Full-length direct RNA sequencing uncovers stress granule-dependent RNA decay upon cellular stress
Figures

RNA shortening upon cellular stress.
(a) Schematic of experimental design (b) Volcano plot for the differential expression of arsenite-treated and unstressed cells. Green color indicates genes with more than two-fold difference and red indicates statistical significance p-value<10–5. (c) Cumulative distribution of read length for arsenite-treated and unstressed cells. (d) Scatter plots of average transcript length for arsenite-treated and unstressed cells stratified by their differential expression change. Downregulated: (-Inf, –0.5), unchanged: (–0.5, 0.5), upregulated (0.5, Inf) fold-change. Only transcripts with at least five aligned reads are shown. Red dotted indicates the y = x line. Color indicates transcripts below (blue) and above (orange) the diagonal. (e) Cumulative distribution of transcript length for arsenite-treated and unstressed cells using only reads with adapter ligated at the 5ʹ end. (f) Schematic of read meta-length calculation. Each annotated transcript is divided into 20 equally sized bins. Each read is then assigned meta-coordinates depending on the bin in which its 5ʹ and 3ʹ templated ends align. The read meta-length is calculated as the difference of the meta-coordinates and presented as a percentage of full length. (g) Scatter plot of average transcript meta-length for arsenite-treated and unstressed cells. Coloring is the same as (d). (h) Scatter plot of average transcript length for heat shock and unstressed cells.

RNA shortening upon cellular stress.
(a) MTS cell proliferation assay showing the percentage of live cells (y-axis) upon arsenite treatment compared to unstressed at different time points (x-axis). Error bars represent std for biological replicates (n = 2). (b) Correlogram of transcript expression for unstressed and arsenite-treated cells. (c) Gene ontology for biological processes for differentially expressed genes in arsenite-treated versus unstressed cells. (d, e) Cumulative distribution plot of average transcript length for coding (d) and noncoding (e) transcripts. Transcripts with less than five reads have been removed. (f) Cumulative distribution of read meta-length for arsenite-treated and unstressed cells. (g) Contour density plot of average transcript meta-length for arsenite-treated and unstressed cells. Only reads with an identified poly(A) tail are used. (h) Cumulative distribution plot of average transcript length for H2O2-treated cells and unstressed. (i) Cumulative distribution plot of average transcript length for heat shock cells and unstressed. (j) Scatter plots of average transcript length for heat shock against unstressed cells stratified by differential expression change. Downregulated: (-Inf, –0.5), unchanged: (–0.5, 0.5), upregulated (0.5, Inf) fold-change. Only transcripts with at least five aligned reads are shown. Red dotted indicates the y = x line. Color indicates transcripts below (blue) and above (orange) the diagonal. (k, l) Cumulative distribution plot of average transcript length for the mouse 3T3 cells treated with arsenite and unstressed. All reads (k) or only reads with ligated adaptor (l) are used.
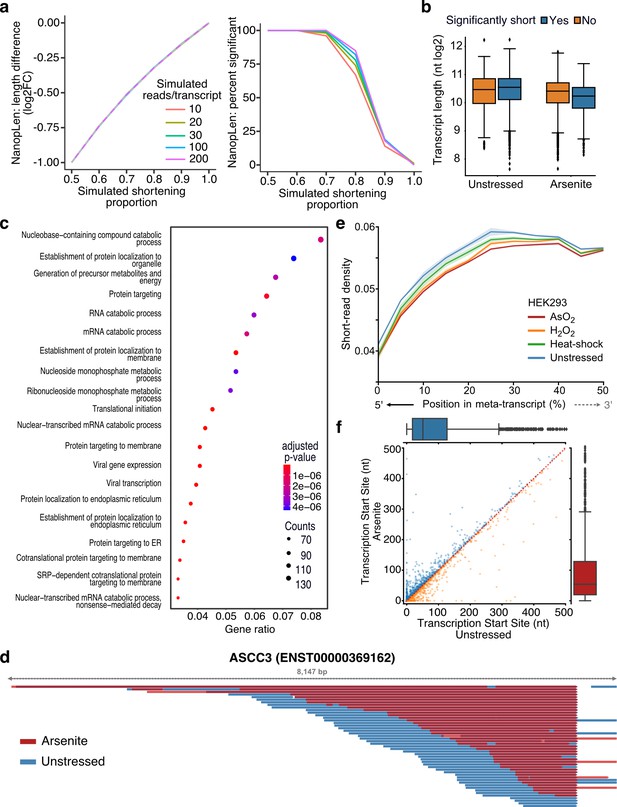
Characterization of stress-induced shortened RNAs identified by NanopLen.
(a) Left: line plot of average length difference estimate for simulated data of varying read depths. True value is depicted by dashed grey line. Right: percentage of simulated genes that are detected as significantly different in length. The null simulation of no shortening corresponds to simulated shortening proportion 1.0. (b) Box plot of average transcript length for statistically significant and nonsignificantly shortened transcripts identified through differential length analysis in arsenite-treated and unstressed cells using NanopLen. (c) Gene ontology analysis for biological processes of significantly shortened transcripts. (d) IGV screenshot of ASCC3 aligned reads for arsenite-treated (red) and unstressed cells (blue). Libraries were randomly downsampled to maximum 50 reads per window and the libraries were overlayed. All reads were used, irrespective of adapter ligation status. (e) Short-read RNA-seq density at the 5ʹ half of transcripts for unstressed, NaAsO2, H2O2 and heat shock-treated HEK293 cells. Shade indicates standard error of the mean for replicates. (f) Scatter plot of transcription start site position for arsenite-treated and unstressed HeLa cells.
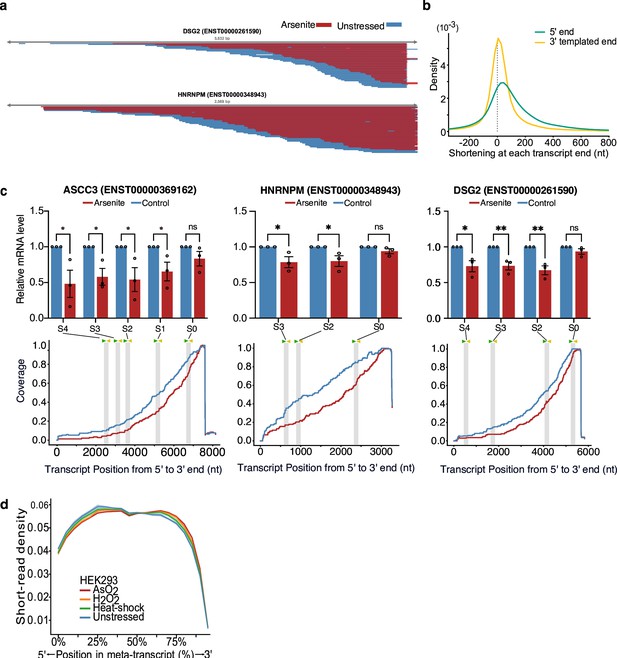
Characterization of stress-induced shortened RNAs identified by NanopLen.
(a) IGV screenshot of aligned reads in arsenite-treated (red) and unstressed cells (blue) for significantly shortened transcripts HNRNPM and DSG2. Libraries were randomly downsampled to maximum 50 reads per window and the libraries were overlayed. All reads were used, irrespective of adaptor ligation status. (b) Density plot of transcript shortening at the 5ʹ and 3ʹ templated end. Positive values on the x-axis indicate that the 5ʹ or 3ʹ of transcripts in arsenite-treated cells are correspondingly downstream or upstream of their unstressed counterparts. (c) RT-qPCR (top) and read coverage (bottom) for transcripts ENST00000369162 of gene ASCC3, ENST00000261590 of gene DSG2 and ENST00000348943 of gene HNRNPM for arsenite-treated and control cells. PCR primer pairs were designed for varying locations along the transcript, and their approximate location is marked on each plot. Signals were normalized against ACTB. Error bars correspond to SEM (N = 3) and individual points are shown. p-Values were calculated using t-test. **p<0.01 and *p<0.05. (d) Meta-plot of short-read RNA-seq density on all transcripts for unstressed, NaAsO2, H2O2, and heat shock -treated HEK293 cells. Shade indicates standard error of the mean for replicates.

The effect of XRN1 knockdown on RNA shortening.
(a) Immunoblot for XRN1, eIF2α-P, and eIF2α for cells transfected with a non-targeting (siCTRL) or XRN1-targeting (siXRN1) siRNA. ACTB is used as control. (b) Cumulative distribution plot of transcript length for XRN1 knockdown (siXRN1) and control (siCTRL) cells with and without arsenite treatment (unstressed). (c) Same as (b) but only reads with ligated 5ʹ end adapter are used. (d) Box plots of differential transcript length in arsenite-treated versus unstressed cells for significantly and nonsignificantly shortened transcripts upon XRN1 knockdown and control. (e, f) Cumulative distribution plot of transcript length for NOT8* D40A E42A and GFP-expressing cells (e) or DCP2* E148Q and GFP-expressing cells (f) with or without (unstressed) arsenite treatment.
-
Figure 3—source data 1
Original files for western blot analysis displayed in Figure 3a.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig3-data1-v1.zip
-
Figure 3—source data 2
File containing original western blots for Figure 3a, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig3-data2-v1.zip
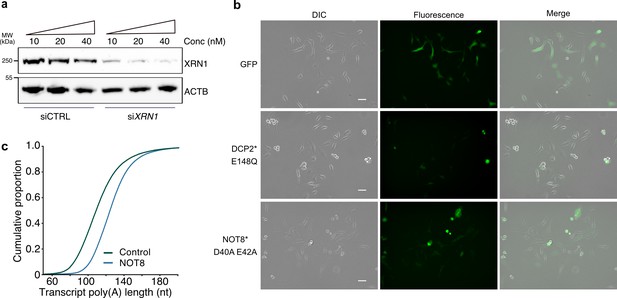
The effect of XRN1 knockdown on RNA shortening.
(a) Immunoblot for XRN1 for cells transfected with non-targeting and XRN1-targeting siRNA at varying concentrations. ACTB is used as control. (b) Visualization by epifluorescence of GFP, GFP-fused DCP2* E148Q and GFP-fused NOT8* D40A E42A expression in HeLa cells. Scale bar 50 µm. (c) Cumulative distribution of average transcript poly(A) tail length for NOT8*-expressing and GFP control cells.
-
Figure 3—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
File containing original western blots for Figure 3—figure supplement 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig3-figsupp1-data2-v1.zip

Association of poly(A) tail length and cis-regulatory elements with RNA shortening.
(a) Nucleotide composition around the 5′ end of reads in arsenite-treated and unstressed cells. All reads were used, irrespective of adapter ligation status. (b–d) Scatter plot of annotated transcript length (b), annotated coding sequence (CDS) length (c) and GC content (d) against transcript differential length in arsenite-treated and unstressed cells. The box plots on the right side summarize the y-axis variable for significant and nonsignificantly shortened transcripts. The Pearson’s correlation coefficient and the Mann–Whitney U test p-value are shown.
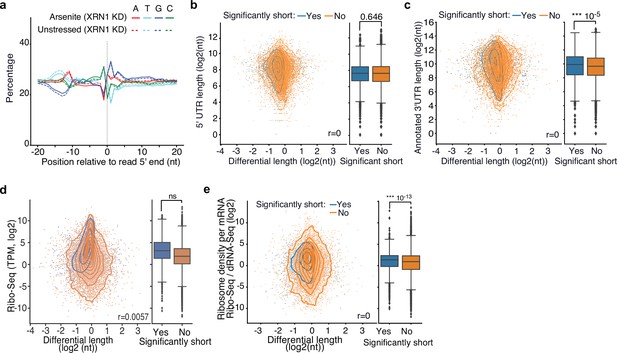
Association of cis-regulatory elements and ribosome occupancy with RNA shortening.
(a) Nucleotide composition around the 5′ end of reads in arsenite-treated and unstressed cells upon XRN1 silencing. All reads were used, irrespective of adaptor ligation status. (b, c) Scatter plot of annotated 5′ (a) and 3ʹ (b) UTR length against transcript differential length in arsenite-treated and unstressed cells. The box plots on the right side summarize the y-axis variable for significant and nonsignificantly shortened transcripts. (d, e) Scatter plot of ribosome profiling footprint (d) and translational efficiency (e) levels against differential length in arsenite-treated and unstressed cells. Transcripts are stratified by significance of shortening. The Pearson’s correlation coefficient and the Mann–Whitney U test p-value are shown.

Translation and RNA shortening.
(a, b) Immunoblot for eIF2α and eIF2α-P upon increasing concentration of arsenite (a) and cell treatment with 200 nM of ISRIB at different arsenite concentrations (b). (c) Stress granules (SGs) visualized by immunofluorescence of HeLa cells treated with indicated concentration of arsenite in the presence or absence of 200 nM ISRIB. A secondary goat anti-rabbit IgG H&L (Alexa Fluor 594) against G3BP1 (SG marker) and DAPI were used for visualization. (d) Ribosome sedimentation curve following cell treatment with ISRIB (200 nM) for arsenite-treated and unstressed cells. (e) Cumulative density plot of transcript length for arsenite-treated cells in the presence or absence of ISRIB. (f) Same as (e) for significantly shortened transcripts only. (g) Box plots of differential transcript length for comparisons indicated on the x-axis. (h–i) Same as (e) and (f) for cycloheximide CHX instead of ISRIB.
-
Figure 5—source data 1
Original files for western blot analysis displayed in Figure 5a and b.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig5-data1-v1.zip
-
Figure 5—source data 2
File containing original western blots for Figure 5a and b, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/96284/elife-96284-fig5-data2-v1.zip
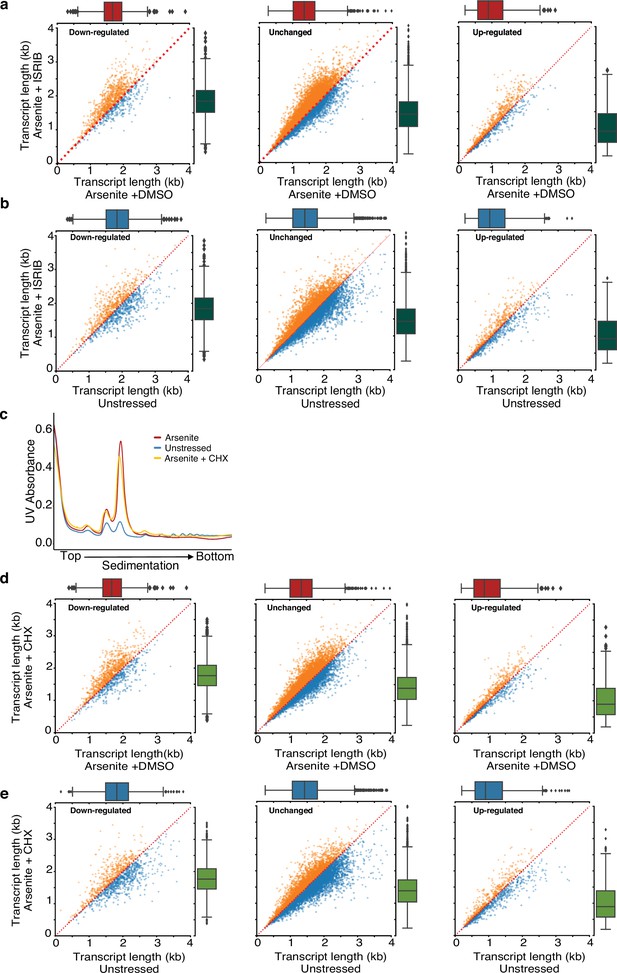
Translation and RNA shortening.
(a) Scatter plots of average transcript length for arsenite-treated cells with and without ISRIB stratified by their differential expression change. Downregulated: (-Inf, –0.5), unchanged: (–0.5, 0.5), upregulated (0.5, Inf) fold-change. Only transcripts with at least five aligned reads are used. Red dotted indicates the y = x line. Color indicates transcripts below (blue) and above (orange) the diagonal. (h) Same as (g) for arsenite and ISRIB treated cells against control. (c) Ribosome sedimentation curve following cell treatment with cycloheximide (CHX) (25 µg/ml) for arsenite-treated and unstressed cells. (d, e) Same as (a) and (b) for cycloheximide CHX instead of ISRIB.
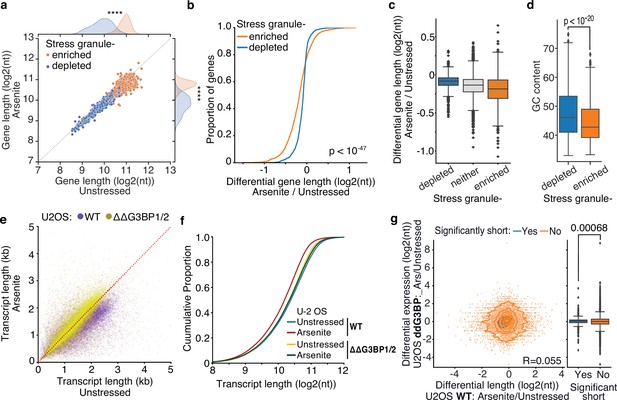
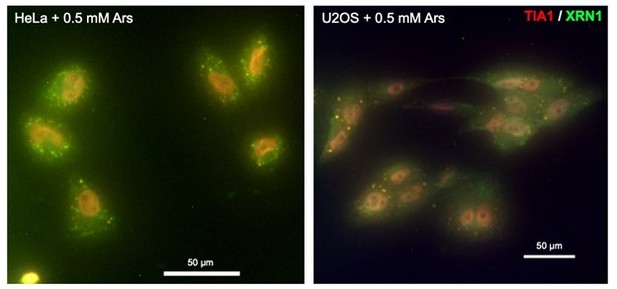
Inhibition of stress granule (SG) formation in cells devoid of G3BP1/2 rescues RNA decay.
(a) Scatter plot of average gene length for arsenite-treated and unstressed cells stratified by gene SG localization. ****p-value<10–172. (b, c) Cumulative distribution and box plots of average gene length difference in arsenite-treated and unstressed cells stratified by gene SG localization. (d) Box plot of GC content percentage for SG-enriched and -depleted gene transcripts. (e) Scatter plot of average transcript length for arsenite-treated and unstressed U-2 OS and ΔΔG3BP1/2 U-2 OS cells. Only transcripts with at least five aligned reads are used. Red dotted indicates the y = x line. (f) Cumulative distribution plot of average transcript length in arsenite-treated and unstressed U-2 OS and ΔΔG3BP1/2 U-2 OS cells. (g) Scatter plot and box plot of differential transcript expression and shortening in arsenite-treated and unstressed ΔΔG3BP1/2 and WT U-2 OS cells.
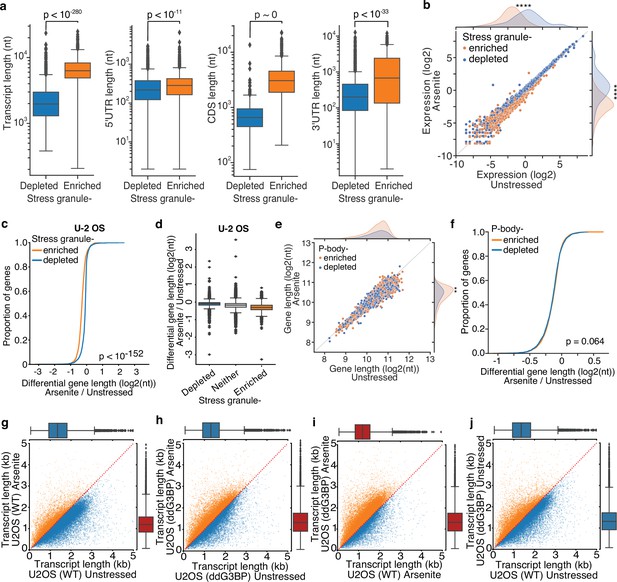
Inhibition of stress granule (SG) formation in cells devoid of G3BP1/2 rescues RNA decay.
(a) Box plots of annotated total, 5′ UTR, coding sequence, and 3′ UTR length for SG-enriched and -depleted transcripts. (b) Scatter plot of upper 90th quantile normalized gene expression for arsenite-treated and unstressed cells stratified by gene SG localization. ****p-value<10–172. (c) Cumulative density plot of transcript differential length for SG-enriched and -depleted gene transcripts in U-2 OS cells. Only transcripts of coding genes are used. (d) Box plots of differential gene length in arsenite-treated and unstressed cells stratified by SG enrichment in U-2 OS cells. (e) Scatter plot of average gene length for arsenite-treated and unstressed cells stratified by gene P-bodies localization. **p-value<0.01. (f) Cumulative distribution plot of average gene length difference in arsenite-treated and unstressed cells stratified by gene P-bodies localization. (g–j) Scatter plot of average transcript length for arsenite-treated and unstressed U-2 OS and ΔΔG3BP1/2 cells. Only transcripts with at least five aligned reads are used. Red dotted indicates the y = x line. Color indicates transcripts below (blue) and above (orange) the diagonal.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Beta actin mouse monoclonal | ProteinTech | Cat # 66009-1-Ig; RRID:AB_2687938 | (1:1000) |
| Antibody | Rabbit-anti XRN1 polyclonal | Thermo Fisher | A300-443A; RRID:AB_2219047 | (1:1000) |
| Antibody | Rabbit-anti eIF2α-P | Cell Signaling | Cat # 9721S; RRID:AB_330951 | (1:1000) |
| Antibody | Rabbit-anti eIF2α | Cell Signaling | Cat # 9722S; RRID:AB_2394335 | (1:1000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Cross-adsorbed Secondary antibody | Thermo Fisher | Cat # 31462; RRID:AB_228338 | (1:10,000) |
| Antibody | Goat anti-Mouse IgG Fc Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher | Cat # 31439; RRID:AB_228292 | (1:10000) |
| Antibody | Anti-G3BP1 polyclonal | Thermo Fisher | Cat # PA5-29455; RRID:AB_2546931 | (1:500) |
| Antibody | Goat anti-rabbit IgG H & L (Alexa Fluor 594) | Abcam | Cat # ab150080; RRID:AB_2650602 | (1:200) |
| Cell line (Homo sapiens) | Human osteosarcoma, U-2 OS (control, WT) | Gift from Paul Anderson lab (PMID:27022092) | ||
| Cell line (H. sapiens) | Human osteosarcoma U-2 OS, U-2 OS (ΔΔG3BP1/2) | Gift from Paul Anderson lab (PMID:27022092) | ||
| Cell line (H. sapiens) | HeLa | ATCC | Cat # HeLa CCL-2; RRID:CVCL_0030 | |
| Cell line (Mus musculus) | Mouse embryonic fibroblasts, NIH/3T3 | ATCC | Cat # CRL-1658; RRID:CVCL_0594 | |
| Chemical compound, drug | DMSO | MilliporeSigma | D8418 | |
| Chemical compound, drug | NaAsO₂ | MilliporeSigma | Cat # S7400-100G | |
| Chemical compound, drug | H2O2 | MilliporeSigma | ||
| Chemical compound, drug | ISRIB | MilliporeSigma | Cat # SML0843-5MG | |
| Chemical compound, drug | CHX | MilliporeSigma | Cat # 239765-1ML | |
| Chemical compound, drug | PMSF | Roche Diagnostics | Cat # 10837091001 | |
| Chemical compound, drug | Protease inhibitor cocktail | Roche Diagnostics | Cat # 11836153001 | |
| Chemical compound, drug | Chemiluminescence | Azure Biosystems | Cat # AC2204 | |
| Chemical compound, drug | Phosphatase inhibitors | MilliporeSigma | Cat # Cocktail 2P5726, Cocktail 3-P0044 | |
| Chemical compound, drug | HALT Protease | Thermo Fisher | Cat # 78740 | |
| Chemical compound, drug | TRIzol reagent | Invitrogen | Cat # 15596-018 | |
| Chemical compound, drug | Bovine serum albumin | MilliporeSigma | Cat # 10735078001 | |
| Chemical compound, drug | Fetal bovine serum | GeminiBio | Cat # 100-106 | |
| Chemical compound, drug | l-Glutamine | Thermo Fisher | Cat # 25030081 | |
| Chemical compound, drug | MEM-nonessential amino acids | Invitrogen | Cat # 11140050 | |
| Chemical compound, drug | 4,6-Diamidino-2-phenylindole (DAPI) | MilliporeSigma | Cat # D8417-1MG | |
| Chemical compound, drug | Dulbecco’s Modified Eagle Medium, DMEM | Thermo Fisher Scientific | Cat # 11965-092 | |
| Commercial assay or kit | Qubit protein assay | Invitrogen | Cat # Q33211 | |
| Commercial assay or kit | MTS Assay, CellTiter 96 AQueous One Solution Cell Proliferation Assay | Promega | Cat # G3582 | |
| Commercial assay or kit | High Sensitivity (HS) RNA Qubit assay | Invitrogen | Cat # Q32852 | |
| Commercial assay or kit | Qubit 1X dsDNA High Sensitivity (HS) assay kit | Thermo Fisher | Cat # Q33231 | |
| Commercial assay or kit | Qubit RNA IQ assay | Thermo Fisher | Cat # Q33222 | |
| Commercial assay or kit | High Sensitivity DNA kit | Agilent Technologies | 5067-4626 | |
| Commercial assay or kit | Universal mycoplasma detection kit | ATCC | Cat # 30-1012K | |
| Commercial assay or kit | Direct RNA sequencing kit | Oxford Nanopore Technologies | SQK-RNA002 | |
| Commercial assay or kit | Oligo d(T)25 Magnetic Beads | New England Biolabs | Cat # s1419S | |
| Commercial assay or kit | T4 RNA ligase | New England Biolabs | Cat # M0204S | |
| Commercial assay or kit | Direct RNA sequencing Flow Cells (MinION) | Oxford Nanopore Technologies | FLO-MIN106 | |
| Commercial assay or kit | Direct RNA sequencing Flow Cells (PromethION) | Oxford Nanopore Technologies | FLO-PRO002 | |
| Commercial assay or kit | Reverse Transcriptase Superscript III First-Strand Synthesis System | Invitrogen | Cat # 18080-051 | |
| Commercial assay or kit | FastStart SYBR Green Master Mix | KAPA Biosystems | Cat # KK4605/07959435001 | |
| Commercial assay or kit | NuPAGE 4–12% Bis-Tris Gel | Invitrogen | Cat # NP0321BOX | |
| Commercial assay or kit | Polyvinylidene fluoride membrane (PVDF) | Millipore | Cat# IPVH00010 | |
| Commercial assay or kit | ProLong Glass Antifade Mountant | Invitrogen | Cat # P36982 | |
| Recombinant DNA reagent | pT7-EGFP-C1-HsNot8-D40AE42A_AH (Plasmid) | Gift from Elisa Izaurralde | Plasmid # 148902; RRID:Addgene_148902 | |
| Recombinant DNA reagent | pT7-EGFP-C1-HsDCP2-E148Q_U (Plasmid) | Gift from Elisa Izaurralde | Plasmid # 147650; RRID:Addgene_147650 | |
| Recombinant DNA reagent | pEGFP-C1 (Plasmid) | Gift from Myriam Gorospe | Available upon request | |
| Sequence-based reagent | Linker-REL5 (Oligo) | IDT | PMID:34428294 | (/5PCBio/rArArUrGrArUrAr CrGrGrCrGrArCrCrArCrCrGr ArGrArUrCrUrArCrArCrUrCr UrUrUrCrCrCrUrArCrArCrGr ArCrGrCrUrCrUrUrCrCrGrArUrCrU) |
| Sequence-based reagent | siRNA: ON-TARGETplus Set of 4 siRNA J-013754-09, XRN1 #1 | Dharmacon | Cat # J-013754-09-0005 | CUUCAUAGUUGGUCGGUAU |
| Sequence-based reagent | siRNA: siGENOME Non-Targeting siRNA #3 | Dharmacon | Cat # D-001210-03-20 | AUGUAUUGGCCUGUAUUAG |
| Software, algorithm | Guppy | https://nanoporetech.com/document/Guppy-protocol | Guppy (3.4.5); RRID:SCR_023196 | |
| Software, algorithm | Cutadapt | DOI: 10.14806/ej.17.1.200 | Cutadapt (2.8); RRID:SCR_011841 | |
| Software, algorithm | Minimap2 | PMID:29750242 | Minimap2 (2.17); RRID:SCR_018550 | |
| Software, algorithm | DESeq2 | PMID:31740818 | DESeq2; RRID:SCR_015687 | |
| Software, algorithm | Nanopolish | https://github.com/jts/nanopolish | Nanopolish (0.14.0); RRID:SCR_016157 | |
| Software, algorithm | ClusterProfiler | PMID:22455463 | ClusterProfiler (4.0); RRID:SCR_016884 | |
| Software, algorithm | STAR | PMID:23104886 | STAR (2.5.3a); RRID:SCR_004463 | |
| Software, algorithm | Pysam | https://github.com/pysam-developers/pysam | Pysam (v0.15.4); RRID:SCR_021017 | |
| Software, algorithm | NanopLen | This paper, https://github.com/maragkakislab/nanoplen | NanopLen | NanopLen is available open source under NIA Public Domain license |
| Software, algorithm | EnhancedVolcano | https://github.com/kevinblighe/EnhancedVolcano | EnhancedVolcano (1.8.0); RRID:SCR_018931 | |
| Software, algorithm | IGV | PMID:36562559 | IGV (2.12.3); RRID:SCR_011793 |
Additional files
-
Supplementary file 1
Sequencing library list and metadata.
- https://cdn.elifesciences.org/articles/96284/elife-96284-supp1-v1.xlsx
-
Supplementary file 2
RT-qPCR primers, siRNAs, antibodies, and oligos.
- https://cdn.elifesciences.org/articles/96284/elife-96284-supp2-v1.xlsx
-
Supplementary file 3
Differential expression analysis for arsenite-treated vs. control libraries.
- https://cdn.elifesciences.org/articles/96284/elife-96284-supp3-v1.xlsx
-
Supplementary file 4
Differential length analysis for arsenite-treated vs. control cells.
- https://cdn.elifesciences.org/articles/96284/elife-96284-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96284/elife-96284-mdarchecklist1-v1.docx