Insights into metabolic heterogeneity of colorectal cancer gained from fluorescence lifetime imaging
Figures
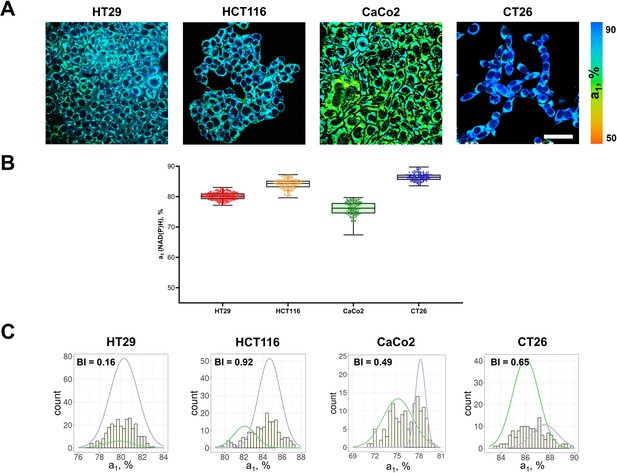
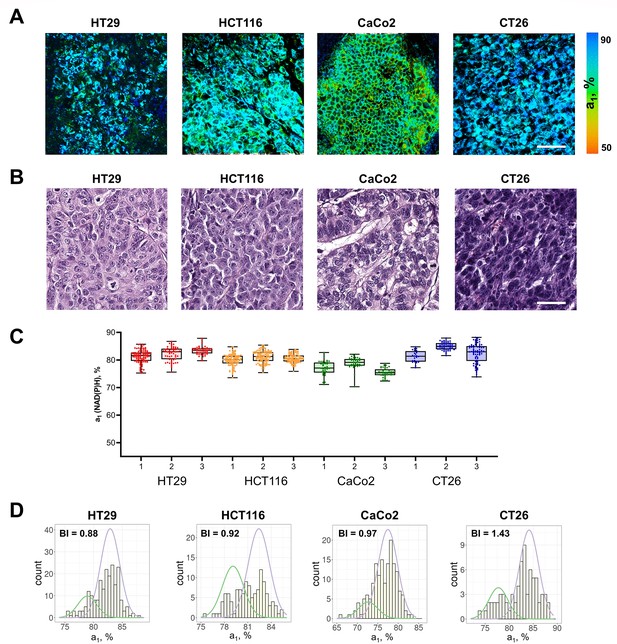
FLIM of NAD(P)H in monolayer cell cultures.
(A) Representative FLIM images of colorectal cancer cell lines. Scale bar = 50 μm. For FLIM: ex. 750 nm, reg. 450–490 nm. (B) The relative contribution of free NAD(P)H (a1, %) in cell cultures. Box shows the median and the quartiles Q1 and Q3, whiskers show minimum and maximum. Dots indicate individual cells (n=280 for HT29 cells, n=185 for HCT116 cells, n=146 for CaCo2 cells, n=138 for CT26 cells). p-values are shown in Supplementary file 1. (C) The distribution of the NAD(P)H-a1 for the cell lines. The bimodality index (BI-a1) is shown on each diagram.
-
Figure 1—source data 1
Original FLIM data for Figure 1A (HT29 cells).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig1-data1-v1.zip
-
Figure 1—source data 2
Original FLIM data for Figure 1A (HCT116 cells).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig1-data2-v1.zip
-
Figure 1—source data 3
Original FLIM data for Figure 1A (CaCo2 cells).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig1-data3-v1.zip
-
Figure 1—source data 4
Original FLIM data for Figure 1A (CT26 cells).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig1-data4-v1.zip
-
Figure 1—source data 5
The dataset (NAD(P)H-a1 values) used to plot the charts shown in Figure 1B.
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig1-data5-v1.xlsx
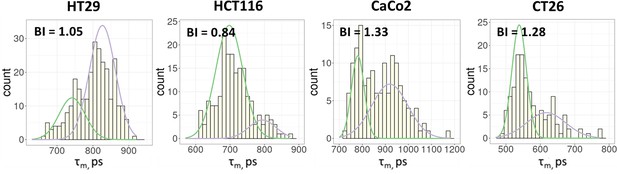
FLIM of NAD(P)H in monolayer cell cultures.
The distribution of the NAD(P)H-τm for the cell lines. The bimodality index (BI-τm) is shown on each diagram.
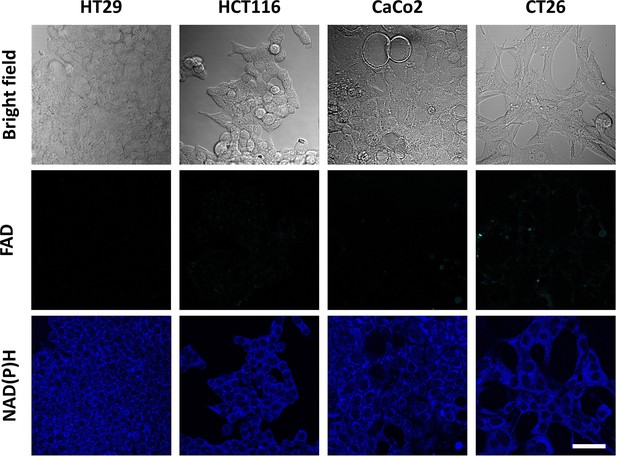
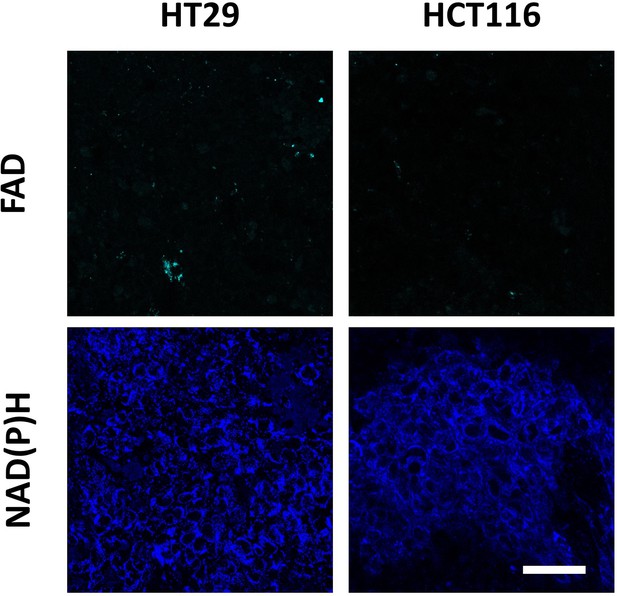
Autofluorescence of cofactors FAD and NAD(P)H in cultured cells HT29, HCT116, CaCo2 and CT26.
Representative fluorescence intensity images of flavins (FAD, ex. 900 nm, em. 500–550 nm) and NAD(P)H (ex. 750 nm, em. 450–490 nm). Scale bar = 50 μm.
FLIM of NAD(P)H in mouse tumors in vivo.
(A) FLIM images of NAD(P)H of tumor cells in mouse models in vivo. Scale bar = 50 μm. For FLIM: ex. 750 nm, reg. 450–490 nm. (B) Representative histological slices of tumors, hematoxylin/eosin (HE) staining, initial magnification 20×. Scale bar = 50 μm. (C) The relative contribution of free NAD(P)H (a1, %) in three representative tumors (numbered 1–3) obtained from different cell lines. Box shows the median and the quartiles Q1 and Q3, whiskers show minimum and maximum. Dots indicate individual cells (n=280 for HT29, n=340 for HCT116, n=160 for CaCo2, n=350 for CT26). p-values are shown in Supplementary file 1. (D) Representative distributions of the NAD(P)H-a1 for each type of tumor. The bimodality index (BI-a1) is shown on the diagrams.
-
Figure 2—source data 1
Original FLIM data for Figure 2A (HT29 tumor).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig2-data1-v1.zip
-
Figure 2—source data 2
Original FLIM data for Figure 2A (HCT116 tumor).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig2-data2-v1.zip
-
Figure 2—source data 3
Original FLIM data for Figure 2A (CaCo2 tumor).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig2-data3-v1.zip
-
Figure 2—source data 4
Original FLIM data for Figure 2A (CT26 tumor).
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig2-data4-v1.zip
-
Figure 2—source data 5
The dataset (NAD(P)H-a1 values) used to plot the charts shown in Figure 2C.
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig2-data5-v1.xlsx
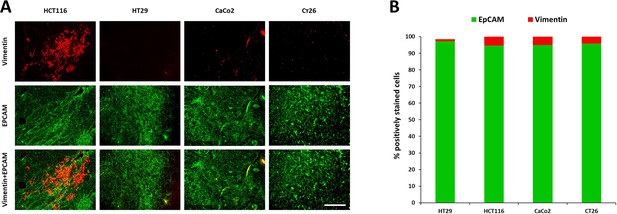
Immunohistochemical analysis of the expression of EpCAM (green, epithelial cells marker) and vimentin (red, mesenchymal cells marker) in mouse tumors.
(A) Immunofluorescence images of HT29, HCT116, CaCo2 and CT26 mouse tumors. Scale bar = 100 μm. (B) The ratio of EpCAM-positive to vimentin-positive areas in tumor sections.
Autofluorescence of cofactors FAD and NAD(P)H in HT29 and HCT116 tumor xenografts in vivo.
Representative fluorescence intensity images of flavins (FAD, ex. 900 nm, em. 500–550 nm) and NAD(P)H (ex. 750 nm, em. 450–490 nm). Scale bar = 50 μm.
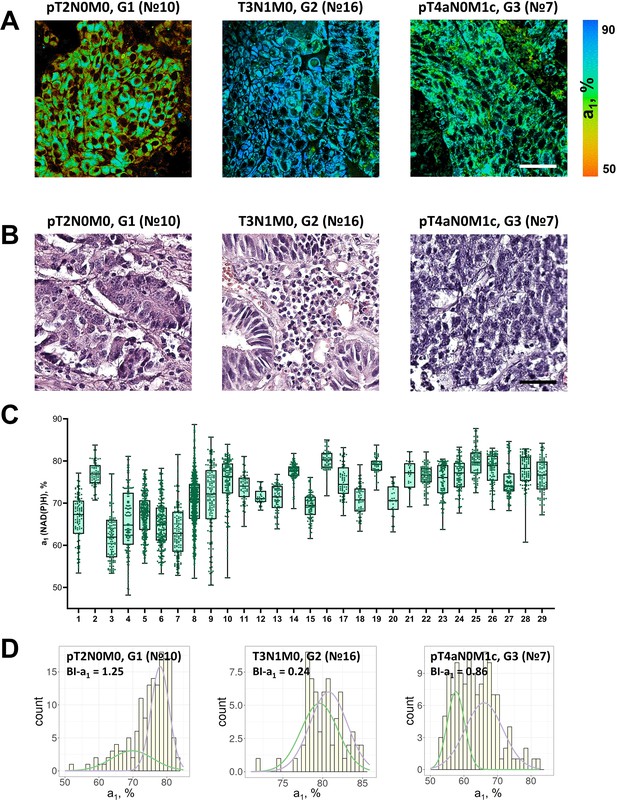
FLIM of NAD(P)H in patients’ tumor samples ex vivo.
(A) Representative FLIM images of patient tumors. Scale bar = 50 μm. For FLIM: ex. 750 nm, reg. 450–490 nm. (B) Histopathology of tumors, hematoxylin/eosin (HE) staining, initial magnification 20×. Scale bar = 50 μm. (C) The relative contribution of free NAD(P)H (a1, %) in patients’ tumors (numbered 1–29). Box shows the median and the quartiles Q1 and Q3, whiskers show minimum and maximum. Dots are the measurements from the individual cells. (D) Representative distributions of the NAD(P)H-a1 for patients’ tumors. The bimodality index (BI-a1) is shown on the diagrams.
-
Figure 3—source data 1
The dataset (NAD(P)H-a1 values) used to plot the charts shown in Figure 3C.
- https://cdn.elifesciences.org/articles/94438/elife-94438-fig3-data1-v1.xlsx
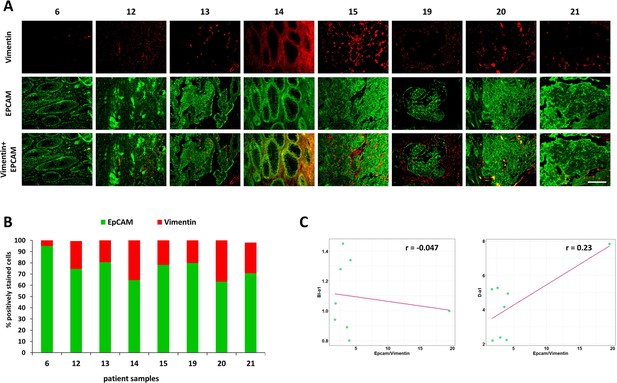
Immunohistochemical analysis of the expression of EpCAM (green, epithelial cells marker) and vimentin (red, mesenchymal cells marker) in patients’ tumors.
(A) Immunofluorescence images of patients’ tumors (numbered from 6 to 21). Scale bar = 100 μm. (B) The ratio of EpCAM-positive to vimentin-positive areas in tumor sections. (C) The bimodality index (BI-a1) and dispersion (D-a1) plotted against the EpCAM/vimentin ratio in patient tumor samples. Pearson correlation r is shown on the plots.
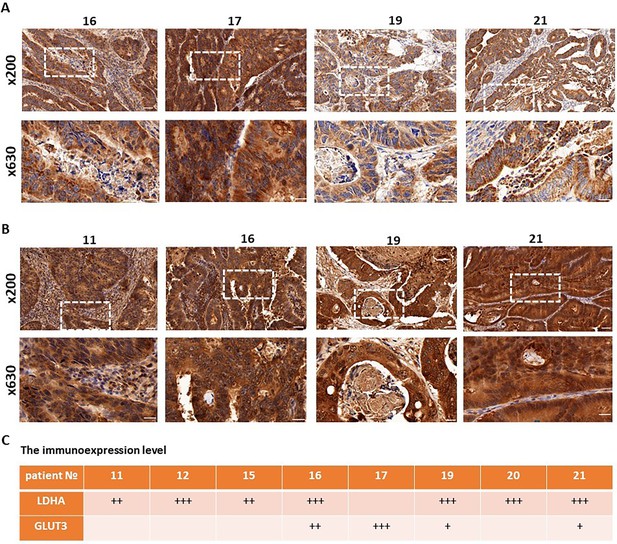
Immunohistochemical analysis of the expression of GLUT3 and LDHA in patients’ tumors.
(A) Representative immunohistochemical images of GLUT3 expression. Scale bar = 50 μm (magnification x200) and 20 μm (magnification x630). (B) Representative immunohistochemical images of LDHA expression. Scale bar = 50 μm (magnification x200) and 20 μm (magnification x630). (С) Semi-quantitative evaluation of the expression level by staining intensity.
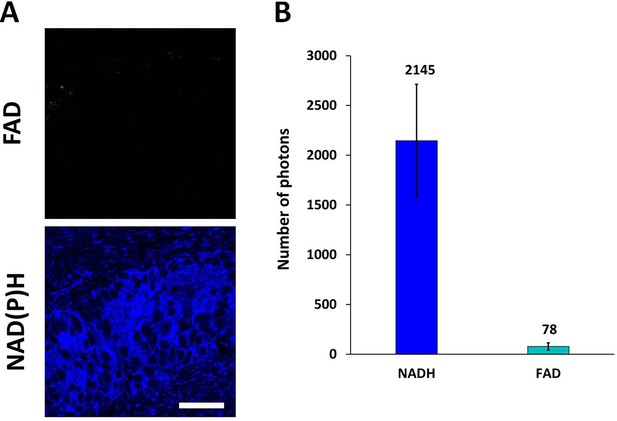
Autofluorescence of cofactors FAD and NAD(P)H in patient tumor ex vivo (№ 11).
(A) Representative fluorescence intensity images of flavins (FAD, ex. 900 nm, em. 500–550 nm) and NAD(P)H (ex. 750 nm, em. 450–490 nm). Scale bar = 50 μm. (B) The number of photons per pixel recorded by FLIM for flavins in ex vivo patient samples. Mean ± SD, n=7 patients.
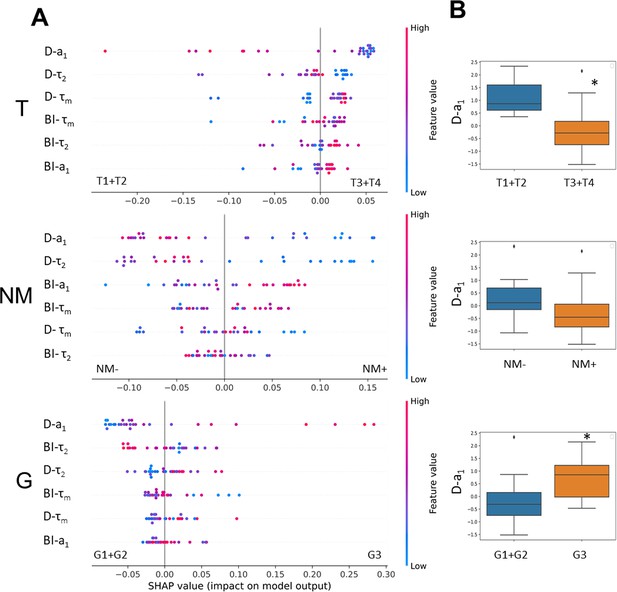
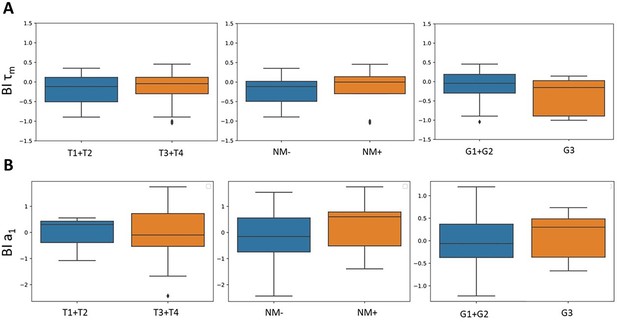
The relationships between metabolic heterogeneity and clinicopathological characteristics of patients’ tumors.
(A) Plots of SHAP analysis for the built decision tree models to determine the importance of dispersion (D) and bimodality index (BI) of the fluorescence decay parameters of NAD(P)H. The higher the value of the variable, the more red the dot is. (B) Box-plots of D-a1 with highest significance, * p-val <0.05.
Tables
NAD(P)H fluorescence decay parameters of colorectal cancer cells in monolayer cultures in vitro and in mouse tumors in vivo.
| Cell line | τm, ns | τ1, ns | τ2, ns | a1, % | BI-τm |
|---|---|---|---|---|---|
| Cell lines in vitro | |||||
| HT29 | 0.80±0.05 | 0.39±0.03 | 2.47±0.17 | 80.11±1.34 | 1.05 |
| HCT116 | 0.71±0.05 | 0.40±0.02 | 2.39±0.20 | 84.13±1.58 | 0.90 |
| CaCo2 | 0.95±0.10 | 0.38±0.05 | 2.53±0.24 | 73.48±2.26 | 1.30 |
| CT26 | 0.57±0.06 | 0.35±0.03 | 2.03±0.22 | 86.48±1.28 | 1.28 |
| Tumors in vivo | |||||
| HT29 | 0.84 [0.81;0.90] | 0.46 [0.42;0.48] | 2.66 [2.54;2.76] | 81.54 [79.93;83.13] | 0.85±0.35 |
| HCT116 | 0.88 [0.85;0.92] | 0.47 [0.45;0.48] | 2.66 [2.56;2.77] | 80.61 [79.32;81.96] | 0.91±0.28 |
| CaCo2 | 1.02 [0.86;1.19] | 0.42 [0.38;0.50] | 2.91 [2.61;3.32] | 76.89 [74.06;78.52] | 1.51±0.71 |
| CT26 | 0.72 [0.67;0.78] | 0.39 [0.37;0.41] | 2.34 [2.21;2.50] | 82.12 [80.96;84.70] | 1.20±0.36 |
-
τm – mean lifetime, τ1 – short lifetime component, τ2 – long lifetime component, a1 – relative contribution of the short lifetime component, BI-τm – bimodality index of the mean lifetime.
The bimodality index BI-a1 and dispersion D-a1 of NAD(P)H in cultured cells, mouse tumors and patients’ tumor samples.
| Cell lines in vitro | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HT29 | HCT116 | CaCo2 | CT26 | |||||||||
| BI-a1 | 0.16 | 0.92 | 0.49 | 0.63 | ||||||||
| D-a1 | 1.83 | 2.01 | 3.41 | 1.67 | ||||||||
| Tumors in vivo | ||||||||||||
| HT29 | HCT116 | CaCo2 | CT26 | |||||||||
| BI-a1 | 1.2±0.32 | 0.93±0.23 | 0.86±0.15 | 1.06±0.54 | ||||||||
| D-a1 | 3.20 | 2.60 | 4.00 | 2.81 | ||||||||
| Patients’ tumors ex vivo | ||||||||||||
| Sample | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| BI-a1 | 1.25 | 0.81 | 1.01 | 1.16 | 1.01 | 1.00 | 0.86 | 0.70 | 1.28 | 1.25 | ||
| D-a1 | 7.77 | 4.18 | 8.68 | 11.99 | 6.93 | 7.83 | 9.33 | 8.25 | 11.51 | 6.96 | ||
| Sample | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| BI-a1 | 0.59 | 1.45 | 1.34 | 1.05 | 0.89 | 0.24 | 1.29 | 1.32 | 0.80 | 0.94 | ||
| D-a1 | 4.5 | 2.37 | 4.94 | 2.19 | 4.16 | 3.34 | 6.10 | 5.41 | 2.23 | 5.19 | ||
| Sample | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |||
| BI-a1 | 1.28 | 0.98 | 1.53 | 1.31 | 0.88 | 1.58 | 1.65 | 0.5 | 1.29 | |||
| D-a1 | 5.27 | 3.29 | 6.54 | 5.77 | 4.87 | 5.83 | 4.14 | 5.67 | 6.36 | |||
Information about patients and their colorectal tumors.
| Characteristics | Number | Percent |
|---|---|---|
| Gender | ||
| Male | 16 | 55.17 |
| Female | 13 | 44.83 |
| Age | ||
| Mean ±SD | 65.28±11.92 y.o. | - |
| Median | 67 y.o. | - |
| Tumor staging | ||
| I | 3 | 10.34 |
| IIA | 6 | 20.69 |
| IIB | 3 | 10.34 |
| IIIB | 11 | 37.93 |
| IIIC | 1 | 3.45 |
| IV | 5 | 17.25 |
| Tumor site | ||
| Cecum colon | 2 | 6.90 |
| Transverse colon | 10 | 34.48 |
| Hepatic flexure | 3 | 10.34 |
| Sigmoid colon | 8 | 27.59 |
| Rectum | 6 | 20.69 |
| Grade | ||
| Low (G1) | 4 | 13.79 |
| Moderate (G2) | 19 | 65.52 |
| High (G3) | 6 | 20.69 |
Additional files
-
Supplementary file 1
Statistical significance of the differences of NAD(P)H a1-% between different cell lines and tumors (p-values).
- https://cdn.elifesciences.org/articles/94438/elife-94438-supp1-v1.docx
-
Supplementary file 2
Clinicopathological characteristics and NAD(P)H fluorescence decay parameters of patients’ tumors.
- https://cdn.elifesciences.org/articles/94438/elife-94438-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/94438/elife-94438-mdarchecklist1-v1.docx