Streptomyces exploration is triggered by fungal interactions and volatile signals
Abstract
It has long been thought that the life cycle of Streptomyces bacteria encompasses three developmental stages: vegetative hyphae, aerial hyphae and spores. Here, we show interactions between Streptomyces and fungi trigger a previously unobserved mode of Streptomyces development. We term these Streptomyces cells ‘explorers’, for their ability to adopt a non-branching vegetative hyphal conformation and rapidly transverse solid surfaces. Fungi trigger Streptomyces exploratory growth in part by altering the composition of the growth medium, and Streptomyces explorer cells can communicate this exploratory behaviour to other physically separated streptomycetes using an airborne volatile organic compound (VOC). These results reveal that interkingdom interactions can trigger novel developmental behaviours in bacteria, here, causing Streptomyces to deviate from its classically-defined life cycle. Furthermore, this work provides evidence that VOCs can act as long-range communication signals capable of propagating microbial morphological switches.
https://doi.org/10.7554/eLife.21738.001eLife digest
Soil is home to many bacteria. In fact, soil gets it characteristic ‘earthy’ smell from a common type of soil bacteria known as Streptomyces. Remarkably, Streptomyces are also the original sources of most of the antibiotics that are prescribed by doctors to treat bacterial infections. Scientists have been studying Streptomyces for over 70 years, and in all this time, there has been unanimous agreement on how these bacteria grow. That is to say that, unlike most other bacteria, Streptomyces grow like plants: they don’t move, and instead produce spores that are dispersed like seeds. This stationary lifestyle makes these bacteria somewhat vulnerable to predators, and so it is thought that Streptomyces make antibiotics to help protect themselves from other bacteria that are able to move around in the soil.
However, this established view of Streptomyces growth has now been turned on its head because Jones et al. have discovered that Streptomyces bacteria can indeed move when grown in the presence of fungi. Specifically, when a species of Streptomyces is grown with yeast, some of the bacteria start to explore their environment, move over top of other bacteria and up hard surfaces to heights that would be the equivalent of humans scaling Mount Everest.
Unexpectedly, Jones et al. also found that these “explorer” Steptomyces can communicate with nearby Streptomyces bacteria with a perfume-like airborne signal and convince their relatives to begin exploring too. Furthermore, while this volatile signal promotes the growth of Streptomyces, it adversely affects other bacteria and makes them sicker such that they are less able to grow and survive.
Together these findings reveal new ways that bacteria and other microbes can interact and communicate with each other. They also emphasise that researchers will need to consider such long-range communication strategies if they hope to better understand microbial communities.
https://doi.org/10.7554/eLife.21738.002Introduction
Our current understanding of microbial growth and development stems largely from investigations conducted using single-species cultures. It is becoming clear, however, that most bacteria and fungi exist as part of larger polymicrobial communities in their natural settings (Scherlach et al., 2013; Traxler and Kolter, 2015). Microbial behavior is now known to be modulated by neighbouring organisms, where interspecies interactions can have profound and diverse consequences, including modifying virulence of human pathogens (Peleg et al., 2010), altering antibiotic resistance profiles of mixed-species biofilms (Oliveira et al., 2015), enhancing bacterial growth (Romano and Kolter, 2005), and increasing production of specialized metabolites by fungi and bacteria (Schroeckh et al., 2009; Stubbendieck and Straight, 2016). Consequently, an important next step in advancing our developmental understanding of microbes will be to expand our investigations to include multi-species cultures, and in doing so, unveil new and unexpected microbial growth strategies.
The soil is a heterogeneous environment that is densely populated with bacteria and fungi, and as such, represents an outstanding system in which to study the effects of bacterial-fungal interactions. Within the polymicrobial communities occupying the soil, Streptomyces represent the largest genus of the ubiquitous actinomycetes group. These Gram-positive bacteria are renowned for both their complex developmental life cycle (Elliot et al., 2008) and their ability to produce an extraordinary range of specialized metabolites having antibiotic, antifungal, antiparasitic, and anticancer properties (Hopwood, 2007).
The Streptomyces life cycle encompasses three developmental stages (Figure 1A). First, a spore germinates to generate one or two germ tubes. These grow by apical tip extension and hyphal branching, ultimately forming a dense vegetative mycelial network that scavenges for nutrients. Second, in response to signals that may be linked to nutrient depletion, non-branching aerial hyphae extend into the air away from the vegetative cells. These aerial hyphae are coated in a hydrophobic sheath that enables escape from the aqueous environment of the vegetative mycelium (Claessen et al., 2003; Elliot et al., 2003), and their emergence coincides with the onset of specialized metabolism within the vegetative cells (Kelemen and Buttner, 1998). Aerial development requires the activity of the ‘bld’ gene products, where mutations in these genes result in colonies lacking the fuzzy/hydrophobic characteristics of wild type. The final developmental stage involves the differentiation of aerial hyphae into spores through a synchronous cell division and cell maturation event. This process is governed by the whi (for ‘white’) gene products, whose mutants fail to form mature, pigmented spores (McCormick and Flärdh, 2012). In addition to being highly stress-resistant, spores also provide a means of dispersing Streptomyces to new environments, as all characterized Streptomyces cell types are non-motile.
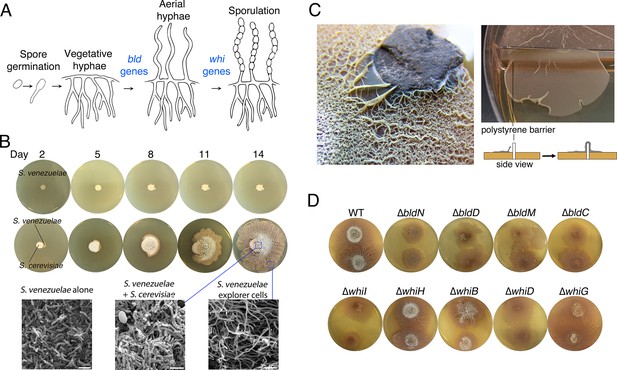
Physical association with yeast triggers Streptomyces exploratory behaviour.
(A) Developmental life cycle of Streptomyces. Germ tubes emerge from a single spore, and grow by apical tip extension and hyphal branching, forming a dense network of branching vegetative hyphae. In response to unknown signals, non-branching aerial hyphae coated in a hydrophobic sheath, escape into the air. Aerial hyphae differentiate into chains of dormant, stress-resistant non-motile spores. The bld gene products are required for the transition from vegetative growth to aerial hyphae formation, while the whi gene products are required for the differentiation of aerial hyphae into spore chains. (B) S. venezuelae grown alone (top row) and beside S. cerevisiae (middle row) on YPD (yeast extract-peptone-dextrose) medium over 14 days. Bottom panels: scanning electron micrographs of S. venezuelae grown alone (left), S. venezuelae on S. cerevisiae (middle), and S. venezuelae beside S. cerevisiae (right) for 14 days on YPD agar medium. White bars: 5 µm. (C) S. venezuelae explorer cells growing up a rock embedded in agar (left), and over a polystyrene barrier within a divided petri dish (right, and schematic below). (D) S. venezuelae wild type and developmental mutants grown beside S. cerevisiae on YPD agar medium for 14 days. Top: S. cerevisiae, together with wild type and ∆bld mutant strains (bld mutants cannot raise aerial hyphae and sporulate). Bottom: S. cerevisiae grown next to ∆whi mutant strains (whi mutants can raise aerial hyphae but fail to sporulate).
In this work, we identify a novel interaction between Streptomyces venezuelae and fungal microbes that induces a previously unknown mode of bacterial growth. We refer to this as ‘exploratory growth’, whereby cells adopt a non-branching vegetative hyphal conformation that can rapidly traverse both biotic and abiotic surfaces. We show that part of the mechanism by which fungi induce exploratory growth involves glucose depletion of the growth medium. Remarkably, this novel mode of growth can be communicated to other – physically separated – streptomycetes through a volatile compound. Volatile signalling further alters cell propagation and survival of other bacteria.
Results
Physical association with yeast stimulates Streptomyces exploratory behaviour
To explore interactions between Streptomyces and fungi, we cultured Streptomyces venezuelae alone or beside the yeast Saccharomyces cerevisiae on solid agar (Figure 1B), and incubated these cultures for 14 days. As expected, during this time S. venezuelae on its own formed a colony of normal size. In contrast, when S. venezuelae was grown beside S. cerevisiae, its growth was radically different. During the first five days, the cells appeared to consume S. cerevisiae, before initiating a rapid outgrowth that led to S. venezuelae colonizing the entire surface of a 10 cm agar plate after 14 days. Remarkably, growth did not cease when physical obstructions were encountered: S. venezuelae cells were able to spread over rocks and polystyrene barriers (Figure 1C).
To gain insight into this phenomenon, we visualized the leading edge of the rapidly migrating S. venezuelae cells (Video 1). We found it initially progressed at a rate of ~1.5 µm/min. This is an order of magnitude faster than would be explained by growth alone, given that hyphal tip extension has been calculated to occur at a rate of 0.13 µm/min (Richards et al., 2012). We refer to this rapid movement as 'exploratory growth', and these spreading cells as 'explorers', based on their ability to effectively transverse both biotic and abiotic surfaces. To further investigate the morphology of these explorer cells, we used scanning electron microscopy (SEM) to visualize S. venezuelae grown alone, S. venezuelae at the yeast interface, and S. venezuelae explorer cells, after 14 days of growth (Figure 1B). We found S. venezuelae alone grew vegetatively, albeit without any obvious branches (branching vegetative cells were observed during growth on other media types, as expected), whereas S. venezuelae growing on S. cerevisiae raised aerial hyphae and sporulated. Microscopic analysis of explorer cells revealed that they failed to branch and were reminiscent of aerial hyphae. Unlike aerial hyphae, however, these filaments were hydrophilic, based on their inability to repel aqueous solutions (Figure 1—figure supplement 1).
Leading edge of S. venezuelae explorer cells over a 17 hr time course.
https://doi.org/10.7554/eLife.21738.007To determine whether exploratory growth required classic developmental regulators (the bld and whi gene products), we grew a suite of S. venezuelae developmental mutants beside S. cerevisiae to evaluate whether these mutations impacted colony spreading (Figure 1D). Four S. venezuelae bld mutants (bldC,D,M,N) and five S. venezuelae whi mutants (whiB,D,G,H,I) were inoculated beside S. cerevisiae. Unexpectedly, all developmental mutant strains displayed a similar exploratory behaviour as wild type after 14 days, although the bldN mutant exhibited slower exploration than the other strains. The mutant strains did, however, differ in their growth on yeast, with the bld mutants failing to raise aerial hyphae, and the whi mutants failing to sporulate. This demonstrated that exploratory growth was distinct from the canonical Streptomyces life cycle, and represented a new form of growth for these bacteria.
To determine whether this exploratory behaviour was unique to S. venezuelae, we inoculated other commonly studied streptomycetes beside S. cerevisiae. We found that well-studied Streptomyces species, including S. coelicolor, S. avermitilis, S. griseus, and S. lividans, failed to exhibit an analogous spreading behaviour when plated next to S. cerevisiae. We next tested 200 wild Streptomyces isolates, growing each beside S. cerevisiae. Of these, 19 strains (~10%) exhibited exploratory growth similar to S. venezuelae. To determine whether this behaviour was confined to a particular Streptomyces lineage, we performed a phylogenetic analysis of these explorer-competent strains using rpoB sequences, and included non-exploratory model Streptomyces species as outgroups (Figure 1—figure supplement 2). We found S. venezuelae and these wild Streptomyces did not form a monophyletic group, suggesting that exploratory growth is wide-spread in the streptomycetes.
We next sought to determine whether Streptomyces exploratory behaviour could be triggered by other fungi. S. venezuelae was inoculated beside laboratory strains of Candida albicans, Candida parapsilosis, and Crypotococcus neoformans, and beside wild soil isolates of S. cerevisiae, Zygosaccharomyces florentinus, Saccharomyces castellii, Pichia fermentans and Debaryomyces hansenii (Figure 1—figure supplement 3). We observed that all species, apart from C. neoformans and P. fermentans, induced S. venezuelae exploratory behaviour. This indicated that a broad range of microbial fungi could trigger exploratory growth.
The yeast TCA cycle must be intact to stimulate S. venezuelae exploratory behaviour
To understand how fungi could stimulate exploration, we took advantage of an S. cerevisiae haploid knockout collection containing 4309 individual knockout strains. Each S. cerevisiae mutant was pinned adjacent to S. venezuelae and after 10 days, S. venezuelae exploratory growth was assessed. We identified 16 mutants that were unable to promote S. venezuelae exploration (Figure 2A). Of these, 13 had mutations affecting mitochondrial function, including eight in genes coding for enzymes in the tricarboxylic acid (TCA) cycle (Figure 2A), three in genes whose products contribute to the mitochondrial retrograde signalling pathway, as well as two whose products are involved in mitochondrial metabolism.
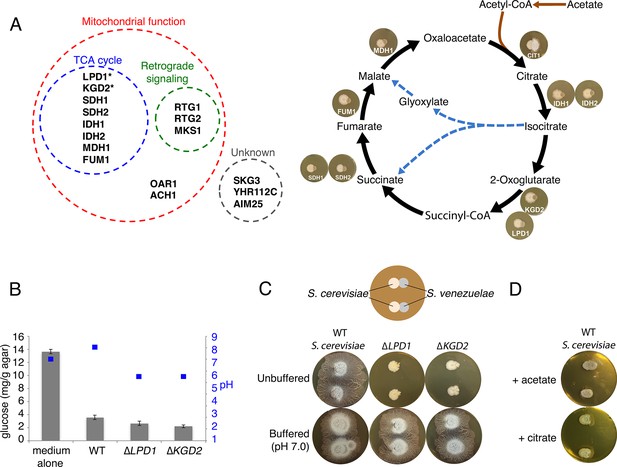
Yeast stimulates S. venezuelae exploratory growth by consuming glucose and inhibits it by acidifying the medium.
(A) S. cerevisiae mutants that fail to stimulate S. venezuelae exploratory growth. Left: functional grouping of the exploration-deficient S. cerevisiae mutations. Asterisks indicate genes also identified in C. albicans as affecting S. venezuelae exploratory growth. Right: Mutations in S. cerevisiae TCA cycle-associated genes affect exploration after citrate production. For each interaction, the indicated S. cerevisiae mutant was grown beside wild type S. venezuelae for seven days on YPD agar medium. (B) Glucose concentration and pH associated with wild type and mutant S. cerevisiae strains grown on YPD agar medium. Glucose concentrations (grey bars) and pH (blue squares) were measured from medium alone, and beneath wild type, ∆LPD1 or ∆KGD2 S. cerevisiae strains grown on YPD medium for seven days. All values represent the mean ± standard error for four replicates. (C) Top: schematic of the experimental set up, with S. cerevisiae grown to the left of S. venezuelae on YPD medium. Two replicates are grown on each agar plate. Bottom: wild type, ∆LPD1, and ∆KGD2 S. cerevisiae strains grown for 14 days beside wild type S. venezuelae on unbuffered YPD agar and YPD agar buffered to pH 7.0 with MOPS. (D) Wild type S. cerevisiae spotted beside wild type S. venezuelae and grown for 14 days on YPD agar medium plates supplemented with acetate or citrate, each buffered to pH 5.5.
We confirmed these mutant effects using Candida albicans strains carrying tetracycline-repressible haploid mutations (Figure 2—figure supplement 1). We grew four mutant strains adjacent to S. venezuelae, and found that two of them, ∆LPD1 and ∆KGD2, also failed to stimulate S. venezuelae exploratory behaviour. As the products of these two genes act in the TCA cycle (Figure 2A), these data collectively suggest that fungal respiration, and in particular TCA cycle function, influences exploratory growth in S. venezuelae.
Exploration is glucose-repressible and pH-dependent
In considering how TCA cycle defects could influence S. venezuelae behaviour, we hypothesized that glucose uptake and/or consumption might play a role. We measured glucose levels of a YPD agar control, and compared this with YPD agar underneath S. cerevisiae. Uninoculated medium had 3.8 times as much glucose as S. cerevisiae-associated agar (Figure 2B), confirming that S. cerevisiae consumed glucose during growth on YPD agar. This suggested that either glucose depletion by yeast, or some product of glucose metabolism, may trigger S. venezuelae exploratory growth.
To test these possibilities, we first asked whether exploratory growth could be triggered by lowering glucose concentrations. We plated S. venezuelae on YP (yeast extract-peptone) in the presence (G+) and absence (G−) of glucose (Figure 2—figure supplement 2). After 10 days, we found growth on G− medium permitted S. venezuelae exploration, irrespective of whether yeast was present. This implied that glucose repressed exploratory growth.
We also tested glucose consumption by the S. cerevisiae LPD1 and KGD2 mutants. The products of these genes, along with that of KGD1, comprise the 2-oxoglutarate dehydrogenase complex responsible for converting 2-oxoglutarate into succinyl-CoA in the TCA cycle (Przybyla-Zawislak et al., 1999) (Figure 2A). We found wild type, ∆LPD1 and ∆KGD2 S. cerevisiae strains consumed similar levels of glucose (Figure 2B), suggesting that other factors must be inhibiting S. venezuelae exploration when grown adjacent to these TCA cycle mutants.
All TCA cycle-associated S. cerevisiae mutants that failed to stimulate S. venezuelae exploratory behaviour were blocked after the production of citrate in the TCA cycle (Figure 2A). We hypothesized that this disruption might result in an accumulation of organic acids, and that S. cerevisiae mutants secreted these acids to maintain a neutral intracellular pH. We measured the pH of wild type, ∆LPD1, and ∆KGD2 strains when grown on YPD (G+) agar, and found wild type S. cerevisiae raised the agar pH from 7.0 to 7.5, whereas both TCA cycle mutants lowered the agar pH to 5.5 (Figure 2B).
To test whether acid secretion by the S. cerevisiae LPD1 and KGD2 mutants prevented S. venezuelae exploratory growth, the two mutants were grown beside S. venezuelae on non-buffered YPD agar, and equivalent medium buffered to pH 7.0 (Figure 2C). After 14 days growth on non-buffered plates, the S. cerevisiae mutants failed to stimulate S. venezuelae exploratory behaviour, whereas the same strains on buffered agar – which would counter the pH-lowering effects of the secreted acids – could now promote S. venezuelae exploration. To further verify this pH-dependent effect, we grew wild type S. cerevisiae beside S. venezuelae on YPD agar supplemented with citrate or acetate (Figure 2D). We found that after 14 days, S. venezuelae spreading was inhibited, confirming that secreted acids inhibited S. venezuelae exploration.
Collectively, these results suggested that S. venezuelae exploratory growth is a glucose- and acid-repressible phenomenon. Consistent with these observations, we also determined that S. venezuelae exploration was associated with a significant rise in pH: as S. venezuelae consumed the yeast, the medium pH rose from 7.0 to 8.0, and once S. venezuelae exploratory growth initiated (day 5), the pH rose further to 9.5 (Figure 3A). This increase in pH was also observed for S. venezuelae grown on G- medium (in the absence of yeast) (Figure 3—figure supplement 1), suggesting that the rise in pH was mediated by the Streptomyces cells. To determine whether high pH was sufficient to promote exploration, we inoculated S. venezuelae cells on YPD agar medium buffered to pH 9.0. Exploration was not induced under these growth conditions (Figure 3—figure supplement 2). These data indicated that alkaline conditions were important but not sufficient for exploration, and further suggested that an adaptation phase was required during the transition to exploratory growth.
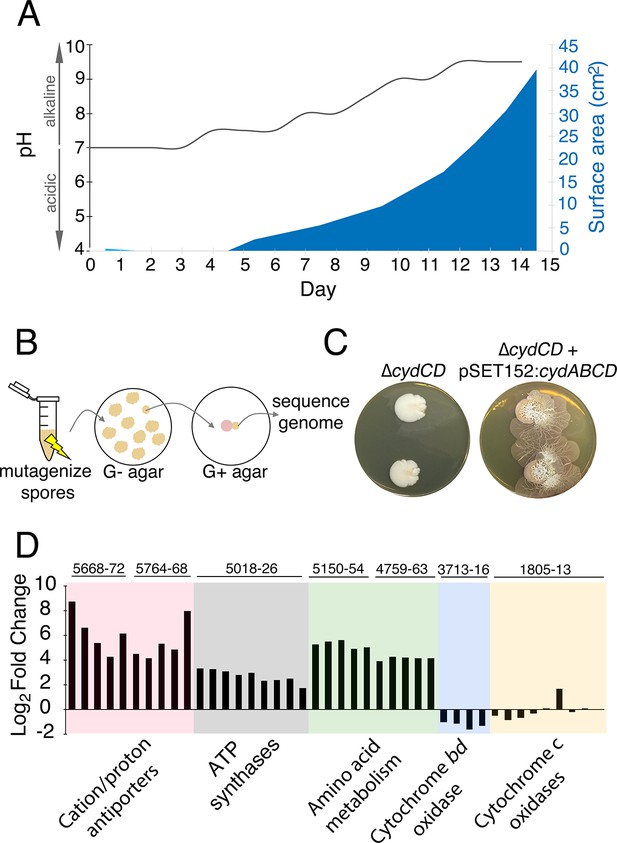
The alkaline stress response is associated with S. venezuelae exploratory behaviour.
(A) The surface area and medium pH associated with S. venezuelae explorer cells beside S. cerevisiae on YPD agar were measured and plotted every day for 14 days. (B) Schematic of the method used to identify genes required for S. venezuelae exploratory growth. S. venezuelae spores were subject to chemical mutagenesis, then screened on G- agar (no glucose, exploration-permissive without S. cerevisiae) for a lack of exploratory growth. Static colonies (beige) were grown beside S. cerevisiae (pink) on YPD medium to confirm a lack of exploratory growth. Genomic DNA was isolated from strains unable to initiate exploratory growth on G- agar, and when inoculated beside S. cerevisiae on YPD medium. Whole genome sequencing was performed to identify mutations responsible for the lack of exploratory growth. (C) Morphology of a mutant cytochrome bd oxidase S. venezuelae strain (∆cydCD) and the corresponding complemented strain grown on YPD agar for 14 days. (D) Transcript levels for alkaline stress-responsive genes in S. venezuelae explorer cells (grown beside S. cerevisiae on YPD medium), divided by levels for non-exploratory S. venezuelae cells (grown alone on YPD medium). Transcript levels were normalized and differential expression was log2-transformed. The associated sven gene numbers are shown above the bar graphs.
S. venezuelae exploration requires an alkaline stress response
To investigate the genetic basis for this phenomenon we employed chemical mutagenesis, and screened for S. venezuelae mutants that failed to display exploratory behaviour when grown on G- medium (where yeast is not required) (Figure 3B). Candidate non-spreading mutant colonies were identified, and were tested in association with S. cerevisiae on YPD (G+) medium to confirm their inability to spread. Of the 48 exploration-defective mutants identified on G− medium, only three were also unable to spread when grown on YPD medium beside S. cerevisiae. This indicated that exploratory growth on G− agar may have distinct genetic requirements from exploratory growth on YPD (G+) medium.
We sequenced the genomes of wild type S. venezuelae and the three non-spreading mutants of interest (those unable to spread on both G- medium alone and YPD (G+) medium beside S. cerevisiae). Each mutant harbored point mutations in the sven_3713-3716 operon. This operon is predicted to encode subunits of the cytochrome bd oxidase complex (cydA/sven_3713 and cydB/sven_3714), along with an ABC transporter required for cytochrome assembly (cydCD/sven_3715) (Brekasis and Paget, 2003). One strain had a mutation in sven_3715 (H673Y), while the other two strains had mutations in sven_3713 (Q186stop) and were likely clonal. To ensure that these mutations were responsible for the exploration-defective phenotype, we complemented the exploratory growth defect in each mutant with a cosmid carrying an intact cydABCD operon, and confirmed that exploration was restored (Figure 3—figure supplement 3). We also deleted cydCD in a wild type S. venezuelae background, and confirmed that this strain was unable to initiate exploration when grown beside S. cerevisiae. As before, spreading could be restored to the mutant after introducing cydABCD on an integrating plasmid vector (Figure 3C). These data indicated that the cytochrome bd oxidase complex was essential for S. venezuelae exploration.
S. venezuelae, like many other bacteria, encodes two cytochrome oxidase complexes. The cytochrome bd oxidase catalyzes terminal electron transfer without a concomitant pumping of protons across the membrane, while the cytochrome bc1-aa3 complex requires proton transfer from the cytoplasm. The cytochrome bd oxidase functions as part of the alkaline stress response in other bacteria (Krulwich et al., 2011). As we had established that alkaline conditions were a prerequisite for S. venezuelae exploration, we questioned whether other alkaline stress-responsive genes might be associated with exploratory growth. Using RNA-sequencing (RNA-seq), we examined the transcription profiles of S. venezuelae alone, compared with S. venezuelae exploratory cells grown beside S. cerevisiae on YPD medium (Figure 3D). The five gene clusters mostly highly upregulated in S. venezuelae explorer cells encoded the ATP synthase complex (sven_5018-26; 7.6-fold increase relative to non-spreading), two predicted cation/proton antiporter complexes (95.9- and 85.3-fold increase relative to non-spreading for sven_5668-72 and sven_5764-68, respectively), and two peptide transporters (17.4- and 38.3-fold increase relative to non-spreading for sven_4759-63 and sven_5150-54, respectively) (Figure 3D).
Higher expression of the cation/proton antiporters, alongside increased ATP synthesis, would be expected to enhance proton uptake into the cell; equivalent genes are upregulated as part of the alkaline stress response in other bacteria (Krulwich et al., 2011). Amino acid catabolism is also upregulated under alkaline growth conditions in other bacteria (Padan et al., 2005). Given the dramatically increased expression of the peptide transporters, we confirmed that exploratory growth required an amino acid source (Supplementary file 1). Collectively, these results suggest that exploration is coupled with a metabolic reprogramming that permits robust growth under highly alkaline conditions.
S. venezuelae explorer cells alkalinize the medium using an airborne volatile organic compound
S. venezuelae exploration is associated with high pH conditions, and our data suggested this rise in pH was promoted by S. venezuelae itself. We hypothesized that this pH effect could be mediated either through the secretion of diffusible basic compounds, or through the release of volatile organic compounds (VOCs). To differentiate between these possibilities, we set up a two-compartment petri plate assay, where S. venezuelae was grown beside S. cerevisiae on YPD agar in one compartment, while the adjacent compartment contained uninoculated YPD agar (Figure 4A). As a negative control, we set up an equivalent set of plates, only with S. venezuelae alone (no yeast) on YPD agar in the first compartment. In each case, the two compartments were separated by a polystyrene barrier. After 10 days, we measured the pH of the uninoculated YPD compartment, and found the compartment adjacent to S. venezuelae alone remained at pH 7.0, whereas the one adjacent to S. venezuelae explorer cells had risen from pH 7.0 to 9.5, indicating the explorer cells produced a basic VOC (Figure 4A).
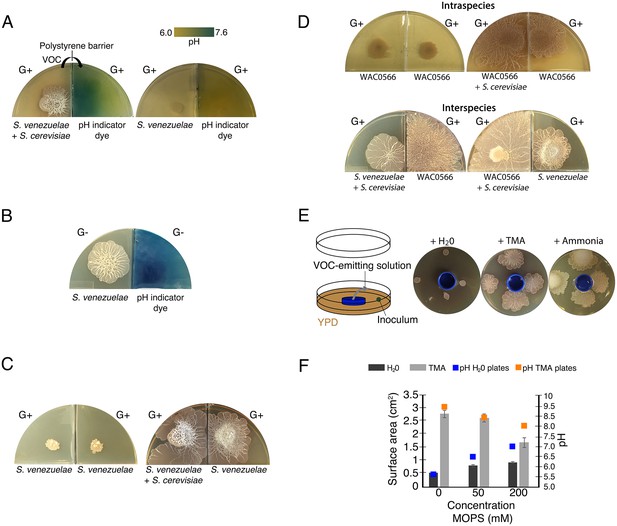
Volatile organic compounds released by S. venezuelae raise the medium pH and induce exploratory growth in physically separated Streptomyces.
(A) Effect of S. venezuelae explorer cells on pH of physically separated medium. Each compartment is separated by a polystyrene barrier. S. venezuelae and S. cerevisiae were grown in the left compartment of one plate (left), while S. venezuelae alone was grown in the left compartment of the other plate (right). After 10 days, bromothymol blue pH indicator dye was spread on the agar in the right compartment of each plate. Blue indicates VOC-induced alkalinity. (B) S. venezuelae was grown alone on YP (G- agar) in the left compartment, while the right compartment contained uninoculated YP (G-) agar. After seven days, the same pH indicator dye as in Figure 4A was spread over the agar in the right compartment. Blue represents a rise in pH above 7.6. (C) Left: S. venezuelae alone was inoculated in each compartment. Right: S. venezuelae was grown beside S. cerevisiae in the left compartment, and S. venezuelae alone was grown in the right compartment. All strains were grown on YPD (G+) agar medium for 10 days. (D) Top left: Wild Streptomyces isolate WAC0566 was grown alone in each compartment. Top right: WAC0566 was grown beside S. cerevisiae in the left compartment, and grown alone in the right compartment. Bottom left: S. venezuelae was grown beside S. cerevisiae in the left compartment, and WAC0566 was grown alone in the right compartment. Bottom right: WAC0566 was grown beside S. cerevisiae in the left compartment, while S. venezuelae was grown alone in the right compartment. All strains were cultured on YPD (G+) agar medium for 10 days. (E) Schematic of the plate-based assay used to assess the effects of volatile-emitting solutions (and controls) on nearby Streptomyces colonies. H2O, TMA, or ammonia solutions were placed in a blue plastic dish, and S. venezuelae was spotted around each dish on YPD medium. Plates were incubated at room temperature for seven days. (F) Surface area and pH of S. venezuelae colonies grown on YPD medium around small dishes containing H2O or TMA solutions, as shown in Figure 4E. S. venezuelae was grown at room temperature for seven days on either unbuffered YPD medium or YPD medium buffered to pH 7.0 using MOPS. All values represent the mean ± standard error for four replicates.
To verify that the VOC was produced by S. venezuelae explorers and not by S. cerevisiae, we repeated the two-compartment assay with S. venezuelae grown alone on G- agar, a condition that also induced exploratory behaviour. We found that S. venezuelae growing alone on G- agar could alkalinize the adjacent YPD compartment. This confirmed that a basic VOC was produced by S. venezuelae explorer cells (Figure 4B).
S. venezuelae exploratory cells use VOCs to induce exploration in other streptomycetes at a distance
Bacterial VOCs can influence a wide range of cellular behaviours. To determine whether the VOC produced by explorer cells represented an exploration-promoting signal for physically separated Streptomyces colonies, we leveraged our two-compartment assay, inoculating one with S. venezuelae beside S. cerevisiae on YPD agar, and the adjacent compartment with S. venezuelae on the same medium (a condition where exploration by S. venezuelae otherwise requires yeast association). As expected, after 10 days, the S. cerevisiae-associated cells were actively spreading. Remarkably, the adjacent S. venezuelae cells (in the absence of yeast) had also initiated exploratory growth (Figure 4C). As a negative control, S. venezuelae alone was grown in both compartments on YPD agar; spreading was not observed for cells grown in either compartment after 10 days (Figure 4C). These data implied that S. venezuelae explorer cells released a VOC that effectively promoted exploratory growth in distantly located S. venezuelae cells. We tested whether our exploration-deficient cydCD mutant was able to respond to this VOC, and observed that despite its inability to explore when grown next to yeast, this mutant was capable of exploration when stimulated by neighbouring explorer cells (Figure 4—figure supplement 1).
To determine whether S. venezuelae explorers used VOCs to potentiate exploration in other species, we again used our two-compartment assay. We cultured S. venezuelae with S. cerevisiae in one compartment, and tested whether these cells could stimulate exploratory growth of the wild Streptomyces isolate WAC0566 in the adjacent compartment (Figure 4D) (WAC0566 initiates exploratory growth when cultured next to yeast, but fails to spread on its own; Figure 4D). Negative control plates were set up in the same way as before, with WAC0566 alone in both compartments. After 10 days, WAC0566 grown adjacent to S. venezuelae explorers initiated exploratory growth, and this was not seen for the negative control (Figure 4D). This indicated that exploratory growth could be communicated to unrelated streptomycetes.
We tested the volatile-mediated communication between these strains in a reciprocal experiment, and found that S. venezuelae exploration could also be stimulated by a VOC produced by yeast-associated WAC0566 (Figure 4D). This inter-species promotion of S. venezuelae exploration was observed for at least 13 other wild Streptomyces strains (Figure 4—figure supplement 2). Importantly, VOC communication of exploratory growth was confined to those species with exploratory capabilities (S. coelicolor failed to respond to the VOC elicitor).
The VOC trimethylamine stimulates Streptomyces exploratory behaviour
We determined that the exploration-promoting VOC could be produced by liquid-grown (G-) cultures, and that it stimulated exploratory growth by both S. venezuelae and WAC0566 (Figure 4—figure supplement 3). To rule out the possibility that any liquid-grown culture could promote exploration, we also grew S. venezuelae and WAC0566 in YPD (G+) liquid medium, and found these cultures were unable to stimulate exploration. This suggested that VOC production was glucose-repressible, and its production correlated with growth conditions that promoted exploration.
To determine the identity of the VOC, we grew S. venezuelae and WAC0566 in G+ and G- liquid culture for three days. We collected the supernatants of each culture, and assayed them using two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS). From this, 1400 unique compounds were identified. To determine which compound(s) were responsible for promoting exploration, we applied a stringent filter, requiring the compound(s) to be: (i) present in at least 50% of S. venezuelae and WAC0566 exploration-inducing (G-) cultures; (ii) present in at least 10-fold greater abundance in exploration-inducing (G-) cultures versus static (G+) cultures; and (iii) have at least a 60% similarity score to known compounds in the 2011 National Institute of Standards and Technology (NIST) Mass Spectral Library. We arrived at a list of 21 candidate compounds (Supplementary file 1). Of these, 12 were not detected in the negative controls (G+ cultures). Within this group of 12, only four were detected in 100% of S. venezuelae and WAC0566 exploration-promoting cultures: trimethylamine (TMA), thiocyanic acid, 6-methyl-5-hepten-2-one, and 2-acetylthiazole. Notably, TMA was >10 fold more abundant than the other three compounds, and thus we focussed our initial investigations on this molecule.
TMA is a volatile nitrogen-containing metabolite with a high pKa (9.81). As we knew S. venezuelae produced a basic VOC, we hypothesized that TMA was responsible for promoting exploration. To test this possibility,we placed commercially-available TMA in a small plastic container at the centre of a YPD (G+) agar plate, and then inoculated S. venezuelae at defined positions around this container (Figure 4E). After seven days, S. venezuelae cultured adjacent to the TMA-emitting solutions had initiated exploratory growth, while those grown next to a water-containing control failed to spread. This implied that TMA was the VOC used by S. venezuelae and WAC0566 to elicit exploratory growth.
TMA production is not well understood, although recent work has revealed two mechanisms by which it can be generated from quaternary amines. Acinetobacter sp. employ a carnitine oxygenase (product of the cntAB gene cluster) in converting L-carnitine into TMA (Zhu et al., 2014), while Desulfovibrio desulfuricans converts choline into TMA using a choline-trimethylamine lyase (encoded by the cutCD genes) (Craciun and Balskus, 2012). S. venezuelae lacks any gene with similarity to cntA, and thus does not use an equivalent pathway to generate TMA. It does possess homologues of cutCD; however, these genes were more highly expressed (~5 fold) in static S. venezuelae cultures (where no TMA was ever detected), than in spreading cultures. This suggested that these gene products may not direct TMA production in S. venezuelae. TMA can also be produced upon biogenic reduction of trimethylamine N-oxide (TMAO) by TMAO reductases. Bacteria known to carry out this reaction typically encode one or more TMAO reducase operons, including some combination of torSTRCAD (or torSTRCADE), torYZ, dmsABC, and ynfEFGH (Dunn and Stabb, 2008; McCrindle et al., 2005). S. venezuelae encodes homologs to some of these genes [specifically torA (top hit: SVEN_1326), dmsAB (top hit: SVEN_3040-3039), and ynfEFG (top hit: SVEN_3040, 3040 and 3039)]. In our RNA seq data, however, all of these genes (along with more divergent homologs) were expressed at extremely low levels, with equivalent levels for each gene being observed in both static and exploratory cultures. This suggested these gene products were unlikely to be involved in converting TMAO to TMA in S. venezuelae.
TMA induces exploratory growth by raising the pH of the growth medium
To confirm that TMA could raise the pH of the growth medium in the same way as explorer cells, we measured the pH of non-inoculated YPD agar around dishes containing TMA, and found the pH rose from 7.0 to 9.5. To test whether TMA induced exploratory growth by raising the pH, we repeated our plate assays described in Figure 4E, and buffered the agar to 7.0 using 50 or 200 mM MOPS (Figure 4F). The pH of these plates rose to 8.0 (as opposed to 9.5 on non-buffered plates), and TMA failed to induce S. venezuelae exploration to the same extent as on non-buffered plates. To further validate the pH-mediated effect of TMA, we tested whether ammonia (another basic VOC) had the same effect (Figure 4E). After seven days, ammonia induced S. venezuelae exploratory growth, suggesting that VOC-mediated alkalinity stimulated Streptomyces exploration.
TMA can reduce the survival of other bacteria
TMA can alter the developmental program of streptomycetes, and is known to modify the antibiotic resistance profiles of bacteria (Letoffe et al., 2014). Given the antibiotic production capabilities of Streptomyces bacteria, we wondered whether the release of TMA might also inhibit the growth of other bacteria. To explore this possibility, we set up a small petri dish of YPD agar inside a larger dish of YPD agar (Figure 5).S venezuelae and S. cerevisiae (exploratory cultures) or S. venezuelae alone (static cultures) were inoculated on the smaller dish, and plates were incubated for 10 days. The soil-dwelling bacteria Bacillus subtilis or Micrococcus luteus were then spread on the larger petri dish. Growth of B. subtilis and M. luteus in association with exploratory or static S. venezuelae cultures, was then assessed after overnight incubation. B. subtilis and M. luteus colony numbers were reduced by an average of 17.4% and 25.1%, respectively, on plates exposed to VOCs produced by exploratory S. venezuelae, relative to those grown adjacent to static cultures. We determined that the pH of medium adjacent to exploratory S. venezuelae had risen to 9.5, suggesting that TMA and its pH-modulatory effects could be responsible for the growth-inhibition of these bacteria.
To directly test the inhibitory potential of TMA, we set up an equivalent assay, where the TMA-producing S. venezuelae-S. cerevisiae combination was substituted with aqueous TMA solutions of varying concentrations. We spread B. subtilis, and M. luteus around the TMA-containing receptacles, and after seven days, quantified growth (Figure 5C). We observed an approximately 50% drop in viable cells when exposed to 0.9% TMA, and in the case of B. subtilis, a further drop in viability was observed as TMA concentrations increased. This confirmed that TMA adversely affected the growth and survival of other soil bacteria.
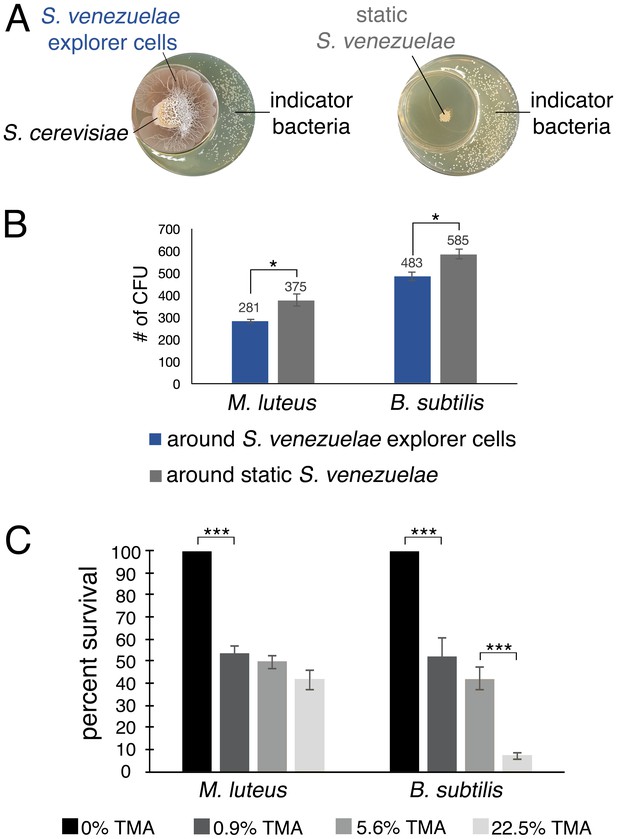
S. venezuelae VOCs inhibit the growth of other bacteria.
(A) S. venezuelae was grown beside S. cerevisiae (left) or alone (right) on YPD agar in a small dish placed within a larger dish containing YPD medium. After 10 days, an indicator strain (B. subtilis or M. luteus) was spread around the dish. (B) Quantification of B. subtilis and M. luteus colonies following growth adjacent to static or explorer S. venezuelae cultures. Values represent the mean ± standard error for three replicates. The asterisk (*) indicates p<0.05, as determined by a Student’s t-test. (C) Quantification of B. subtilis and M. luteus survival following incubation around small dishes containing TMA solutions at concentrations ranging from 0–22.5%. Plates were incubated at room temperature for two days. Percent survival indicates the OD600 of strains around wells containing 0.9%, 5.6%, or 22.5% TMA solutions compared to the OD600 of strains around wells containing H2O (100% survival). Values represent the mean ± standard error for three biological replicates, and each biological replicate is the average of four technical replicates. The asterisks (***) indicate p<0.005, as determined by a Student’s t-test.
Discussion
The canonical multicellular lifecycle of Streptomyces bacteria begins with fungus-like hyphal growth, and ends with sporulation (Figure 1A). In this system, spore dispersal is the sole means by which these bacteria can establish themselves in new environments. Here, we demonstrate a new developmental behavior for Streptomyces that provides them with an alternative means of colonizing new habitats. In response to fungal neighbours and nutrient (glucose) depletion, Streptomyces can escape the confines of their classically defined lifecycle, and initiate exploratory growth. Exploratory growth is remarkably relentless: explorer cells are not limited by inanimate barriers, and can grow over abiotic surfaces. Explorer cells alter their local environment through the release of the alkaline, volatile compound TMA. Emitting TMA not only promotes exploratory behaviour by the producing cells, it also functions as an airborne signal that elicits an exploratory response in physically distant streptomycetes, and provides further fitness benefits by inhibiting the growth of other bacteria.
Metabolic cues trigger a developmental switch
S. venezuelae exploration is triggered by two key metabolic cues: glucose depletion and a rise in pH. We observed exploratory growth under low glucose conditions. In low-glucose areas of the soil, Streptomyces may initiate exploratory growth in an attempt to colonize environments with more readily available nutrients, whereas in high-glucose areas (e.g. near plant roots, or in association with fruit) (Kliewer, 1965; Lugtenberg et al., 1999; Romano and Kolter, 2005), exploration may be less advantageous, initiating only after nearby fungi – or other microbes – consume the existing glucose supply. Microbial alteration of nutrient profiles is likely to be common in the soil environment (e.g. Romano and Kolter, 2005), and we expect that the exploratory growth away from glucose-depleted areas would provide a benefit analogous to that of motility systems in other bacteria. Although the mechanism underlying exploration remains to be elucidated, it may be linked to sliding motility given its apparently passive nature (no appendages involved), and the fact that Streptomyces are known surfactant producers.
S. venezuelae exploration is also promoted by a self-induced rise in extracellular pH. Alkaline growth conditions trigger morphological switches in a range of fungi, including the human pathogens C. albicans, C. neoformans, and Aspergillus fumigatus (Bertuzzi et al., 2014; Davis et al., 2000; O'Meara et al., 2014). This is this first time this phenomenon has been observed in bacteria.
Volatile compounds promote communication and enhance competition
Exploratory growth by Streptomyces cells is coordinated by the airborne compound TMA. TMA can further induce exploration in physically distant streptomycetes. Importantly, this volatile signal is not limited to S. venezuelae, and can be both transmitted and sensed by other Streptomyces species. Consequently, it is possible for Streptomyces to respond to TMA produced by other bacteria and initiate exploratory growth under conditions where glucose concentrations are high and/or glucose-titrating organisms are absent. Developmental switching in response to VOC eavesdropping has not been previously reported, but exploiting community goods in this way is not unprecedented. For example, quorum signals and siderophores produced by one organism can be taken up or used by others (Lyons and Kolter, 2015; Traxler et al., 2012). The VOC repertoire of microorganisms appears to be vast (Chuankun et al., 2004; Insam and Seewald, 2010; Kai et al., 2009; Schöller et al., 2002; Schulz and Dickschat, 2007; Wilkins and Schöller, 2009). Volatile compounds have historically been implicated in the ‘avoidance responses’ of fungi, promoting their growth away from inanimate objects (Cohen et al., 1975; Gamow and Böttger, 1982). Increasingly, these compounds are now being found to have important roles in communication between physically separated microbes (Audrain et al., 2015; Bernier et al., 2011; Briard et al., 2016; Kim et al., 2013; Letoffe et al., 2014; Schmidt et al., 2015, 2016; Tyc et al., 2015; Wang et al., 2013; Wheatley, 2002). A range of fungi use the volatile alkaline compound ammonia to induce morphological switches in other fungi, and to mediate inhibition of neighbouring colonies (Palková et al., 1997). Our observations suggest that VOCs may also be key bacterial morphological determinants, communicating developmental switches both within and between different microbial species.
In addition to serving as communication signals, VOCs may also provide their producing organisms with a competitive advantage in the soil. Volatile molecules can modulate the antibiotic resistance profiles of bacteria (Letoffe et al., 2014), and can themselves have antifungal or antibacterial activity (Schmidt et al., 2015). TMA is a particularly potent example. Here, we show that exposure of other bacteria to TMA inhibits their growth, while previous work has revealed that TMA exposure increases bacterial sensitivity to aminoglycoside antibiotics. Notably, Streptomyces synthesize an extraordinary range of antibiotics, including many aminoglycosides. Thus in the soil, Streptomyces-produced TMA may have direct antibacterial activity, in addition to sensitizing bacteria to the effect of Streptomyces-produced antibiotics. The ability of Streptomyces to modulate the growth of other soil-dwelling bacteria during exploratory growth would maximize their ability to colonize new environments, and exploit whatever nutrients are present.
Ecological implications for exploratory growth within microbial communities
Exploratory growth represents a powerful new addition to the Streptomyces developmental repertoire, and one that appears to be well-integrated into the existing life cycle. When grown next to yeast, explorer cells emerge from a mass of sporulating cells (Figure 6). This functional differentiation represents an effective bet-hedging strategy, whereby spreading explorer cells scavenge nutrients for the group, while the sporulating cells provide a highly resistant genetic repository, ensuring colony survival in the event of failed exploration. Explorer cells resemble vegetative hyphae, in that their surface is hydrophilic; however, unlike traditional vegetative hyphae, explorer cells do not appear to branch. We presume that explorer cells dispense with frequent branching as a trade-off for the ability to rapidly spread to new environments. Exploratory growth also occurs independently of the typical bld- and whi-developmental determinants, supporting the notion that this is a unique growth strategy. It is possible, however, given the slower exploration observed for bldN mutants (where bldN encodes a sigma factor), that BldN regulon members help to facilitate the exploration process.
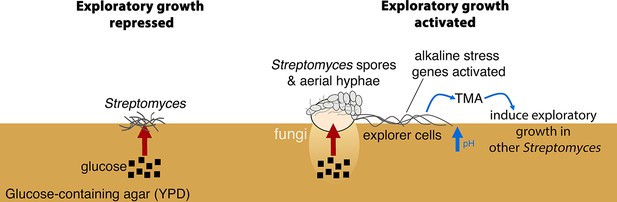
New model for Streptomyces development.
When S. venezuelae is grown alone on glucose-rich medium S. venezuelae exploratory growth is repressed (left). When S. venezuelae is grown beside S. cerevisiae or other yeast on glucose-rich medium (right), the yeast metabolizes glucose, relieving the repression of S. venezuelae exploration. S. venezuelae explorer cells produce the volatile pheromone TMA, which raises the pH of the medium from 7.0 to 9.5. Explorer cells activate alkaline stress genes to withstand the alkaline pH. TMA, and its associated medium alkalinisation, can induce exploratory growth in physically separated Streptomyces.
While we observed exploratory growth in a subset of Streptomyces species, it is possible that this capability is more broadly conserved and is stimulated by different conditions than those investigated here. Indeed, microbes are abundant in the soil, and interactions between different organisms within these communities are likely to be more the norm than the exception. Our work illustrates the importance of inter-species interactions in bacterial development, as a key to revealing novel growth strategies. It also emphasises the need to consider long-range communication strategies, in the form of volatile compounds, which may play widespread roles in regulating development and metabolic activities in microbial communities.
Materials and methods
Strains, plasmids, media and culture conditions
Request a detailed protocolStrains, plasmids and primers used in this study are listed in Supplementary file 1. S. venezuelae ATCC 10712 was grown on MYM (maltose-yeast extract-malt extract) agar medium for spore stock generation. Spreading was investigated during growth on the surface of YPD (yeast extract-peptone-dextrose/glucose) agar, glucose-deficient YP (G-) agar, yeast extract agar supplemented with different amino acid sources (tryptone or 2% casamino acids) or YPD/G- agar medium supplemented with citrate, acetate, borate or MOPS buffer. All strains were grown at 30°C, apart from the TMA experiments which were conducted at room temperature in a fume hood. S. cerevisiae strain BY4741 (MATa; his3∆1; leu2∆0 ura3∆0 met15∆0) was grown on the same spreading-investigative media at 30°C or room temperature. Prior to plating S. venezuelae and S. cerevisiae together, S. venezuelae was cultured in liquid MYM at 30°C, while S. cerevisiae was grown in liquid YPD at 30°C overnight. Three microliters of S. venezuelae cultures were applied to the right of 3 µL S. cerevisiae on the surface of YPD agar medium, and plates were then incubated at 30°C or room temperature for up to 14 days
Scanning electron microscopy (SEM) and light microscopy
Request a detailed protocolSEM was used to examine strains grown on YPD or MYM agar for up to 14 days. Samples were prepared and visualized using a TEMSCAN LSU scanning electron microscopy as described previously (Haiser et al., 2009). To monitor the rate of exploratory growth (Video 1), an Olympus SZX12 Sterioscope and CoolSNAP HQ photometric camera were used to capture 70 frames of growth over the course of 17 hr.
Phylogenetic analyses
Request a detailed protocolrpoB (Guo et al., 2008) was amplified from each of the 19 exploration-competent wild isolates using primers RpoBPF and RpoBPR (Supplementary file 1), before being sequenced using RpoBF1 and RpoBR1 (Supplementary file 1). Trimmed rpoB sequences were aligned using Mafft version 7.2.6.6. A maximum likelihood tree was built using RAxML version 8.2.4 (Stamatakis, 2006), using a GTRGAMMA model of nucleotide substitution, with 500 bootstrap replicates to infer support values of nodes. Outputs were visualized using FigTree.
Yeast library screening
Request a detailed protocolOvernight cultures of S. venezuelae were spotted onto rectangular plates containing YPD agar (OmniTray: Nunc International) using a 384-pin replicator. Each strain of a S. cerevisiae BY4741 haploid deletion library was inoculated beside an individual S. venezuelae colony using a 384-pin replicator. Plates were grown for five days at 30°C and screened for an absence of S. venezuelae exploratory growth. Yeast mutants unable to stimulate S. venezuelae exploratory growth were re-tested on individual YPD agar plates. For C. albicans deletion screens, C. albicans GRACE collection tetracycline repressible deletion mutants (Roemer et al., 2003) were inoculated beside S. venezuelae on YPD agar plates. Mutants were induced using 1 or 5 µg/mL tetracycline, which is below the minimum inhibitory concentration of tetracycline for S. venezuelae.
Glucose assays and measurement of pH
Request a detailed protocolMeasurements of glucose levels beneath S. cerevisiae colonies and in YPD alone were performed using a Glucose (GO) Assay Kit (Sigma). For all experiments, pH levels of solid agar were measured using one or a combination of pH sticks and the pH indicator dye bromothymol blue (Sigma, St Louis, MO).
Chemical mutagenesis and whole-genome sequencing
Request a detailed protocolApproximately 108 S. venezuelae spores were added to 1.5 mL 0.01 M KPO4 at pH 7.0. Spores were centrifuged and resuspended in 1.5 mL 0.01 M KPO4 at pH 7.0. The spores were then divided into two 750 µL aliquots in screw-cap tubes. As a control, 25 µL H2O was added to one aliquot, while 25 µL ethyl methanesulfonate (EMS, Sigma, M0880) was added to the other aliquot. Tubes were vortexed for 30 s, and incubated shaking at 30°C for 1 hr, with an additional inversion being performed every 10 min. Spores were centrifuged at 3381 ×g for 3 min at room temperature, prior to being resuspended in 1 mL freshly made and filter-sterilized 5% w/v sodium thiosulfate solution. Spores were washed twice in 1 mL H2O, after which they were resuspended in 1 mL H2O. For each tube, a dilution series ranging from 10−4 to 10−8 was made using H2O, and 100 µL of each dilution was then spread onto MYM agar plates and incubated for three days at 30°C. Individual colonies were counted to ensure that survival of the EMS-treated spores was, at most, 50% that of the untreated (H2O) control. Colonies were collected from plates inoculated with EMS-treated spores, and were screened for loss of spreading capabilities on G- agar plates. Select mutants were then tested for their inability to spread when plated next to yeast; those mutants that also failed to initiate spreading in the presence of S. cerevisiae were grown in liquid MYM, and chromosomal DNA was extracted using the Norgen Biotek Bacterial Genomic DNA Isolation kit for downstream sequencing.
Using the Illumina Nextera XT DNA sample preparation kit, DNA libraries were prepared for three non-exploratory S. venezuelae mutants, alongside their wild type S. venezuelae parent. Whole genome-sequencing was performed on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) using 150 bp paired-ends reads. Reads were aligned to the S. venezuelae reference genome using Bowtie 2 (Langmead and Salzberg, 2012) and were converted to BAM files using SAMtools (Li et al., 2009). Single nucleotide polymorphisms (SNPs) were called using SAMtools mpileup and bcftools, and SNP locations, read depth, and identities were generated using VCFtools (Danecek et al., 2011).
Construction of cydCD (cytochrome bd oxidase) deletion strain and mutant complementation
Request a detailed protocolAn in-frame deletion of sven_3715-3716 was generated using ReDirect technology (Gust et al., 2003). The coding sequence was replaced by an oriT-containing apramycin resistance cassette. The gene deletion was verified by PCR, using combinations of primers located upstream, downstream and internal to the deleted genes (see Supplementary file 1). The cydCD mutant phenotype was complemented using a DNA fragment encompassing the WT genes, sven_3713-3714, and associated upstream and downstream sequences (see Supplementary file 1), cloned into the integrating plasmid vector pSET152. To control for any phenotypic effects caused by plasmid integration, pSET152 alone was introduced into wild type and the cydCD mutant strains, and these strains were used for phenotypic comparison with the complemented mutant strain.
RNA isolation, library preparation and cDNA sequencing
Request a detailed protocolRNA was isolated as described previously from two replicates of S. venezuelae explorer cells growing beside S. cerevisiae for 14 days, and two replicates of S. venezuelae alone grown for 24 hr on YPD agar plates (we were unable to isolate high quality RNA from S. venezuelae alone at later time points). For all four replicates, ribosomal RNA (rRNA) was depleted using a Ribo-zero rRNA depletion kit. cDNA and Illumina library preparation were performed using a NEBnext Ultra Directional Library Kit, followed by sequencing using unpaired-end 80 base-pair reads using the HiSeq platform. Reads were aligned to the S. venezuelae genome using Bowtie 2 (Langmead and Salzberg, 2012), then sorted, indexed, and converted to BAM format using SAMtools (Li et al., 2009). BAM files were visualized using Integrated Genomics Viewer (Robinson, 2011), and normalization of transcript levels and analyses of differential transcript levels were conducted using Rockhopper (McClure et al., 2013). RNA-seq data has been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE86378 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=idmrgcmexranpun&acc=GSE86378).
Analysis of volatile metabolites via GC×GC-TOFMS
Request a detailed protocolS. venezuelae and WAC0566 were grown in liquid YPD (G+) or YP (G-) for three days. For each strain and condition, six biological replicates were grown, and for each, three technical replicates were analyzed. Four milliliters of each culture supernatant were transferred to 20 mL air-tight headspace vials, which were stored at −20°C prior to volatile analysis. Headspace volatiles were concentrated on a 2 cm triphasic Divinylbenzene/Carboxen/Polydimethylsiloxane (DVB/CAR/PDMS) solid-phase microextraction (SPME) fiber (Supelco, Bellenfonte, PA) (30 min, 50°C, 250 rpm shaking). Volatile molecules were separated, identified, and relatively quantified using two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS), as described previously (Bean et al., 2012; Rees et al., 2016). The GC×GC-TOFMS (Pegasus 4D, LECO Corporation, St. Joseph, MI) was equipped with a rail autosampler (MPS, Gerstel, Linthicum Heights, MD) and fitted with a two-dimensional column set consisting of an Rxi−624Sil (60 m × 250 μm×1.4 μm (length × internal diameter × film thickness); Restek, Bellefonte, PA) first column followed by a Stabilwax (Crossbond Carbowax polyethylene glycol; 1 m × 250 μm×0.5 μm; Restek, Bellefonte, PA) second column. The main oven containing column one was held at 35°C for 0.5 min, and then ramped at 3.5 °C/min from 35°C to 230°C. The secondary oven containing column 2, and the quad-jet modulator (2 s modulation period, 0.5 s alternating hot and cold pulses), were heated in step with the primary oven with +5°C and +25°C offset relative to the primary oven, respectively. The helium carrier gas flow rate was 2 mL/min. Mass spectra were acquired over the range of 30 to 500 a.m.u., with an acquisition rate of 200 spectra/s. Data acquisition and analysis was performed using ChromaTOF software, version 4.50 (LECO Corp.).
Identification of candidate volatile signals
Request a detailed protocolChromatographic data was processed and aligned using ChromaTOF. For peak identification, a signal-to-noise (S/N) cutoff was set at 100, and resulting peaks were identified by a forward search of the NIST 2011 Mass Spectral Library. For the alignment of peaks across chromatograms, maximum first and second-dimension retention time deviations were set at 6 s and 0.15 s, respectively, and the inter-chromatogram spectral match threshold was set at 600. Analytes that were detected in greater than half of exploration-promoting Streptomyces cultures (grown in G- medium) and not detected in media controls or S. venezuelae grown in G+ medium (failed to promote exploration), were considered candidate molecules associated with the phenotype of interest.
Assays for volatile-mediated phenotypes
Request a detailed protocolAqueous solutions (1.5 mL) of commercially available TMA solutions (Sigma), ammonia solutions (Sigma) or water (negative control) were added to small, sterile plastic containers and placed in a petri dish containing 50 mL YPD agar. TMA solutions were typically diluted to 11.5% w/v, although concentrations as low as 0.9% were able to promote spreading and inhibit the growth of other bacteria. Ammonia solutions of 0.1–1 M were used, and all were able to induce spreading. S. venezuelae was inoculated around the small vessels, after which the large petri dish was closed and incubated in the fume hood at room temperature for up to 10 days. For buffering experiments, YPD plates were supplemented with 50 or 200 mM MOPS buffer (pH 7.0). Medium pH was measured as above, while colony surface areas were measured using ImageJ (Abràmoff et al., 2004). For bacterial survival assays around TMA-containing vessels, B. subtilis and M. luteus strains were grown overnight in LB medium, before being subcultured to an OD600 of 0.8. One hundred microliters of each culture were then spread on YPD agar plates, adjacent to water or TMA-containing vessels. For assays to measure how S. venezuelae explorer VOCs affect the survival of other bacteria, S. venezuelae was grown alone or beside S. cerevisiae in a small petri dish containing YPD agar. This small dish was placed inside a larger dish containing YPD agar. Plates were grown for 10 days, before B. subtilis and M. luteus were subcultured to an OD600 of 0.8, and diluted 1/10 000. Fifty microliters of each culture were then spread on the larger plate containing YPD agar, and colonies were quantified after overnight growth.To test the effect of TMA on B. subtilis and M. luteus growth, these indicator strains were grown overnight in LB medium, before being subcultured to an OD600 of 0.8. One hundred microlitres were spread around wells containing 1.5 mL solutions of TMA at different concentrations on YPD (water control, 0.9%, 5.6% and 22.5%). Plates were incubated for two days at room temperature in the fume hood, before cells were scraped into 2 mL YPD and vigorously mixed. Dilution series were used to measure the OD600 of the resulting cell suspensions. Error bars indicate standard error of three biological replicates, and four technical replicates of each.
Data availability
-
Streptomyces exploration is triggered by fungal interactions and volatile signalsPublicly available at NCBI Gene Expression Omnibus (accession no: GSE86378).
References
-
Airborne bacterial interactions: Functions out of thin air?Frontiers in Microbiology 6:1–5.https://doi.org/10.3389/fmicb.2015.01476
-
A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2)The EMBO Journal 22:4856–4865.https://doi.org/10.1093/emboj/cdg453
-
Soil volatile fungistasis and volatile fungistatic compoundsSoil Biology and Biochemistry 36:1997–2004.https://doi.org/10.1016/j.soilbio.2004.07.020
-
Avoidance response, house response, and wind responses of the sporangiophore of PhycomycesThe Journal of General Physiology 66:67–95.https://doi.org/10.1085/jgp.66.1.67
-
Microbial conversion of choline to trimethylamine requires a glycyl radical enzymeProceedings of the National Academy of Sciences 109:21307–21312.https://doi.org/10.1073/pnas.1215689109
-
The variant call format and VCFtoolsBioinformatics 27:2156–2158.https://doi.org/10.1093/bioinformatics/btr330
-
Candida albicans RIM101 pH response pathway is required for host-pathogen interactionsInfection and Immunity 68:5953–5959.https://doi.org/10.1128/IAI.68.10.5953-5959.2000
-
Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114Journal of Bacteriology 190:5814–5823.https://doi.org/10.1128/JB.00227-08
-
BookMulticellular development in StreptomycesIn: Whitworth D, editors. Myxobacteria: Multicellularity and Differentiation. ASM Press. pp. 419–439.https://doi.org/10.1128/9781555815677.ch24
-
Avoidance and rheotropic responses in phycomyces. Evidence for an 'avoidance gas" mechanismThe Journal of General Physiology 79:835–848.https://doi.org/10.1085/jgp.79.5.835
-
A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematicsInternational Journal of Systematic and Evolutionary Microbiology 58:149–159.https://doi.org/10.1099/ijs.0.65224-0
-
Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolorJournal of Bacteriology 191:6501–6512.https://doi.org/10.1128/JB.00767-09
-
Volatile organic compounds (VOCs) in soilsBiology and Fertility of Soils 46:199–213.https://doi.org/10.1007/s00374-010-0442-3
-
Bacterial volatiles and their action potentialApplied Microbiology and Biotechnology 81:1001–1012.https://doi.org/10.1007/s00253-008-1760-3
-
Initiation of aerial mycelium formation in StreptomycesCurrent Opinion in Microbiology 1:656–662.https://doi.org/10.1016/S1369-5274(98)80111-2
-
Changes of concentration of glucose, fructose and total soluble solids in flowers and berries of Vitis viniferaAmerican Journal of Enology and Viticulture 16:101–110.
-
Molecular aspects of bacterial pH sensing and homeostasisNature Reviews Microbiology 9:330–343.https://doi.org/10.1038/nrmicro2549
-
Fast gapped-read alignment with Bowtie 2Nature Methods 9:357–359.https://doi.org/10.1038/nmeth.1923
-
The sequence alignment/Map format and SAMtoolsBioinformatics 25:2078–2079.https://doi.org/10.1093/bioinformatics/btp352
-
On the evolution of bacterial multicellularityCurrent Opinion in Microbiology 24:21–28.https://doi.org/10.1016/j.mib.2014.12.007
-
Computational analysis of bacterial RNA-Seq dataNucleic Acids Research 41:e140.https://doi.org/10.1093/nar/gkt444
-
Signals and regulators that govern Streptomyces developmentFEMS Microbiology Reviews 36:206–231.https://doi.org/10.1111/j.1574-6976.2011.00317.x
-
Microbial dimethylsulfoxide and trimethylamine-N-oxide respirationAdvances in Microbial Physiology 50:147–198.https://doi.org/10.1016/S0065-2911(05)50004-3
-
The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the hostMolecular and Cellular Biology 34:673–684.https://doi.org/10.1128/MCB.01359-13
-
Alkaline pH homeostasis in bacteria: New insightsBiochimica Et Biophysica Acta (BBA) - Biomembranes 1717:67–88.https://doi.org/10.1016/j.bbamem.2005.09.010
-
Medically important bacterial-fungal interactionsNature Reviews Microbiology 8:340–349.https://doi.org/10.1038/nrmicro2313
-
Genetic and biochemical interactions involving tricarboxylic acid cycle (TCA) function using a collection of mutants defective in all TCA cycle genesGenetics 152:153–166.
-
The volatile metabolome of Klebsiella pneumoniae in human bloodJournal of Breath Research 10:27101.https://doi.org/10.1088/1752-7155/10/2/027101
-
Mechanistic basis of branch-site selection in filamentous bacteriaPLoS Computational Biology 8:e1002423.https://doi.org/10.1371/journal.pcbi.1002423
-
Molecular bacteria-fungi interactions: effects on environment, food, and medicineAnnual Review of Microbiology 67:375–397.https://doi.org/10.1146/annurev-micro-092412-155702
-
Volatile affairs in microbial interactionsThe ISME Journal 9:1–7.https://doi.org/10.1038/ismej.2015.42
-
Microbial small talk: Volatiles in fungal-bacterial InteractionsFrontiers in Microbiology 6:1495.https://doi.org/10.3389/fmicb.2015.01495
-
Bacterial volatiles: the smell of small organismsNatural Product Reports 24:814–842.https://doi.org/10.1039/b507392h
-
Volatile Metabolites from ActinomycetesJournal of Agricultural and Food Chemistry 50:2615–2621.https://doi.org/10.1021/jf0116754
-
Multifaceted interfaces of bacterial competitionJournal of Bacteriology 198:2145–2155.https://doi.org/10.1128/JB.00275-16
-
Natural products in soil microbe interactions and evolutionNatural Products Reports 32:956–970.https://doi.org/10.1039/C5NP00013K
-
Interspecies modulation of bacterial development through iron competition and siderophore piracyMolecular Microbiology 86:628–644.https://doi.org/10.1111/mmi.12008
-
Volatiles in Inter-Specific bacterial interactionsFrontiers in Microbiology 6:1412.https://doi.org/10.3389/fmicb.2015.01412
-
Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1FEMS Microbiology Letters 341:45–51.https://doi.org/10.1111/1574-6968.12088
-
The consequences of volatile organic compound mediated bacterial and fungal interactionsAntonie Van Leeuwenhoek 81:357–364.https://doi.org/10.1023/A:1020592802234
-
Volatile organic metabolites from selected streptomyces strainsActinomycetologica 23:27–33.https://doi.org/10.3209/saj.SAJ230202
Article and author information
Author details
Funding
Natural Sciences and Engineering Research Council of Canada (Vanier Scholarship)
- Stephanie E Jones
Ontario Government (Ontario Graduate Scholarship)
- Louis Ho
Canadian Institutes of Health Research (MOP133636)
- Justin R Nodwell
Natural Sciences and Engineering Research Council of Canada (RGPIN-2015-04681)
- Marie A Elliot
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We would like to extend our thanks to Dr. Leah Cowen and Teresa O’Meara for access to the S. cerevisiae and GRACE yeast library collections, Dr. J.P Xu and Aaron Vogen for access to environmental yeast strains, Dr. Gerry Wright for access to his wild Streptomyces library, Dr. Mark Buttner and Maureen Bibb for the S. venezuelae developmental mutants, Dr. Chris Yip and Amine Driouchi for microscopy assistance, Ben Furman for assistance with the Streptomyces phylogeny, David Crisante for artistic assistance, and Dr. Heather Bean, Dr. Mark Buttner, Dr. Erin Carlson, Andy Johnson and Matt Moody for helpful discussions.
Copyright
© 2017, Jones et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 17,100
- views
-
- 2,144
- downloads
-
- 177
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 177
- citations for umbrella DOI https://doi.org/10.7554/eLife.21738





