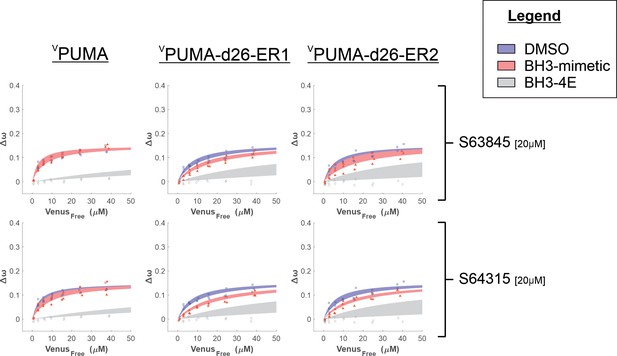
Binding curves generated from qF3 data demonstrate that VPUMA, VPUMA-d26, VPUMA-d26-ER1, and VPUMA-d26-ER2 bind to CBCL-XL (blue lines) in a BH3-dependent manner (BH3-4E, grey lines, indicate primarily collisions).
The addition of ABT-263 or A-1331852 (red lines) is sufficient to displace VPUMA-d26, VPUMA-d26-ER1, and VPUMA-d26-ER2 but not VPUMA. As expected, ABT-263 was less effective than A-13311852. Data points are averages from independent experiments. Line was fit to the data points from the three independent experiments.
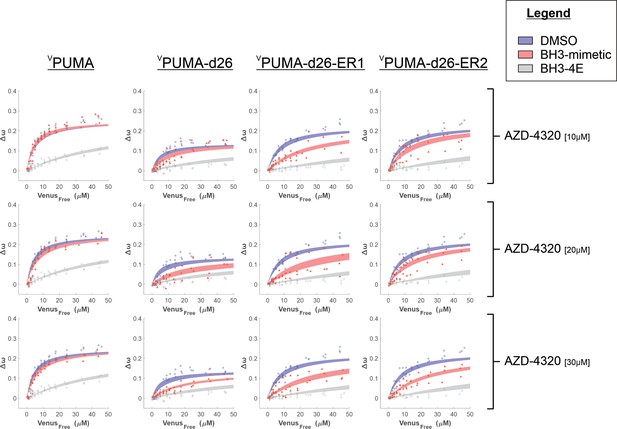
Binding curves generated from qF3 data demonstrate that VPUMA, VPUMA-d26, VPUMA-d26-ER1 and VPUMA-d26-ER2 bind to CBCL-XL (blue lines) in a BH3-dependent manner (BH3-4E, grey lines, indicate primarily collisions).
The addition of AZD-4320 (red lines) is sufficient to displace VPUMA-d26, VPUMA-d26-ER1, and VPUMA-d26-ER2 but not VPUMA. Data points are averages from independent experiments. Line was fit to the data points from the three independent experiments.
Binding curves generated from qF3 data demonstrate that VPUMA, VPUMA-d26-ER1 and VPUMA-d26-ER2 bind to CMCL-1 (blue lines) in a BH3-dependent manner (BH3-4E, grey lines).
The maximum values of ∆ω are lower for CMCL-1 due to the increased distance between the fluorescence proteins that results from the extended amino terminus of MCL-1. The addition of either S63845 or S64315 (red lines) resulted in a slight decrease in ∆ω and a change in the shape of the curve that results in a higher calculated Kd in drug treated cells for VPUMA-d26-ER1 (middle column) and VPUMA-d26-ER2 (right column) binding to CMCL-1 in comparison to VPUMA binding to CMCL-1 (Left column), indicating protein displacement. Data points are averages from independent experiments. Line was fit to the data points from the 3 independent experiments.
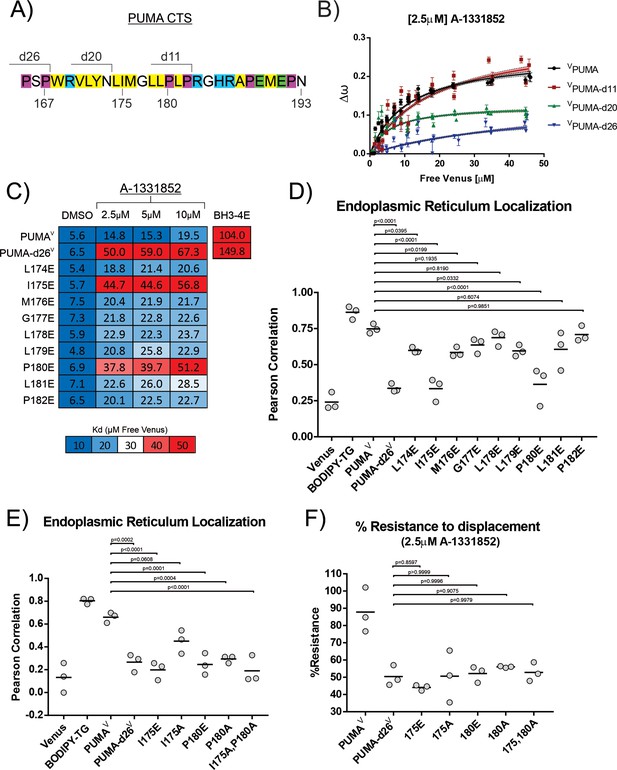
Residues I175 and P180 in the PUMA CTS contribute to both ER localization and BH3-mimetic resistance.
(A) Amino acid composition of the PUMA CTS. Color depicts amino acid chemical properties (yellow = hydrophobic, purple = alpha-helix breaking residue, blue = positively charged, green = negatively charged). Lines indicate deletion points. Residue numbers are indicated below the sequence. (B) Binding curves generated by qF3 for the indicated mutants. VPUMA and VPUMA-d11 resisted BH3-mimetic displacement from BCL-XL while VPUMA-d26 was displaced (higher Kd value). VPUMA-d20 binding was at least altered such that the distance between the donor and acceptor was increased, as shown by the dramatically reduced values for ∆ω,a measure directly related to FRET efficiency and bound fraction. (C) Calculated Kd values determined by qF3 for PUMAV mutants containing a single-glutamic acid substitution in the PUMA CTS displayed as a heat map with calculated values in the heatmap cells. Averages from 3 biological replicates are shown and suggest that residues I175 and P180 are required for resistance to BH3-mimetics. The color scale was changed for this figure to visually differentiate the effect of the point mutations I175E and P180E from the change in binding that resulted from substituting residues at other locations with a glutamic acid residue. (D) Pearson’s correlation coefficients calculated from confocal micrographs for co-localization of PUMAV and the indicated PUMAV mutants with the ER localization marker CBIK suggest that residues I175 and P180 are most important for PUMA localization at the ER. (E) Pearson’s correlation coefficients calculated from confocal micrographs for co-localization indicate a more conservative mutation to alanine at position I175 (PUMAV I175A) results in increased localization to the ER, while PUMAV P180A was not localized at membranes. (F) Mutation of residues I175 and P180 abrogated resistance to displacement by the BH3-mimetic A1331852 equivalent to deletion of the entire CTS. % Resistance to displacement of PUMAV mutants (indicated below) from CBCL-XL calculated from FLIM-FRET binding curves. Data points are averages from independent experiments. Line indicates the mean of the data points shown. P values in panels (D,E,F) were calculated using an ordinary one-way ANOVA method GraphPad Prism 9.5.0 to examine the differences in the mean Pearson Correlation Coefficient values between the tested group and the reference groups (PUMAV for panels D and E, PUMA-d26V for panel F).
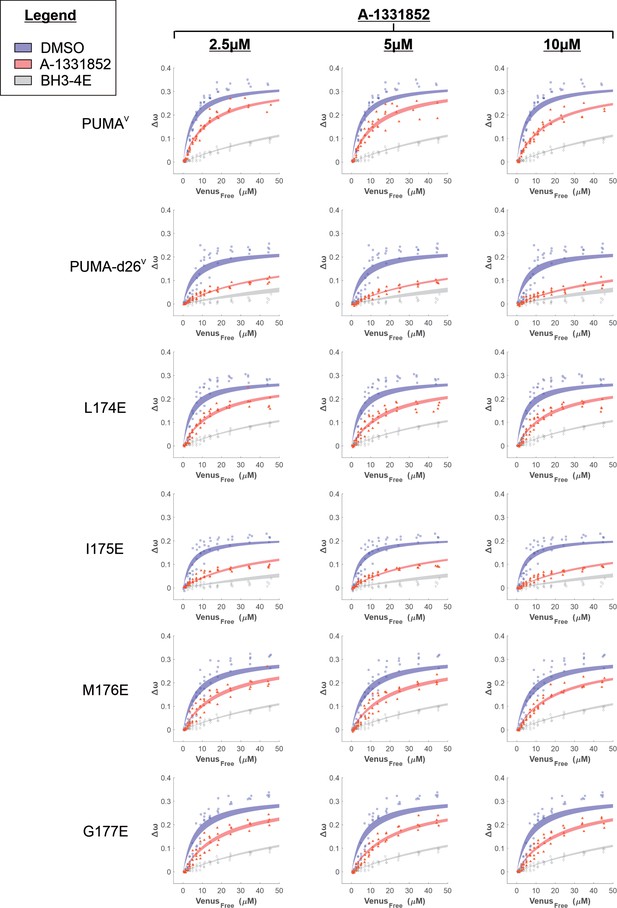
Binding curves generated from qF3 data demonstrate that PUMAV and PUMA-d26V bind to CBCL-XL (blue points) in a BH3-dependent manner (BH3-4E, grey points).
The addition of BH3-mimetic A-1331852 (red points) most effectively displaced PUMA-d26V and PUMAV mutant I175E. Data from 4 independent experiments (data points) were fit to a Hill equation (lines) with shaded areas representing the 95% confidence interval for the best fit.
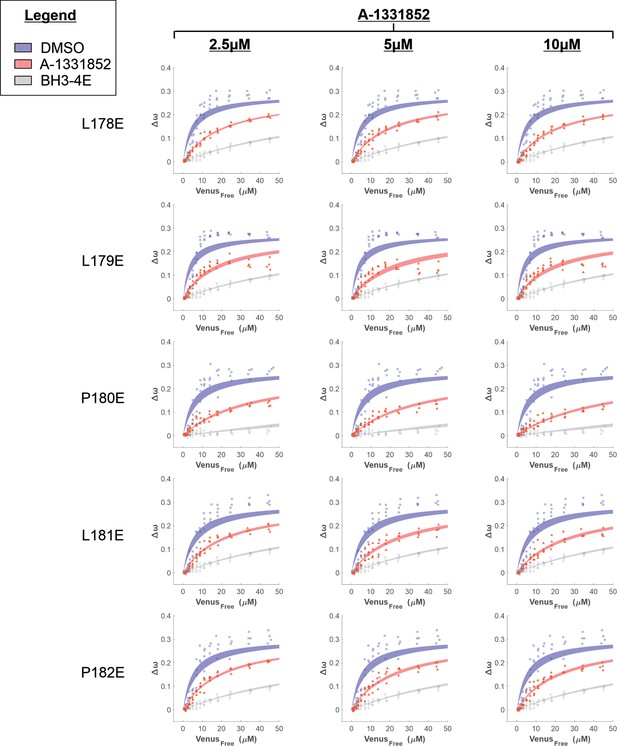
Binding curves generated from qF3 data demonstrate that PUMAV mutants bind to CBCL-XL (blue points) in a BH3-dependent manner (negative control, BH3-4E, grey points).
The addition of BH3-mimetic A-1331852 (red points) displaced PUMAV mutants from CBCL-XL but was most effective for PUMAV mutant P180E. Data from 4 independent experiments (data points) were fit to a Hill equation with a slope of 1 (lines) with shaded areas representing the 95% confidence interval for the best fit.
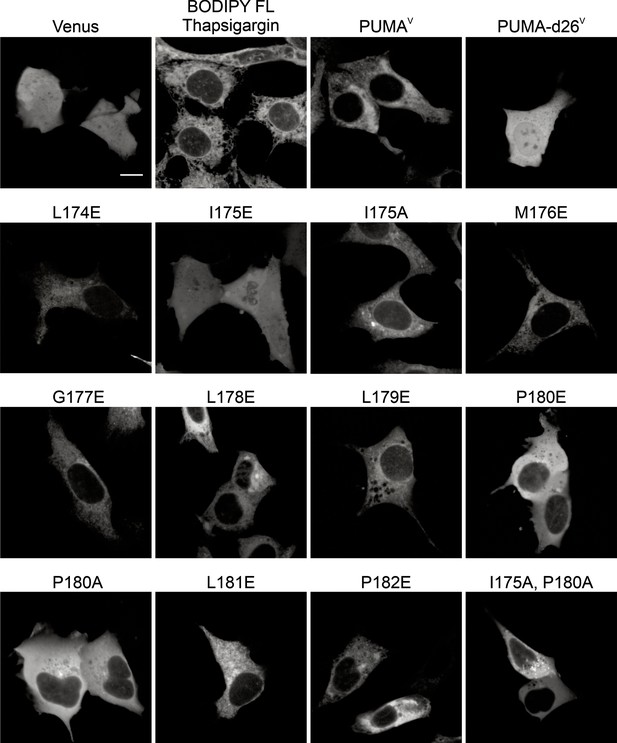
Representative confocal micrographs of the Venus fluorescence from BMK-dko cells expressing the proteins indicated above or the PUMAV mutants indicated by the amino acid substitution.
Proteins were expressed by transient transfection. White scale bar indicates 5 μm. Note cytoplasmic expression pattern for the PUMAV mutants I175E, P180E, P180A and the double mutant I715A, P180A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Antibody to PUMA (polyclonal) | Cell Signaling Technology | Cat. #: 4976 S; RRID: AB_2064551 | Dilution (1:400) |
| Antibody | Alexa-488 anti-Rabbit IgG secondary antibody (polyclonal) | Abcam | Cat: # ab150077; RRID: AB_2630356 | Dilution (1:1000) |
| Cell line (M. musculus) | Baby Mouse Kidney (BMK)- WT | PMID:11836241 | Dr. Eileen White (Rutgers University) | |
| Cell line (M. musculus) | Baby Mouse Kidney (BMK)- DKO | PMID:11836241 | Dr. Eileen White (Rutgers University) | |
| Cell line (M. musculus) | NMuMG | RRID: CVCL_0075 | Dr. Jeff Wrana (University of Toronto) | |
| Cell line (H. sapiens) | HEK293 | Other (Graham et al., 1977) | RRID: CVCL_0045 | Provided by Dr. Frank Graham (McMaster University). |
| Cell line (H. sapiens) | HCT-116 | Other (Polyak et al., 1996) | RRID: CVCL_0291 | Provided by Dr. Bert Vogelstein (John Hopkins University). |
| Cell line (H. sapiens) | MCF-7 | PMID:3790748 | RRID: CVCL_0031 | Provided by Dr. Ronald N. Buick (University of Toronto) |
| Chemical compound, drug | DRAQ5 | ThermoFisher Scientific, Molecular probes | Cat. #62251 | Far red nucleic acid specific fluorescent dye for cell imaging |
| Chemical compound, drug | MitoTracker Red | ThermoFisher Scientific, Molecular probes | Cat. #: M22425 | Mitochodria specific fluorescent dye |
| Chemical compound, drug | BODIPY FL thapsigargin | MedChemExpress | Cat. #: HY-D1608 | ER specific fluorescent dye |
| Chemical compound, drug | Alexa 647-maleimide | ThermoFisher Scientific, Molecular probes | Cat. #: A20347 | Thiol reactive fluorescent dye |
| Chemical compound, drug | Alexa568-maleimide | ThermoFisher Scientific, Molecular probes | Cat. #. A20341 | Thiol reactive fluorescent dye |
| Chemical compound, drug | PC (L-α-phosphatidylcholine) | Avanti Polar Lipids | Cat. #:840051 C | for making liposomes, used 48% PC |
| Chemical compound, drug | DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine) | Avanti Polar Lipids | Cat. #: 840035 C | for making liposomes, used 10% DOPS |
| Chemical compound, drug | PI (L-α-phosphatidylinositol) | Avanti Polar Lipids | Cat. #:840042 C | for making liposomes, used 10% PI |
| Chemical compound, drug | PE (L-α-phosphatidylethanolamine) | Avanti Polar Lipids | Cat. #: 841118 C | for making liposomes, used 28% PE |
| Chemical compound, drug | TOCL, (18:1 Cardiolipin) | Avanti Polar Lipids | Cat. #: 710335 C | for making liposomes, used 4% TOCL |
| Chemical compound, drug | A-1331852 | Chemietek | Cat. #: CT-A115 | in DMSO |
| Chemical compound, drug | S-63845 | Chemietek | Cat. #: 1799633-27-4 | in DMSO |
| Chemical compound, drug | S-64315 also named “MIK665” | ChemieTek | Cat. # CT-MIK665 | in DMSO |
| Chemical compound, drug | Navitoclax; ABT-263 | Selleckchem | Cat. #: S1001 | in DMSO |
| Chemical compound, drug | AZD4320 | ChemieTek | Cat. #: CT-A4320 | in DMSO |
| Chemical compound, drug | Thapsigargin | Sigma-Aldrich | Cat. #:T9033 | in DMSO |
| Chemical compound, drug | Etoposide | Sigma-Aldrich | Cat. #: 33419-42-0 | in DMSO |
| Chemical compound, drug | Tunicamycin | Sigma-Aldrich | Cat. #: T7765 | in DMSO |
| Commercial assay or kit | TransIT-X2 | Mirus | Cat. #: Mir 6003 | Transfection reagent |
| Other | Cell Carrier-384, Ultra | PerkinElmer | Cat. #: 6057300 | for live cell imaging |
| Other | Non-binding surface, 96-well plate, black with clear bottom | Corning | Cat. #: 3881 | For recombinant protein and liposome assays critical to use non-binding plate |
| Other | Opera Phenix | PerkinElmer | Cat. #: HH14000000 | Automated spinning disc confocal microscope |
| Other | INO-FHS microscope | PMID:35442739 | Custom built by INO for Dr. David Andrews' lab | |
| Other | The Infinite M1000 | Tecan | Platereader for in vitro assays with recombinant proteins and liposome assays | |
| Gene (H. sapiens) | Bax | PMID:14522999, | GI: L22473.1 | For recombinant protein |
| Gene (H. sapiens) | Bcl-XL | PMID:18547146 | GI: Z23115.1 | For recombinant protein |
| Gene (H. sapiens) | PUMA | PMID:35442739 | GI: 27113 Addgene plasmid# 166739 | |
| Gene (M. musculus) | BimL | PMID:30860026 | GI: AAD26594.1 | For recombinant BimL purification |
| Gene (M. musculus) | tBid | PMID:22464442 | GI: NM_007544.4 | for expression of VtBid in cells |
| Gene (M. musculus) | Bid | PMID:19062087 | GI: NM_007544.4 | Jean-Claude Martinou, SeronoPharmaceutical Institutte For purification of Bid |
| Software, algorithm | GraphPad Prism, version 6 | San Diego, California | RRID: SCR_002798 | Scientific graphing program, used to perform statistical analysis |
| Software, algorithm | MATLAB with toolboxes: Signal Processing, Curve Fitting, Image Processing, Version R2020a | https://doi.org/10.5683/SP3/ZKXQW8 | RRID:SCR_00162 | https://www.mathworks.com/products/matlab.html |
| Software, algorithm | INO software package including INO-FHS Acquisition, INO_FHS_Analysis, and INO_FHS_Batch Analysis | INO Client Release_r10357 package | ||
| Transfected Construct | mVenus-pEGFP-C1 | other | GI: KU341334.1 | Dr. Ray Truant (McMaster University). Backbone EGFP-C1 (Clonetech) |





