Opposing chemosensory functions of closely related gustatory receptors
Figures
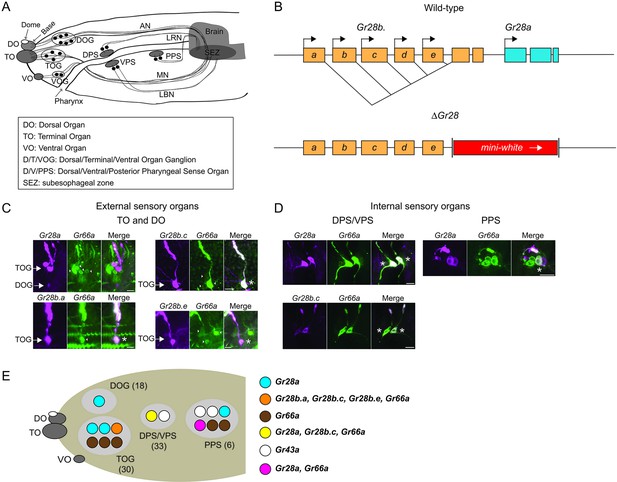
Expression of the Gr28 genes in the larval sensory organs.
(A) Schematic representation of the larval chemosensory system. Three external sensory organs (dorsal organ [DO], terminal organ [TO], and ventral organ [VO]) hold collectively the dendritic extensions of neuronal cell bodies in the respective ganglia (dorsal organ ganglia [DOG], terminal organ ganglia [TOG], and ventral organ ganglia [VOG]). Three clusters of sensory neurons (dorsal pharyngeal sense organ [DPS], ventral pharyngeal sense organ [VPS], and posterior pharyngeal sense organ [PPS]) reside along the pharynx. The antennal nerve (AN) connects the DOG neurons to the subesophageal zone (SEZ). The TOG and VOG neurons project along the maxillary nerve (MN) to the SEZ. The DPS and PPS neurons project axons to the SEZ via the labral nerve (LRN). The VPS neurons project to the SEZ through the labial nerve (LBN). Note that olfactory neurons are omitted in the schematic here and in (E) (below). (B) Structure of the Gr28 locus. The six Gr28 genes are clustered within 10 kilobases. The five Gr28b genes are transcribed from distinct promotors, with first unique exons that are spliced to common second and third exons. Gr28a is downstream of the Gr28b genes. The Gr28 mutation (ΔGr28) used in this study lacks the shared common exons of the Gr28b genes and the entire Gr28a gene. (C) Expression of the Gr28 genes in the external sensory organs. Note that images show only one of the bilaterally symmetrical organs. Co-expression between Gr66a (a marker for bitter taste gustatory receptor neurons [GRNs]) and different Gr28 genes (in ‘Merge’ panel) was assessed using GAL4 and LexA drivers for Gr28 genes and Gr66a, respectively. For Gr28b.a, Gr28b.c and Gr28b.e co-expression was observed in each case. However, Gr66a and Gr28a are expressed in different GRNs. Asterisks refer to a GRN expressing both Gr66a and the indicated Gr28 gene. Scale bars are 5 μm. (D) Expression of the Gr28 genes in internal sensory organs. Note that the bilaterally symmetrical halves of the DPS/VPS are physically close to each other, and the images includes both halves, while the image of the PPS shows only one side of the bilaterally symmetrical organ. Gr28b.c, but none of the other Gr28b genes, is co-expressed with Gr66a in the DPS/VPS. In the PPS, none of the Gr28b genes is found, while Gr28a and Gr66a are partially co-expressed, but each gene is also expressed exclusively in a subset of GRNs. Asterisks refer to a GRN expressing both Gr66a and the indicated Gr28 gene. Scale bars are 5 μm. (E) Expression summary: only relevant neurons in one of the paired taste organs are shown, with total number of neurons indicated in parenthesis. The cartoon summarizes the immunostainings shown in (C) and (D). The VOG is not shown as none of the Gr28-GAL4 drivers is expressed there. Immunostaining was performed on whole-mount preparations from larvae heads of the following genotypes: UAS-mCD8:RFP lexAop-rCD2:GFP;Gr66a-LexA/Gr28a-GAL4, UAS-mCD8:RFP lexAop-rCD2:GFP;Gr66a-LexA/Gr28b.c-GAL4, UAS-mCD8:RFP lexAop-rCD2:GFP;Gr66a-LexA/Gr28b.e-GAL4 and UAS-mCD8:RFP lexAop-rCD2:GFP;Gr66a-LexA/+; Gr28b.a-GAL4/+. The Gr43aGAL4 GRNs, which do not overlap with Gr28a-GAL4 neurons (Mishra et al., 2018), are shown for reference to experiments described in (Figure 2).
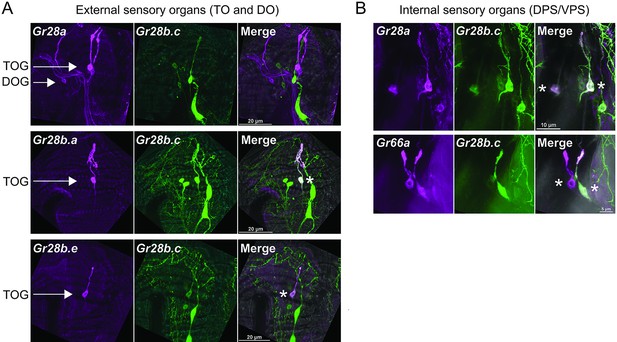
Co-expression analysis between Gr28 genes in larval sensory organs.
Immunostaining with anti-GFP (green) and anti-mCD8 (red) antibodies on whole-mount preparations of the heads from larvae of the genotypes: (1) UAS-mCD8:RFP lexAop-rCD2:GFP;Gr28a-GAL4 or Gr28b.e-GAL4 or Gr66a-GAL4/+;Gr28b.c-LexA/+: (2) UAS-mCD8:RFP lexAop-rCD2:GFP;+; Gr28b.a-GAL4/Gr28b.c-LexA. Asterisks refer to gustatory receptor neuron (GRN) expressing both Gr28b.c and the indicated Gr66a or Gr28 gene. Note that images show only one of the bilaterally symmetrical external organs in the dorsal organ ganglia (DOG)/terminal organ ganglia (TOG) (A) and both bilaterally symmetrical organs in the dorsal pharyngeal sense organ (DPS)/ventral pharyngeal sense organ (VPS) (B).
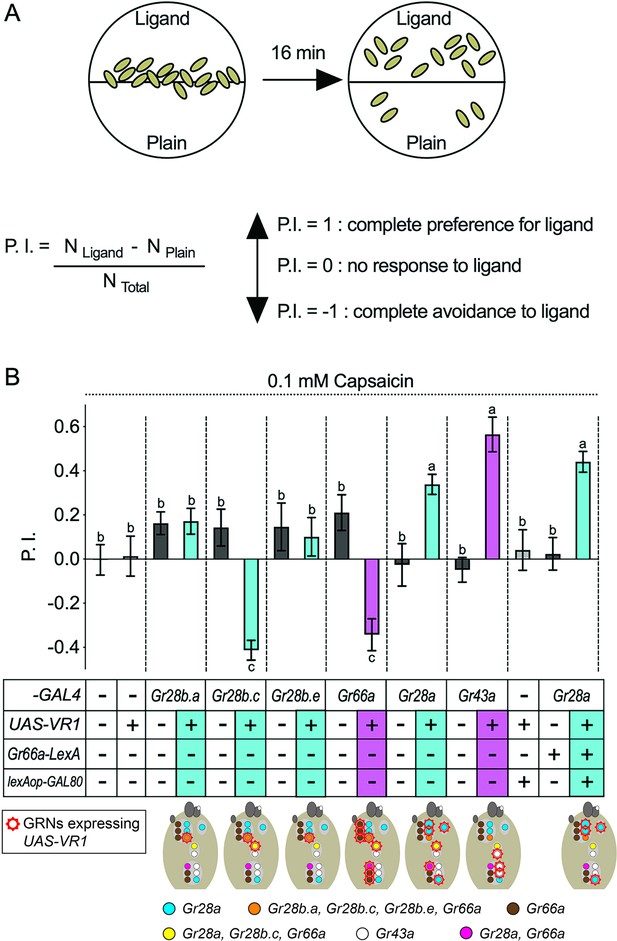
Intrinsic valence of different Gr28 gustatory receptor neurons (GRNs).
(A) Behavioral two-choice preference assay to quantify appetitive and avoidance of larvae for chemical ligands. Fifteen feeding stage, third-instar larvae are placed along the demarcation of a 1% agarose containing dish (35 mm), with one side plain, and the other side containing ligand. The preference index (P.I.; see ‘Materials and methods’) is calculated after counting location of larvae after 16 min. (B) Gr28a GRNs mediate capsaicin preference, while Gr28b.c GRNs elicit capsaicin avoidance in larvae. GRNs expressing the capsaicin receptor VR1 are marked with a red crown in diagrams below. w1118, reporter gene only (UAS-VR1/+), and respective GAL4 driver only (Gr-GAL4/+) larvae serve as negative controls (white panels) and show neither preference for nor avoidance to 0.1 mM capsaicin. Experimental larvae expressing VR1 in Gr28-GAL4 neurons are shown in blue. Positive control larvae (purple panels) expressing VR1 in bitter taste GRNs (Gr66a-Gal4) or appetitive, sweet GRNs (Gr43aGAL4) show expected avoidance to or preference for capsaicin. Experimental larvae (blue panels) expressing VR1 in Gr28a-GAL4 GRNs display robust preference for capsaicin, which is still observed when expression is further restricted to Gr28aonly GRNs (Gr66a-LexA/UAS-VR1; Gr28a-GAL4/lexAop-GAL80; right panel). In contrast, when VR1 is expressed in Gr28b.c-GAL4 GRNs, larvae strongly avoid capsaicin. Neither avoidance nor preference was observed when VR1 is expressed in the Gr28b.a-GAL4 or Gr28b.e-GAL4 GRNs. Each bar represents the mean ± SEM of P.I. (n = 10–22 assays). The taste behavior of Gr-GAL4>UAS-VR1 larvae is compared to three controls (w1118, UAS-VR1/+ and Gr-GAL4/+) using one-way ANOVA with Bonferroni’s multiple comparison tests (p<0.05), whereby different letters indicate a statistically significant difference. Dashed lines delineate groups for ANOVA. All control and experimental larvae are in the w1118 background, carry one copy of the indicated transgene(s), and were generated from crosses of respective strains listed in ‘Materials and methods’.
-
Figure 2—source data 1
Taste preference assay for 0.1 mM capsaicin of larvae expressing VR1 in different GRNs.
Taste preference assay of larvae with expression of VR1 in Gr28aonly GRNs using lexAop-GAL80 under control of Gr66a-LexA for 0.1 mM capsaicin.
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig2-data1-v1.xlsx
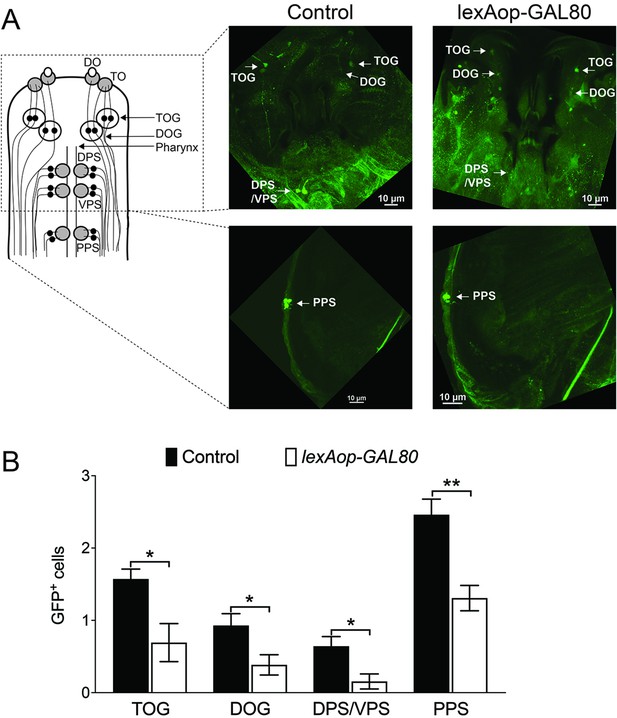
Suppression of GAL4 in a subset of Gr28a-GAL4 neurons.
(A) Expression of GCaMP6m was restricted to a subset of Gr28a-GAL4 GRNs by inclusion of Gr66a-LexA and lexAop-GAL80. Immunostaining with anti-GFP (green) antibody on whole-mount preparations of the heads from larvae. The ‘control’ genotype shown on the left is w1118; Gr66a-LexA/UAS-GCaMP6m;Gr28a-GAL4/+. The ‘lexAop-GAL80’ genotype on the right is w1118;Gr66a-LexA/UAS-GCaMP6m;Gr28a-GAL4/lexAop-GAL80. Note the reduced number GFP-positive GRNs in the ‘lexAop-GAL80’ genotype compared to the ‘control.’ (B) Quantification of GFP-positive GRNs in the different sensory organs. Each bar represents the mean ± SEM of GFP-expressing cells with 13–14 larvae. Asterisks indicate a significant difference between control larvae and larvae expressing lexAop-GAL80 by under control Gr66a-LexA driver (lexAop-GAL80) (two-tailed, Mann–Whitney U test, **p<0.01, *p<0.05).
-
Figure 2—figure supplement 1—source data 1
Expression of GCaMP6m limited to a subset of Gr28a GRNs using lexAop-GAL80 under control of Gr66a-LexA.
(B) Quantification of GFP positive GRNs recorded from images of Gr28a GRNs expressing UAS-GCaMP6m (A).
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Quantification of GFP positive GRNs recorded from images of Gr28a GRNs expressing UAS-GCaMP6m (A).
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig2-figsupp1-data2-v1.xlsx
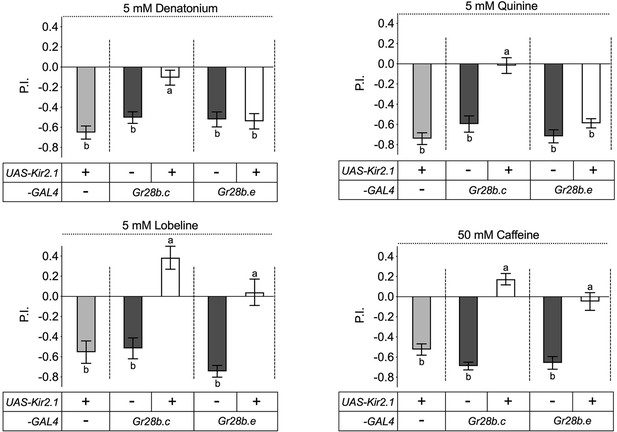
The Gr28b.c neurons mediate avoidance behavior to bitter compounds.
Inactivation of Gr28b.c gustatory receptor neurons (GRNs) (Gr28b.c-GAL4/UAS-Kir2.1) elicits significantly reduced avoidance of larvae to all four bitter compounds tested – denatonium, quinine, lobeline, and caffeine – while control larvae, carrying either the driver or the reporter only, showed strong avoidance of these compounds. In contrast, larvae with inactivated Gr28b.e GRNs (Gr28b.e-GAL4/UAS-Kir2.1) still avoid denatonium and quinine (top), but no longer avoid lobeline and caffeine (bottom). Each bar represents the mean ± SEM of preference index (P.I.) (n = 11–20 assays). The taste behavior of Gr28b.c-GAL4; UAS-Kir2.1 and Gr28b.e-GAL4; UAS-Kir2.1 larvae was compared to two controls (UAS-Kir2.1/+ and Gr28b.c -GAL4/+ or Gr28b.e-GAL4/+) using Kruskal–Wallis test by ranks with Dunn’s multiple comparison tests (p<0.05). Bars with different letters are significantly different. Dashed lines delineate groups for ANOVA. Fly genotypes: w1118; UAS-Kir2.1/+ (light gray), w1118; Gr28b.c-GAL4/+ (dark gray), w1118; Gr28b.c-GAL4/UAS-Kir2.1 (white), w1118; Gr28b.e-GAL4/+ (dark gray), w1118; Gr28b.e-GAL4/UAS-Kir2.1 (white).
-
Figure 3—source data 1
Taste preference assay for bitter compounds of larvae with inactivated Gr28b.c or Gr28b.e GRNs using expression of UAS-Kir2.1.
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig3-data1-v1.xlsx
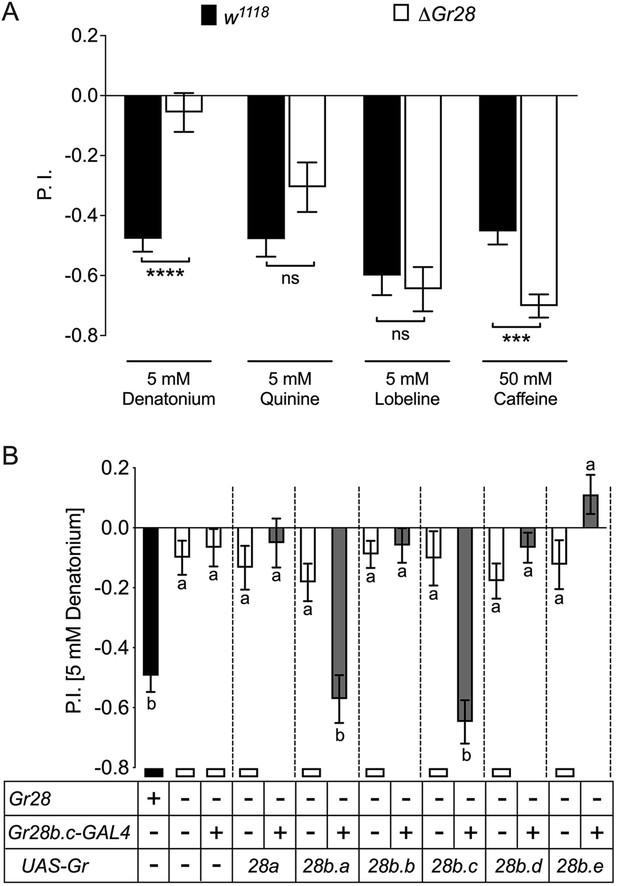
Role of individual Gr28 genes in bitter taste avoidance.
(A) Gr28 genes are required for sensing denatonium. Wild-type (w1118) larvae, but not Gr28 mutant larvae (w1118;ΔGr28/ΔGr28), strongly avoid denatonium. In contrast, w1118;ΔGr28/ΔGr28 larvae do not show significantly reduced avoidance to quinine, lobeline and caffeine. Each bar represents the mean ± SEM of preference index (P.I.) (n = 12–22 assays). Asterisks indicate a significant difference between w1118; ΔGr28/ΔGr28 and w1118 larvae (two-tailed, Mann–Whitney U test, ****p<0.0001, ***p<0.001, ns, not significant). (B) Single Gr28b genes can rescue avoidance response to denatonium when expressed in Gr28b.c neurons of Gr28 mutant larvae. The behavior of w1118;ΔGr28/ΔGr28 larvae expressing UAS-Gr28 transgenes under control of the Gr28b.c-GAL4 driver was compared to Gr28+ control (w1118, black bar), and three Gr28 mutant controls (ΔGr28, ΔGr28 plus driver and ΔGr28 plus respective UAS-Gr28 transgene, white bar) using Kruskal–Wallis test by ranks with Dunn’s multiple comparison tests (p<0.05). Each bar represents the mean ± SEM of P.I. (n = 11–22 assay). Bars with different letters are significantly different. Dashed lines delineate groups for ANOVA. Fly genotypes: wild-type: w1118 (black), mutants: w1118;ΔGr28/ΔGr28, w1118; ΔGr28/ΔGr28 Gr28b.c-GAL4, w1118;ΔGr28/ΔGr28; UAS-Gr28 (indicated Gr28 genes)/+ and w1118; ΔGr28/ΔGr28 UAS-GCaMP6m; UAS-Gr28 (for Gr28b.b or Gr28b.c genes)/+ (white), rescues: w1118; ΔGr28/ΔGr28 Gr28b.c-GAL4; UAS-Gr28 (indicated Gr28 genes)/+ and w1118; ΔGr28 UAS-GCaMP6m/ΔGr28 Gr28b.c-GAL4; UAS-Gr28 (for Gr28b.b or Gr28b.c genes)/ + (gray).
-
Figure 4—source data 1
Taste response to bitter compounds of Gr28 mutant larvae.
(A) Taste preference assay of wild-type and Gr28 mutant larvae for bitter compounds. (B) Taste preference assay of Gr28 mutant larvae for denatonium expressing single Gr28 genes in Gr28b.c GRNs.
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig4-data1-v1.xlsx
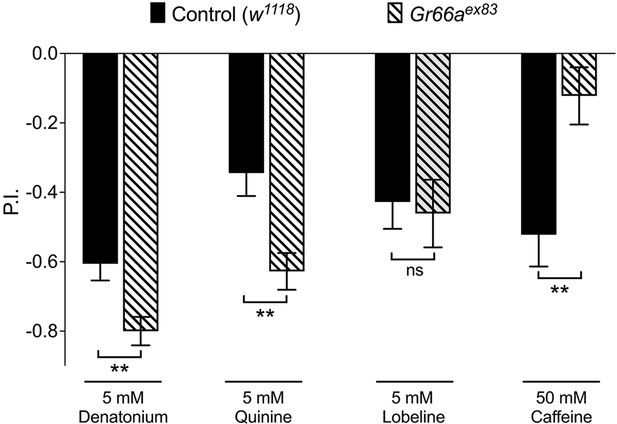
Gr66a is necessary for caffeine avoidance of larvae.
Gr66a null mutant larvae are unable to avoid of caffeine but none of other bitter compounds. Each bar represents the mean ± SEM of P.I. (n = 11–12 assays). Asterisks indicate a significant difference between the Gr66a mutant (Gr66aex83) and control larvae (w1118) (two-tailed, Mann–Whitney U test, **p<0.01, ns, not significant).
-
Figure 4—figure supplement 1—source data 1
Taste response to bitter compounds of Gr66a mutant larvae.
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig4-figsupp1-data1-v1.xlsx
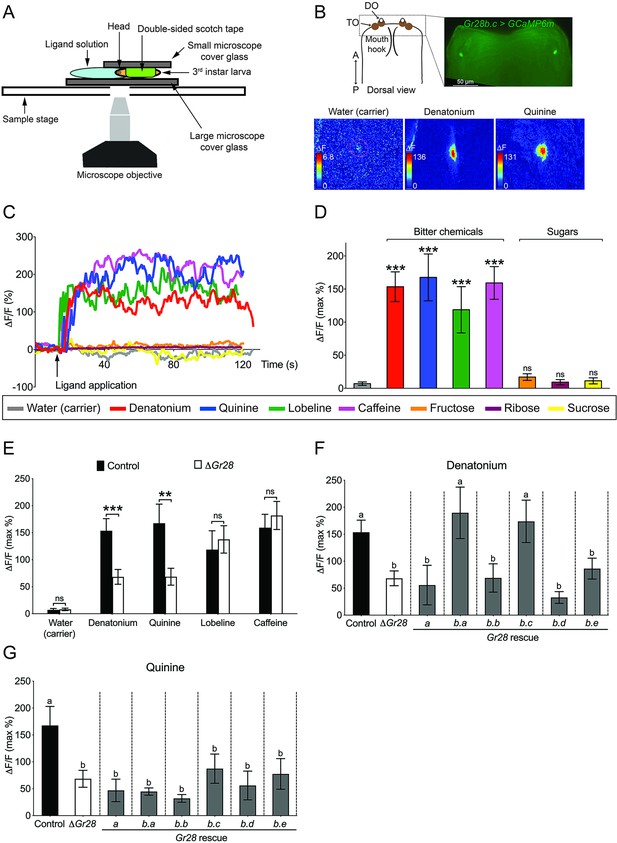
Cellular Ca2+ responses of Gr28b.c gustatory receptor neurons (GRNs) to select bitter compounds requires Gr28b.a or Gr28b.c.
(A) Diagram of Ca2+ imaging experimental set up. (B) Representative still images of Ca2+ response in the Gr28b.c expressing GRN of the TOG. Ca2+ responses of the Gr28b.c GRNs upon stimulation with indicated ligands. ΔF indicates the changes in fluorescence light intensity of the cell body after ligand application. (C, D) Representative traces (C) and quantified Ca2+ responses (D) of the Gr28b.c GRNs after stimulation with indicated ligands. Fly genotype: w1118; Gr28b.c-GAL4/UAS-GCaMP6m. Each bar represents the mean ± SEM of Ca2+ imaging with 12–16 larvae. Asterisks indicate a significant difference between carrier (water) and indicated ligands (two-tailed, Mann–Whitney U test, ***p<0.001, ns, not significant). (E) Neurons of larvae lacking the Gr28 genes exhibit significantly reduced responses to denatonium and quinine. Gr28b.c-expressing GRNs in the TOG of Gr28 mutant larvae (ΔGr28) have significantly reduced Ca2+ responses to denatonium and quinine but not to lobeline or caffeine when compared to Gr28b.c-expressing GRNs of wild-type controls. Larvae genotypes: Gr28+ control (black bar): w1118; Gr28b.c-GAL4/UAS-GCaMP6m. ΔGr28 control (white bar): w1118; ΔGr28 Gr28b.c-GAL4/ΔGr28 UAS-GCaMP6m. Each bar represents the mean ± SEM with 13–16 larvae. Asterisks indicate a significant difference between Gr28+ and ΔGr28 larvae (two-tailed, Mann–Whitney U test, ***p<0.001, **p<0.01; ns, not significant). (F, G) Gr28b.c or Gr28b.a transgenes rescue denatonium responses in Gr28b.c-GAL4 neurons of ΔGr28 larvae. Expression of Gr28b.c or Gr28b.a is under control of Gr28b.c-GAL4 restores responses to denatonium, but not quinine in TOG GRNs of ΔGr28 larvae. Each bar represents the mean ± SEM of Ca2+ imaging with 12–17 larvae. The Ca2+ responses of Gr28 mutant larvae expressing UAS-Gr28 transgenes under Gr28b.c-GAL4 driver is compared to Gr28+ (black) and ΔGr28 (white) controls using Kruskal–Wallis test by ranks with Dunn’s multiple comparison tests (p<0.05). Bars with different letters are significantly different. Dashed lines delineate groups for ANOVA. Fly genotypes: Gr28+ control (black bar): w1118; Gr28b.c-GAL4/UAS-GCaMP6m. ΔGr28 control (white bar): w1118; ΔGr28 Gr28b.c-GAL4/ΔGr28 UAS-GCaMP6m. Gr28 rescues (gray bar): w1118; ΔGr28 Gr28b.c-GAL4/ΔGr28 UAS-GCaMP6m; UAS-Gr28 (indicated Gr28 genes)/+. Concentration of ligands was 100 mM for sugars, 50 mM for caffeine, and 5 mM for denatonium, quinine, and lobeline.
-
Figure 5—source data 1
Ca2+ imaging experiments with Gr28b.c GRNs in the TOG.
(D) Ca2+ responses of Gr28b.c GRNs in the TOG to bitter compounds and sugars.
(E) Ca2+ responses of Gr28 mutant larvae to bitter compounds. (F, G) Ca2+ responses of Gr28 mutant larvae to denatonium (F) or quinine (G) expressing single Gr28 genes in Gr28b.c GRNs.
- https://cdn.elifesciences.org/articles/89795/elife-89795-fig5-data1-v1.xlsx
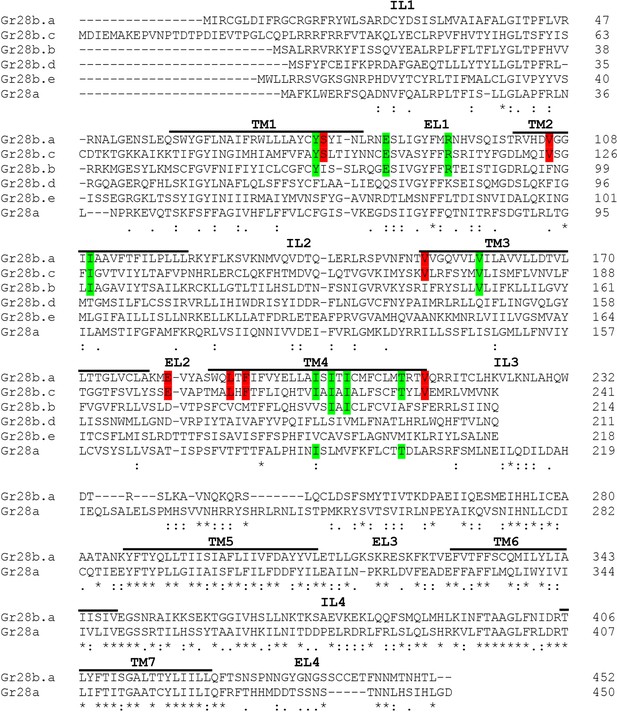
Amino acid alignment of the six Gr28 proteins.
The sequence alignment was generated using Clustal Omega tool from ClustalW2 (https://www.ebi.ac.uk/Tools/msa/clustalo/). IL and EL indicate intracellular loop and extracellular loop, respectively. Note that the C terminal region starting at the IL3 is identical in the Gr28b proteins. Red highlighted letters indicate amino acids identical only in Gr28b.a and Gr28b.c. Green highlighted letters indicate amino acids conserved in Gr28b.a, Gr28b.c, and one other Gr28 protein. Asterisks below the sequences indicate residues identical in all Gr28 proteins, colons indicate conserved residue (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW), and periods indicate moderately conserved residue (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY). TM1-7 indicate helical transmembrane segments predicted using HMMTOP 2.0 software.
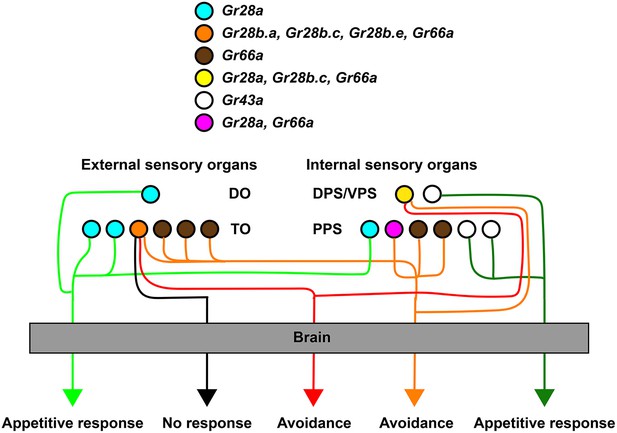
Role of different gustatory receptor neuron (GRN) subsets in taste behavior of larvae.
GRNs sufficient for mediating avoidance behavior can be defined by Gr28b.c-GAL4, while GRNs sufficient for mediating appetitive behavior are defined by a subset of Gr28a-GAL4 GRNs (Gr28aonly GRNs; see also Figure 2B and Figure 2—figure supplement 1). Note that each ensemble is composed of at least a pair of neurons located in the external taste organs and a pair of neurons in the internal taste organs. Also, a larger set of avoidance neurons (Gr66a-GAL4) might function independently of any Gr28b.c neurons, and one set of fructose sensing neurons (Gr43aGAL4) distinct from Gr28a-GAL4 GRNs mediates appetitive behavior.
Tables
Conserved amino acids in the amino termini of Gr28b.a and Gr28b.c.
The seven amino acid residues identical in the amino-terminal region of Gr28b.c and Gr28b.a are shown in bold (residue number is taken from Gr28b.c). The nine additional amino acids also identical in one additional Gr28 proteins are also listed. These residues are considered potentially critical for recognition of denatonium since only Gr28b.a and Gr28b.c can rescue response to denatonium when expressed in Gr28b.c neurons of ΔGr28/ΔGr28 mutant larvae.
| Location | Conserved in Gr28b.c/Gr28b.a | Other (if applicable) |
|---|---|---|
| EL1 | E103 | Gr28b.b |
| R111 | Gr28b.b | |
| EL2 | E200 | None |
| TM1 | Y94 | Gr28b.b |
| S95 | None | |
| TM2 | V124 | None |
| I128 | Gr28b.b | |
| TM3 | V170 | None |
| V177 | Gr28b.b | |
| TM4 | L207 | None |
| F209 | None | |
| I218 | Gr28a | |
| I220 | Gr28b.b | |
| I222 | Gr28b.b | |
| T229 | Gr28a | |
| V232 | none |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-GFP (rabbit polyclonal) | Thermo Fisher Scientific | Cat# A6455, RRID:AB_221570 | IF (1:1000) |
| Antibody | Anti-mCD8 (rat monoclonal) | Thermo Fisher Scientific | Cat# MCD0800, RRID:AB_10392843 | IF (1:200) |
| Antibody | Anti-rabbit Alexa 488 (goat polyclonal) | Thermo Fisher Scientific | Cat# A11070, RRID:AB_2534114 | IF (1:500) |
| Antibody | Anti-rat Cy3 (goat polyclonal) | Jackson ImmunoResearch Laboratories Inc | Cat# 112-165-072, RRID:AB_2338248 | IF (1:300) |
| Chemical compound, drug | Caffeine | MilliporeSigma | C0750 | |
| Chemical compound, drug | Capsaicin | MilliporeSigma | M2028 | |
| Chemical compound, drug | Denatonium benzoate | MilliporeSigma | D5765 | |
| Chemical compound, drug | Lobeline hydrochloride | MilliporeSigma | 141879 | |
| Chemical compound, drug | D-(-)-ribose | MilliporeSigma | R7500 | |
| Chemical compound, drug | Quinine hydrochloride dihydrate | MilliporeSigma | Q1125 | |
| Chemical compound, drug | Fructose | Spectrum Chemical | F1092 | |
| Chemical compound, drug | Agarose | Apexbio | 20-102 | |
| Chemical compound, drug | Sucrose | Macron Fine Chemicals | 8360-06 | |
| Chemical compound, drug | Charcoal | J.T. Baker | 1560-01 | |
| Genetic reagent (Drosophila melanogaster) | w1118 | Bloomington Drosophila Stock Center | BDSC: 3605; FLYB: FBst0003605 | |
| Genetic reagent (D. melanogaster) | Gr28a-GAL4 | Thorne and Amrein, 2008 | FLYB: FBtp0056017 | FlyBase symbol: w*; P{Gr28a-GAL4.T}SF36S |
| Genetic reagent (D. melanogaster) | Gr28a-GAL4 | Thorne and Amrein, 2008 | FLYB: FBtp0056017 | FlyBase symbol: w*; P{Gr28a-GAL4.T}SF36B1 |
| Genetic reagent (D. melanogaster) | Gr28b.a-GAL4 | Thorne and Amrein, 2008 | FLYB: FBtp0054526 | FlyBase symbol: w*; P{Gr28b.a-GAL4}NT42aC51a |
| Genetic reagent (D. melanogaster) | Gr28b.c-GAL4 | Thorne and Amrein, 2008 | FLYB: FBtp0054528 | FlyBase symbol: w*; P{Gr28b.c-GAL4}NT21B1 |
| Genetic reagent (D. melanogaster) | Gr28b.e-GAL4 | Scott et al., 2001 | FLYB: FBtp0014672 | FlyBase symbol: w*; P{Gr28b.e-GAL4.4.245}Gr28a3AII |
| Genetic reagent (D. melanogaster) | ΔGr28/ΔGr28 | Mishra et al., 2018 | FLYB: FBab0049019 | FlyBase symbol: w*; Df(2L)ΔGr28 |
| Genetic reagent (D. melanogaster) | Gr66a-GAL4 | Scott et al., 2001 | FLYB: FBtp0014661 | FlyBase symbol: w*; P{Gr66C1-GAL4.3.153} |
| Genetic reagent (D. melanogaster) | UAS-Gr28a | Ni et al., 2013 | FLYB: FBal0344045 | FlyBase symbol: w*; P{UAS-Gr28a.G}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Gr28b.a | Ni et al., 2013 | FLYB: FBal0291410 | FlyBase symbol: w*; P{UAS-Gr28b.A}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Gr28b.b | Ni et al., 2013 | FLYB: FBal0291412 | FlyBase symbol: w*; P{UAS-Gr28b.B}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Gr28b.c | Ni et al., 2013 | FLYB: FBal0291411 | FlyBase symbol: w*; P{UAS-Gr28b.C}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Gr28b.d | Ni et al., 2013 | FLYB: FBal0291409 | FlyBase symbol: w*; P{UAS-Gr28b.D}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Gr28b.e | Ni et al., 2013 | FLYB: FBal0291408 | FlyBase symbol: w*; P{UAS-Gr28b.E}attP2 |
| Genetic reagent (D. melanogaster) | Gr43aGAL4 | Miyamoto et al., 2012 | BDSC:93447; FLYB:FBst0093447 | FlyBase symbol: w1118;Ti{GAL4}Gr43aGAL4 |
| Genetic reagent (D. melanogaster) | UAS-VR1E600K | Marella et al., 2006 | FLYB: FBal0215202 | FlyBase symbol: w1118;P{UAS-VR1E600K} |
| Genetic reagent (D. melanogaster) | lexAop-rCD2:GFP | Lai and Lee, 2006 | FLYB: FBst0066687 | FlyBase symbol: w*; P{lexAop-rCD2-GFP} |
| Genetic reagent (D. melanogaster) | UAS-mCD8:RFP | Bloomington Drosophila Stock Center | BDSC: 32220; FLYB: FBti0131987 | FlyBase symbol: y1w*;P{10XUAS-IVS-mCD8::RFP}su(Hw)attP8 |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6m | Bloomington Drosophila Stock Center | BDSC: 42748; FLYB: FBti0151346 | FlyBase symbol: w1118; P{20XUAS-IVS-GCaMP6m}attP40 |
| Genetic reagent (D. melanogaster) | UAS-Kir2.1-GFP | Baines et al., 2001; Paradis et al., 2001 | FLYB: FBst0006596 | FlyBase symbol: w*; P{UAS-Hsap\KCNJ2.EGFP}1 |
| Genetic reagent (D. melanogaster) | Gr66a-LexA | Thistle et al., 2012 | BDSC: 93024; FLYB: FBst0093024 | FlyBase symbol: w1118; P{Gr66a-lexA.S}2;TM2/TM6B |
| Genetic reagent (D. melanogaster) | lexAop-GAL80 | Thistle et al., 2012 | FLYB: FBtp0079728 | FlyBase symbol: w1118; P{lexAop-GAL80. T} |
| Genetic reagent (D. melanogaster) | Gr28b.c-LexA | This paper | FlyBase symbol: w1118;P{Gr28b.c-LexA}#8 | |
| Sequence-based reagent | Gr28b.c_F | This paper | PCR primers | 5′-AATCTAGGTACCCCGGCTGCTCGTCTCCCTGGATGT-3′ |
| Sequence-based reagent | Gr28b.c_R | This paper | PCR primers | 5′-CGTCAAACTAGTGACCGCTTCGTTTGAGCTTCAACC-3′ |
| Recombinant DNA reagent | LexA vector CMC105 (plasmid) | This paper Larsson et al., 2004 | Insect expression vector | |
| Software, algorithm | NIS-Elements | Nikon | N/A | |
| Software, algorithm | Prism software 10.1.0 (264) | GraphPad Software | N/A | |
| Software, algorithm | Adobe pPhotoshop 2022 | Adobe | N/A | |
| Other | Normal goat serum | SouthernBiotech | Cat# 0060-01 | IF (5%) ‘Materials and methods’ |
| Other | Nikon Eclipse Ti inverted microscope | Nikon | N/A | ‘Materials and methods’ |
| Other | Nikon A1R confocal microscope system | Nikon | N/A | ‘Materials and methods’ |
| Other | PertriPetri dish, 60 × 15 mm | Falcon | REF353004 | ‘Materials and methods’ |
| Other | Microscope cover glass, 24 × 50 mm | VWR | 16004-098 | ‘Materials and methods’ |
| Other | Microscope cover glass, 12CIR-1 | Thermo Fisher Scientific | 1254580 | ‘Materials and methods’ |





