Differences in size and number of embryonic type II neuroblast lineages correlate with divergent timing of central complex development between beetle and fly
Figures
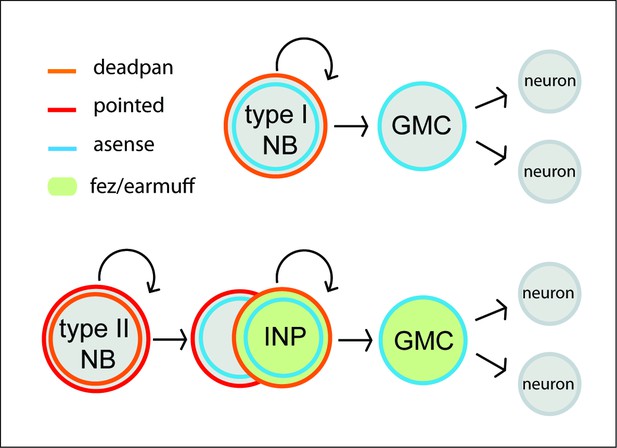
Neuroblasts (NBs) of the Drosophila nervous system.
Type I NBs (top panel) and type II NBs (lower panel). Type I NBs undergo stem cell-like divisions, with each round producing a ganglion mother cell (GMC) that divides one more time to produce two neurons or glia cells. In Drosophila, all type I NBs express dpn and ase, whereas GMCs express only ase. Type II NBs express dpn and pnt but not ase. They also undergo repeated divisions, with each round producing an intermediate progenitor cell (INP), which is also a proliferating progenitor and expresses ase. INPs undergo a maturation process during which pnt is still expressed initially, but not dpn. Mature INPs express dpn, ase, and fez/erm, whereas GMCs express ase and fez/erm. Type II NB lineages are only found in the anterior brain; they produce central complex neurons and glia (Bayraktar and Doe, 2013; Southall and Brand, 2009).
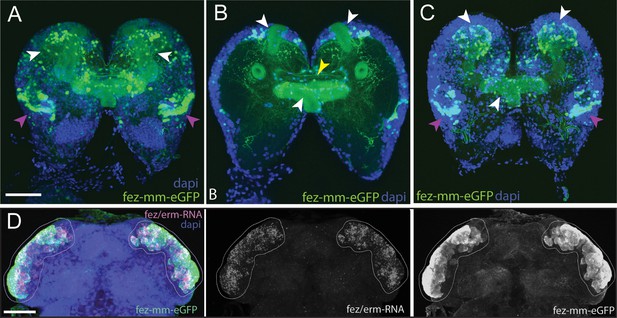
Magic mushrooms (fez-mm-eGFP) reporter line.
(A–C) eGFP-expression in a larval instar brain. (D) EGFP antibody labeling in combination with Tc-fez/erm RNA in situ in the embryonic head. (A–D) DAPI labeling of nuclei. Scale bars in (A) and (D) are 50 µm. (A) Full projection of whole brain scan (fourth-instar larvae): fez-mm-eGPF expression marks many cells of the mushroom bodies (white arrowheads) and of the larval optic lobe (purple arrowheads). Note that we use the terms dorsal and ventral with respect to the neuraxis of the larva. See Koniszewski et al., 2016 for orientation. (B) Central part of the whole brain scan (projection of substack, same brain as in A): mm-eGFP marks the peduncles of the mushroom bodies (white arrowheads) and some central complex ensheathing cells (probably glia cells; yellow arrowhead). (C) Projection of the dorsal part of same brain as in (A) shows calyces and peduncles of mushroom bodies (white arrowheads), and optic neuropile (purple arrowheads). (D) Extensive co-localization of fez-mm-eGFP with Tc-fez/erm-RNA in white encircled area, which gives rise to protocerebral structures. Projection of all planes where expression was detected, embryonic stage NS11. Figure 2—figure supplement 1 and Supplementary file 1a and b give details of the generation and sequence-based characterization of this line.
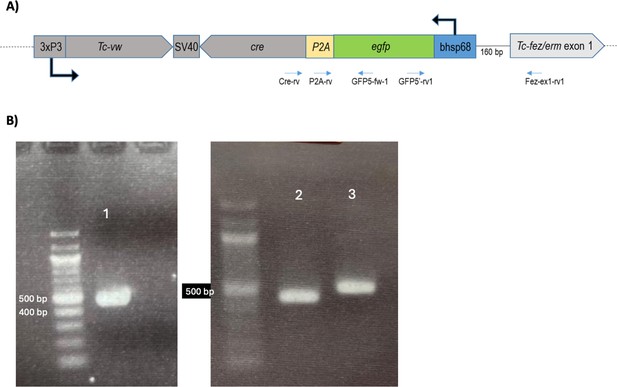
Tc-fez/erm locus with inserted reporter construct.
(A) Scheme of inserted construct in fez-mm-eGFP line, upstream the Tc-fez/erm gene. The orientation of egfp is in opposite direction compared to Tc-fez/erm. Distance between the bhsp68 promoter and the Tc-fez/erm TSS is 160 bp, which corresponds to the distance between CRISPR guide sequence Tc-fez/erm upstream 2 (see Supplementary file 1a) and the Tc-fez/erm TSS. (B) Gel electrophoresis with products from PCR using fez-mm-eGFP genomic DNA with primers given in Supplementary file 1b and mapped in (A). Gel images were extracted from Figure 2—figure supplement 1—source data 1 and 2.
-
Figure 2—figure supplement 1—source data 1
Uncropped image of agarose DNA-Gel including the part shown in Figure 2—figure supplement 1B (left).
- https://cdn.elifesciences.org/articles/99717/elife-99717-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Uncropped image of agarose DNA-Gel including the part shown in Figure 2—figure supplement 1B (right).
- https://cdn.elifesciences.org/articles/99717/elife-99717-fig2-figsupp1-data2-v1.zip
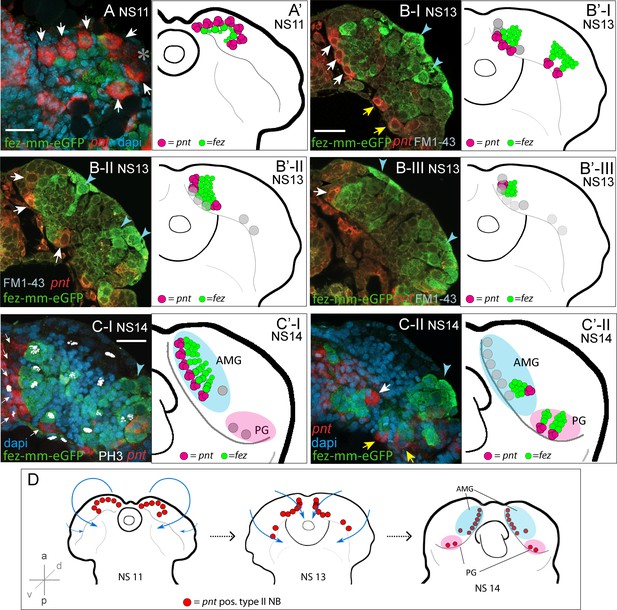
Developmental occurrence and progression of Tc-pnt-clusters and associated fez-mm-eGFP cells (type II neuroblast [NB] lineages).
(A–C) Anti-GFP antibody staining reflecting fez-mm-eGFP expression in combination with Tc-pnt RNA in situ hybridization and DAPI labeling of nuclei (A, C) or FM1-43 membrane staining (B), single planes from confocal z-stacks of right head lobes. Scalebars are 25 µm. (A’–C’) Schematic drawing of the head lobe at the respective stages; clusters out of focus depicted by gray circles. (D) Schematic depiction of the localization of type II NBs during Tribolium head morphogenesis. (A) At stage NS11, seven Tc-pnt-clusters (white arrows) are found in an anterior-medial horseshoe-like arrangement with fez-mm-eGFP cells in the center. Asterisk indicates the position of one Tc-pnt-cluster, which is outside the focal plane of this image. (B) At stage NS13, the seven anterior-medial clusters have been moved to the medial margin of the developing brain by morphogenetic movements of the head lobes (white arrows). The clusters are situated in different dorsoventral levels shown in the different images (B-I) to (B-III). Two additional posterior Tc-pnt-clusters have appeared (yellow arrows). Blue arrows point to fez-mm-eGFP-positive cells that are not part of the type II NB lineages. (B-I) Ventral level; (B-II) mid-level; (B-III) dorsal level. (C) At stage NS14, a group of six clusters are arranged in one plane anterior-medially in the brain lobe (white arrows) (C-I). Anti-PH3 labeling marks mitotic cells within the lineages (white signal) and clusters out of the focal plane are indicated by gray circles in (C’-I). (C-II) One cluster of the anterior median group (white arrow) is in a deeper plane, as well as the two posterior clusters (yellow arrows). Blue arrowheads point to lateral fez-mm-eGFP expression not associated with type II NBs. (A’-C’) AMG = anterior-medial group (blue), PG = posterior group (pink). Gray circles in (B’-C’) indicate position of type II NBs in different focal planes. (D) Tc-pnt-expressing cells identified as type II NBs first appear bilaterally in the anterior-most part of the embryonic head (stage NS11). During head morphogenesis, this anterior tissue folds over so that the type II NBs end up in a more medial position. Two more type II NBs emerge posteriorly (stage 13). At the end of embryogenesis (stage NS14), the type II NBs can be divided in an AMG (blue; including type II NBs 1–7, numbering starting from anterior) and a PG (pink, including type II NBs 8–9).
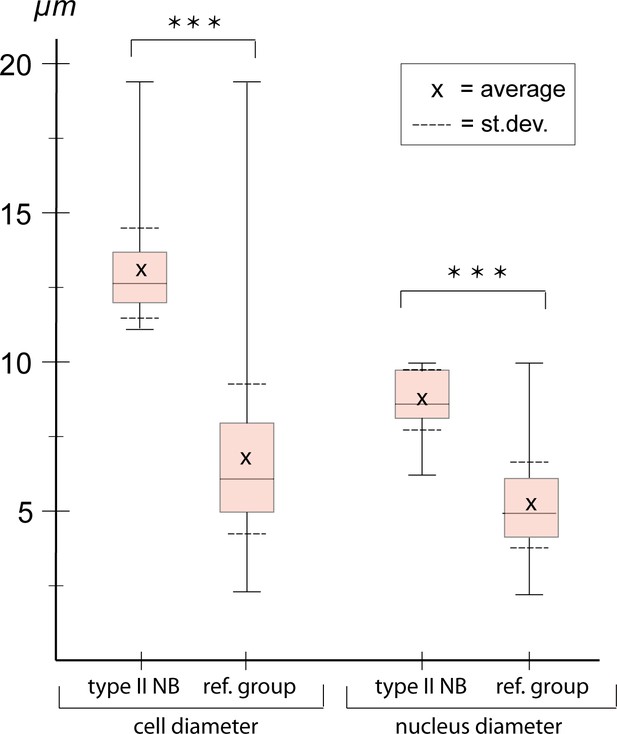
Cell and nuclear diameter of type II neuroblasts (NBs) compared to a control group.
Stages NS13-14. Left: diameter of Tc-pnt-pos. type II NBs (average diameter = 12.98 µm; n=43) compared to the control group (average diameter = 6.75, n=1579). Right: average nuclear diameter of type II NBs (=8,73 µm, n=17) compared to the average nuclear diameter of the control group (=5.21, n=1080). Differences between groups are significant (t-test; *** ≘p<0.001).
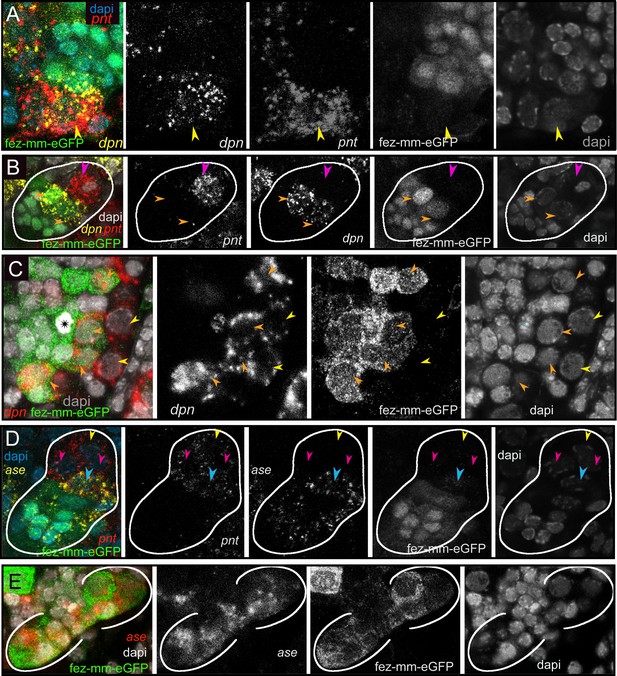
Differential expression of the neural markers Tc-pnt, Tc-fez/erm, Tc-dpn, and Tc-ase in type II neuroblasts (NBs), intermediate progenitors (INPs), and ganglion mother cells (GMCs).
(A, B, D) Anti-GFP staining visualizing fez-mm-eGFP expression in combination with HCR-labeling of further factors. (C, E) GFP antibody staining in combination with RNA in situ hybridization. (A–E) DAPI staining of nuclei. All panels are single planes from confocal z-stacks. (A) Stage NS12, expression of Tc-pnt and Tc-dpn in type II NBs (yellow arrowhead) with adjacent fez-mm-eGFP-positive INPs. (B) Stage NS13, white line marks one lineage. Expression of Tc-dpn is absent in the immature-I Tc-pnt + INPs (pink arrowhead) but present in mature INPs (orange arrowheads), white line marks one lineage. Type II NB not in focus of this image. (C) Stage NS14, expression of Tc-dpn in type II NBs (yellow arrowheads), and mature INPs (orange arrowheads). Asterisk marks mitosis in a fez-mm-eGFP cell visible through the condensation of chromatin stained by DAPI. (D) Stage NS13, Tc-ase- and Tc-pnt-expression in type II NBs and INPs, white line marks one lineage. Yellow arrowhead: type II NBs (Tc-pnt+), pink arrowheads: immature-I INP (Tc-pnt+), blue arrowhead: immature-II INP (Tc-fez+/ Tc-ase+/ Tc-pnt+). (E) Stage NS13, expression of Tc-ase in fez-mm-eGFP-positive cells, location of type II NBs and immature-I INPs is outside the focus.
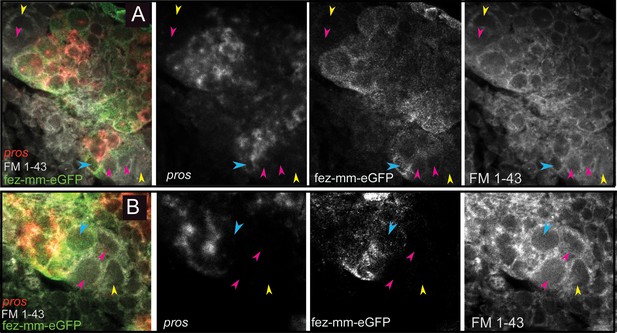
Expression of Tc-pros in fez-mm-eGFP cells.
(A, B) Anti-GFP antibody staining in combination with Tc-pros RNA in situ hybridization. Membranes stained with FM 1–43. Single planes from confocal z-stacks. (A) Stage NS13, two type II NB lineages producing INPs and GMCs in opposing directions. Expression of Tc-pros in mature INPs and GMCs. Cluster of type II NBs (yellow arrowheads) and immature-I INPs (pink arrowheads). Immature-II INPs express Tc-fez/erm but not Tc-pros (blue arrowhead). (B) Stage NS13, detail of Tc-pros-negative type II NBs (yellow arrowhead) and immature-I INPs (pink arrowheads) and immature-II INP (blue arrowhead). Note that distinction between type II NB and immature INPs in (A) and (B) is only made based on position.
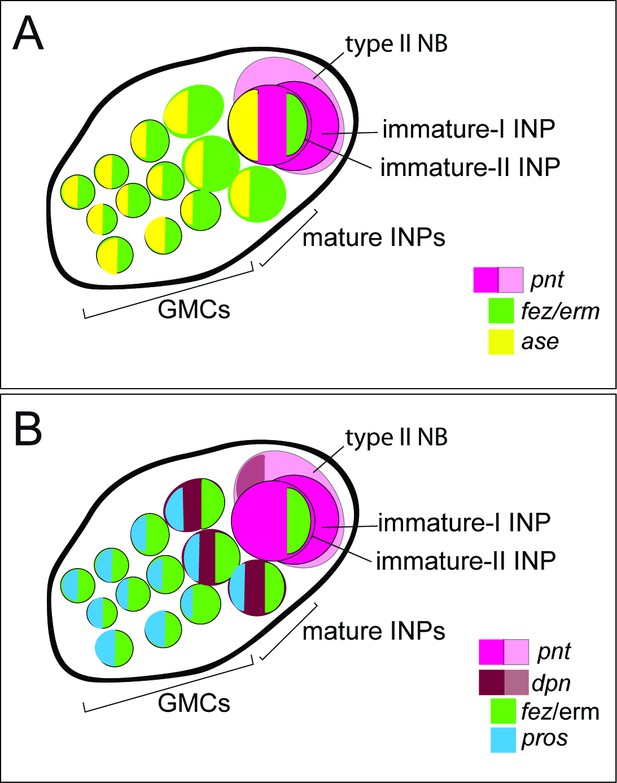
Schematic drawing of expression of different markers in a type II neuroblast (NB) lineage.
(A) and (B) show the same lineage with Tc-fez/erm and Tc-pnt expression but with different additional markers mapped on top. (A) Subclassification into type II NBs (Tc-pnt+), immature-I INPs (Tc-pnt+), immature-II INPs (Tc-pnt+, Tc-fez/erm+, Tc-ase+) and mature INPs and GMCs (Tc-fez/erm+, Tc-ase+) (based on stainings shown in Figure 5). (B) Type II NB (Tc-pnt+, Tc-dpn+), immature-I (Tc-pnt+), and immature-II (Tc-pnt+, Tc-fez/erm+). Mature INPs express Tc-dpn, Tc-pros, and Tc-fez/erm, whereas ganglion mother cells (GMCs) express only Tc-fez/erm and Tc-pros (based on staining shown in Figures 5 and 6). Note that of the selected markers immature-I INPs express Tc-pnt only.
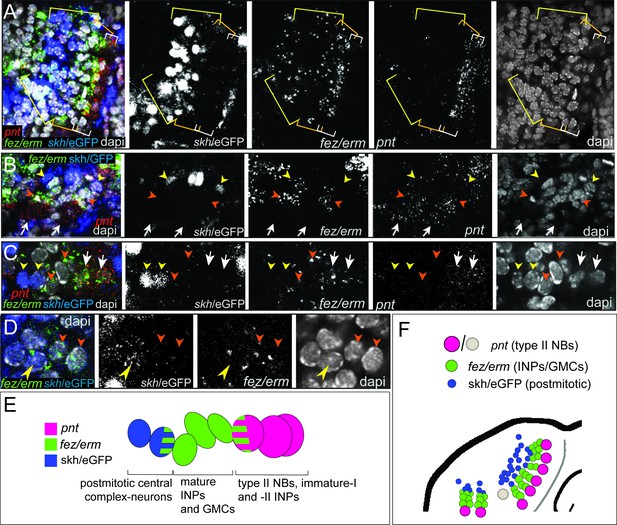
Expression of the transgenic central complex reporter skh/eGFP in relation to type II neuroblast (NB) lineages.
(A–D) Antibody labeling of eGFP, which is expressed in the pattern of skh (Garcia-Perez et al., 2021) in combination with HCR visualizing Tc-fez/erm and Tc-pnt, DAPI labeling of nuclei, single planes of confocal z-stacks, all stage NS14. (A) Left head lobe, anterior group of Tc-pnt-positive type II NBs, Tc-fez/erm-expressing intermediate progenitors (INPs)/ganglion mother cells (GMCs) and skh/eGFP-positive postmitotic cells. (B) Left head lobe, posterior group of type II NB lineages. Tc-pnt+ type II NBs or immature-I/-II INPs (white arrows), Tc-fez/erm+ INPs/GMCs (orange arrowheads) and skh/eGFP postmitotic cells (yellow arrowheads). (C) Detail of one lineage including type II NB cluster (Tc-pnt+, white arrows), INPs/GMCs (fez+, orange arrowheads), and skh/eGFP cells at the end of the lineage (yellow arrowheads). (D) Detail of transition from Tc-fez/erm+ cells (orange arrowheads) to skh/eGFP cells reveals a small area of co-expression of both factors (yellow arrowhead). (E) Schematic drawing of type II NB lineage showing relative positions and markers of type II NBs, INPs/GMCs, and postmitotic central complex forming cells. (F) Schematic overview of the arrangement of the type II NB lineages including skh/eGFP+ central complex forming cells in the head lobe of stage NS14 (immature-I/-II INPs not shown for simplicity). No data on skh-GFP expression in lineage of type II NB 7 (marked in gray) is available.

Size comparison of Tribolium and Drosophila type II neuroblast (NB) lineages.
Anteriormedian lineages from Tribolium stages 13 and 14 (quantified in this work, see Tables 1 and 2) compared to Drosophila stages 15 and 16 (pooled data from Walsh and Doe, 2017). Left: lineage sizes comprising type II NBs, intermediate progenitors (INPs), and ganglion mother cells (GMCs) (Tribolium n=4; Drosophila n=8). Right: number of Tc-dpn-positive mature INPs (Tribolium n=5; Drosophila n=8). t-test p-value significance level (t-test; *** ≘p<0.001, ** ≘p<0.01).
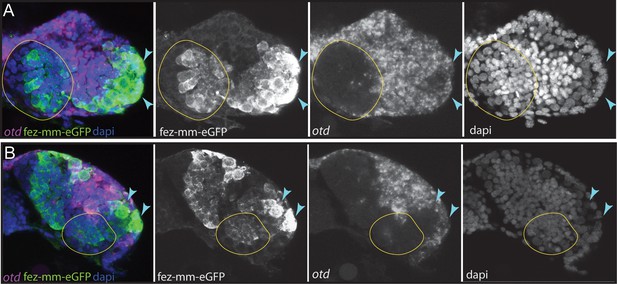
Expression of Tc-otd in relation to the fez-mm-eGFP lineages.
(A/B) RNA in situ hybridization for Tc-otd in combination with GFP antibody staining and DAPI labeling of nuclei. (A) and (B) show projections of different levels of NS14 embryos, right head lobe. (A) Tc-otd is neither expressed in type II neuroblasts (NBs) nor the in the fez-mm-eGFP-positive intermediate progenitors (INPs) of the anterior-medial group (both positioned within encircled area) but it is expressed in the directly surrounding tissue. (B) Likewise, Tc-otd is not expressed in type II NBs or fez-mm-eGFP cells of the posterior group (encircled area). A lateral area of Tc-fez/erm and Tc-otd co-expression (A/B, blue arrow) likely is part of the eye anlagen (Posnien et al., 2011b).
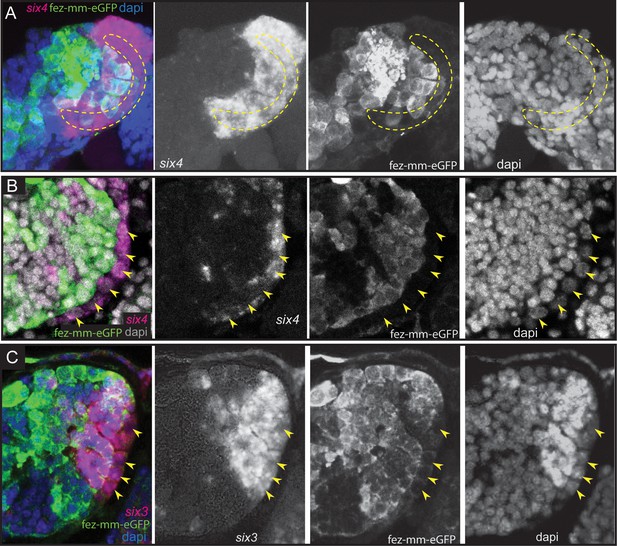
Expression of Tc-six4 and Tc-six3 in type II neuroblasts (NBs) and intermediate progenitors (INPs).
(A–C) RNA in situ hybridizations of Tc-six4 and Tc-six3 in combination with GFP antibody staining reflecting fez-mm-eGFP expression, DAPI labeling of nuclei, left head lobes shown. (A) Stage NS13; Tc-six4 is expressed in an area encompassing type II NBs (positioned in encircled area) and mm-eGFP expressing INPs. (B) Stage NS14; Tc-six4 is now restricted to type II NBs and their youngest progeny that do not express fez-mm-eGFP (yellow arrowheads). (C) Stage NS14; Tc-six3 specifically marks the four anterior-most lineages. It is expressed in the type II NBs at the base of the lineages (yellow arrowheads) and in the fez-mm-eGFP-positive INPs and possibly also in GMCs. It is absent from the remaining lineages of the anterior-medial group and is also not seen in the posterior group (not shown).
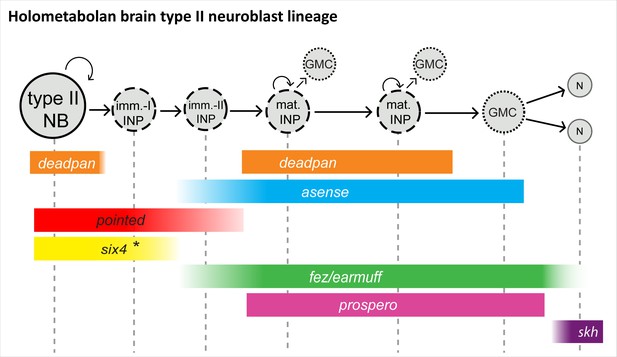
Conserved aspects of gene expression in type II neuroblast (NB) lineages of the holometabolan models Drosophila and Tribolium.
Overview on the different cell types within one lineage (type II NBs, intermediate progenitors [INPs], ganglion mother cells [GMCs], and central complex neurons), their mitotic activity, and summary of the expression of key factors in the respective cell types. (I) NPs are divided into immature-I, immature-II, and mature INPs, with each subtype having a unique expression profile. Note that each lineage can have several mature INPs. We strongly assume that glia are also produced from these lineages in Tribolium, as they are in Drosophila (Bayraktar and Doe, 2013). * In the Tribolium embryo Tc-six4 is only expressed in the anterior-medial lineages, and its expression is overlapping with Tc-fez/erm at an earlier stage (NS12), whereas it is restricted to type II NBs and immature-I INPs, mutually exclusive with Tc-fez/erm, at a later embryonic stage (NS14). Drosophila six4 was found in all eight larval lineages (Chen et al., 2021). The scheme is based on this work and Bayraktar and Doe, 2013; Garcia-Perez et al., 2021; Weng et al., 2010; Xie et al., 2016; Chen et al., 2021. imm.-I/-II INP = immature-I/-II intermediate progenitor; mat. N=neuron; skh = shaking hands (enhancer trap) (Garcia-Perez et al., 2021).
Tables
Tc-fez/erm-positive cell number (type I neuroblasts, intermediate progenitors, and ganglion mother cells).
| Embryo #/stage | Total number of cells of seven anterior lineages | % dividing cells (PH3 antibody) | Cells per lineage (average) |
|---|---|---|---|
| 1/NS 13 | 146 | 8.2 | 20.9 |
| 2/NS 13 | 111 | 6.3 | 15.9 |
| 3/NS 14 | 139 | 5.0 | 19.9 |
| 4/NS 14 | 96 | 10.5 | 13.7 |
Mature intermediate progenitors (Tc-fez/erm and Tc-dpn positive).
| Embryo #/stage | Total number of mature intermediate progenitors of seven anterior lineages | % dividing cells (PH3 antibody) | Number of mature intermediate progenitors per lineage (average) |
|---|---|---|---|
| 1/NS 13 | 24 | n.d. | 3.4 |
| 2/NS 13 | 12 | n.d. | 1.7 |
| 3/NS 13 | 23 | n.d. | 3.3 |
| 4/NS 14 | 27 | 3.7 | 3.9 |
| 5/NS 14 | 30 | 13.3 | 4.3 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Tribolium castaneum) | vermillionwhite | Bucher lab stock, University of Göttingen | vermillion-white (vw) | For transgenesis, mutant eye color (white) is rescued to black by 3XP3-vw |
| Genetic reagent (T. castaneum) | skh GFP enhancer trap line | Garcia-Perez et al., 2021, Bucher lab stock, University of Göttingen | G10011-GFP | Central complex reporter line |
| Genetic reagent (T. castaneum) | fez/erm GFP enhancer trap line | This paper, Bucher lab stock, University of Göttingen | fez-mm-eGFP | New CRISPR-Cas9 generated line analyzed in this work |
| Recombinant DNA reagent | [3xP3:Tc’v-SV40-Cre-2A-EGFP:bhsp68-eb] | He et al., 2019, Addgene plasmid #124068 | NHEJ repair template | |
| Recombinant DNA reagent | [bhsp68-Cas9] | Gilles et al., 2015, Addgene plasmid #65959 | Cas9 helper-plasmid | |
| Recombinant DNA reagent | [U6b-BsaI-gRNA] | Gilles et al., 2015, Addgene plasmid #65956 | Tribolium U6b promoter with BsaI cloning site to insert guide RNA sequence | |
| Sequence-based reagent | Tc-fez/erm upstream 1 | This paper | CRISPR guide RNA | GTGATTACGTGCCGCCGAAG |
| Sequence-based reagent | Tc-fez/erm upstream 2 | This paper | CRISPR guide RNA | GCGCTTGCTCGGTTCTCAGT |
| Sequence-based reagent | Tc-fez/erm upstream 3 | This paper | CRISPR guide RNA | GCCGTCGTGAGTGAAACGCC |
| Sequence-based reagent | Dm-ebony | He et al., 2019 | CRISPR guide RNA | GAACCGGGCAGCCCGCCTCC |
| Sequence-based reagent | Dm-yellow | He et al., 2019 | CRISPR guide RNA | GCGATATAGTTGGAGCCAGC |
| Sequence-based reagent | GFP-5'-rv1 | This paper | Primer | TGAACTTGTGGCCGTTTACG |
| Sequence-based reagent | fez-exon1-rv1 | This paper | Primer | AACATTAGGTGAGCAGGGCC |
| Sequence-based reagent | P2A-rv | This paper | Primer | TCTTCCACGTCTCCTGCTTG |
| Sequence-based reagent | GFP-fw-1 | This paper | Primer | TTCTTCAAGGACGACGGCAA |
| Sequence-based reagent | Cre-rv1 | This paper | Primer | GTTGCATCGACCGGTAATGC |
| Sequence-based reagent | Tc-fez/earmuff (TC004673) RNA probe | Posnien et al., 2011b | Gene-specific forward and reverse primers for probe template | fw:CAAGCCCTCCATCGTGACCC rv:GAATCGGAGGCGGAAGTACT |
| Sequence-based reagent | Tc-pointed (TC034783) RNA probe | This paper | Gene-specific forward and reverse primers for probe template | fw:GACCGCTTTATTTGCATTGT rv:TGCTTCTCGTAGTTCATCTTC |
| Sequence-based reagent | Tc-asense (TC008437) RNA probe | Posnien et al., 2011b | Gene-specific forward and reverse primers for probe template | fw:CGTCAGTGTGGTATCCCCTC rv:GCTGTTCCCACCACTGCATGA |
| Sequence-based reagent | Tc-deadpan (TC005224) RNA probe | This paper | Gene-specific forward and reverse primers for probe template | fw:CTCGAGTAACTTACCATTT rv:CTACCACGGCCTCCACATA |
| Sequence-based reagent | Tc-prospero, (TC010596) RNA probe | This paper | Gene-specific forward and reverse primers for probe template | fw:ACACAGGGATTCTCGGTCTC rv:TTGCGTGTCCAAGCAAGAA |
| Sequence-based reagent | Tc-six4, (TC003852) RNA probe | Posnien et al., 2011a | Gene-specific forward and reverse primers for probe template | fw:AAGTCGGCGCGAAAGAACGG rv:CTAAATTTATGGTACTTGAT |
| Sequence-based reagent | TC-six3, (TC000361) RNA probe | Posnien et al., 2011b | Gene-specific forward and reverse primers for probe template | fw:ATATGGCGCTCGGACTCGGC rv:CTCGTACGGTATATATCACG |
| Sequence-based reagent | Tc-otd, (TC003354) RNA probe | Posnien et al., 2011b | Gene-specific forward and reverse primers for probe template | fw:ATGTGGCCTCCAGAGGCAGT rv:TTAAGCCATATTTGCAAACT |
| Sequence-based reagent | Tc-fez/erm (TC004673) label B1 probe | Molecular Instruments | Probe for hybridization chain reaction | |
| Sequence-based reagent | Tc-deadpan (TC005224) label B2 probe | Molecular Instruments | Probe for hybridization chain reaction | |
| Sequence-based reagent | Tc-asense (TC008437) label B3 probe | Molecular Instruments | Probe for hybridization chain reaction | |
| Sequence-based reagent | Tc-pointed (TC034783) label B4 probe | Molecular Instruments | Probe for hybridization chain reaction | |
| Sequence-based reagent | B1- Alexa Fluor 488, 546 | Molecular Instruments | Hairpin | |
| Sequence-based reagent | B2- Alexa Fluor 456, 514 | Molecular Instruments | Hairpin | |
| Sequence-based reagent | B3- Alexa Fluor 647, 488 | Molecular Instruments | Hairpin | |
| Sequence-based reagent | B4- Alexa Fluor 594, 647 | Molecular Instruments | Hairpin | |
| Antibody | Anti-GFP primary antibody (chicken polyclonal) | Abcam | ab13970 | Dilution: 1/1000 v/v |
| Antibody | Anti phospho-histone 3 primary antibody (rabbit polyclonal) | Sigma-Aldrich | H0412 | Dilution: 1/100 v/v |
| Antibody | Secondary antibody coupled with Alexa Fluor 488 (goat anti-chicken, polyclonal) | Thermo Fisher Scientific | A-11039 | Dilution: 1/1000 v/v |
| Antibody | Secondary antibody coupled with Alexa Fluor 647 (goat anti-rabbit, polyclonal) | Thermo Fisher Scientific | A-21244 | Dilution: 1/500 v/v |
| Antibody | Anti-Digoxigenin-POD (poly), Fab fragments (sheep polyclonal) | Roche | 11633716001 | Dilution: 1/2000 v/v |
| Antibody | Anti-Fluorescein-POD, Fab fragments (sheep polyclonal) | Roche | 11426346910 | Dilution: 1/2000 v/v |
| Chemical compound, drug | Tyramide conjugate Alexa Fluor 555 | Invitrogen | B40955 | Dilution: 1/250 v/v |
| Chemical compound, drug | DAPI nuclear dye | Invitrogen | D1306 | Dilution: 1/1000 (of 1 µg/µl stock sol.) |
| Chemical compound, drug | FM1-43 | Invitrogen | T3163 | Dilution: 1/1000 (of 5 µg/µl stock sol.) |
| Chemical compound, drug | VECTASHIELD Antifade Mounting Medium | Vector Laboratories | VEC-H-1000 |
Additional files
-
Supplementary file 1
CRISPR guide RNA and primer sequences.
(a) Target sites for CRISPR-Cas9-mediated non-homologous end joining knock-in of the eGFP containing transgene (see Figure 2—figure supplement 1A). Guide sequences 1–3 are within 2.6 kb upstream of the transcription start site (TSS). (b) Primer sequences for line fez-mm-eGFP insertion site testing. See Figure 2—figure supplement 1A for primer binding sites.
- https://cdn.elifesciences.org/articles/99717/elife-99717-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/99717/elife-99717-mdarchecklist1-v1.docx





