Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence
Figures
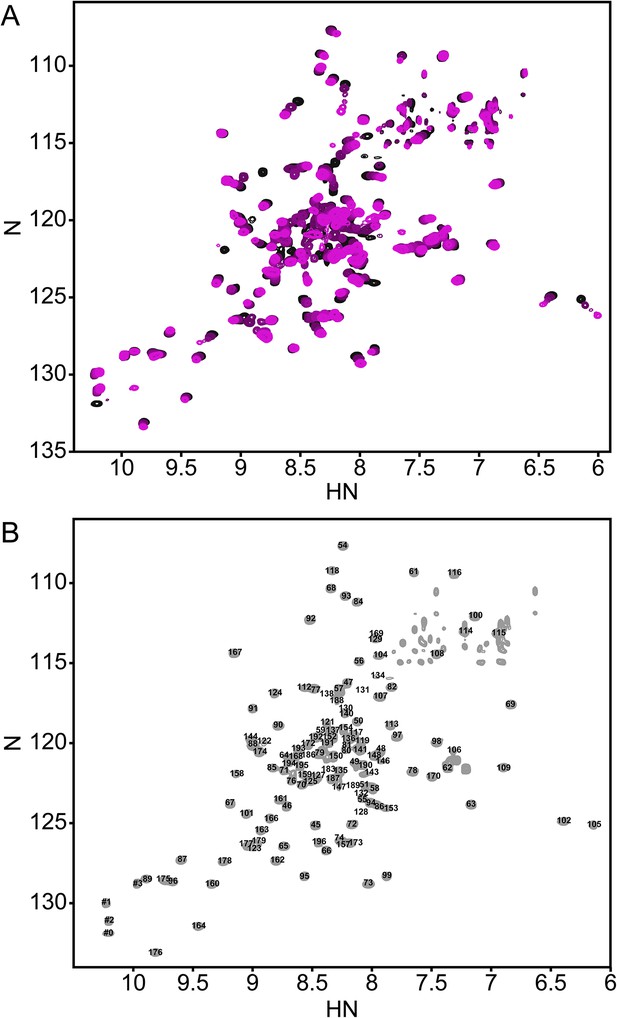
The annotated PhoQ PD (1H, 15N)-HSQC-NMR spectrum reveals significant peak shifting and broadening during pH titration.
(A) (1H, 15N)-HSQC-NMR spectra of neutral to acidic pH-titration of the PhoQ PD. The pH-titration is represented as a magenta (pH 6.5) to black (pH 3.5) color gradient. The pH-titration spectra include pH 6.5, 6.0, 5.5, 4.9, 4.1, and 3.5. (B) The assigned (1H, 15N)-HSQC-NMR spectra of the S. enterica Typhimurium PhoQ PD at pH 3.5. Residue numbers are labeled proximal to their corresponding peak.
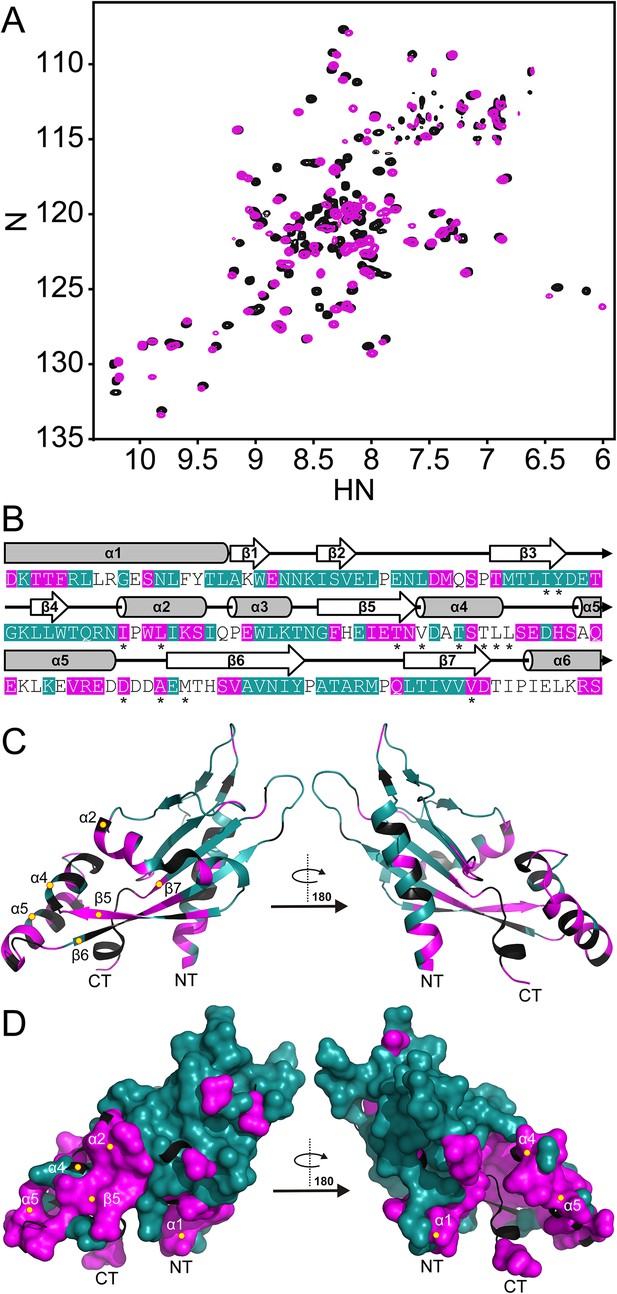
The PhoQ PD experiences significant pH-dependent perturbations which map to α4 and α5 and the α/β-core.
(A) Comparison of (1H, 15N)-HSQC-NMR spectra of the PhoQ PD at pH 6.5 (magenta) and pH 3.5 (black). (B) Residues that experience CSPs >0.08 ppm and/or peak broadening determined from the spectral comparison in panel A are mapped onto the S. enterica Typhimurium PhoQ PD (residues 45–188) primary and secondary structures (pH-sensitive residues, magenta; pH-insensitive residues, teal; ambiguous or non-assigned residues, no color). The locations of activating mutations from Figure 2—figure supplement 1 are indicated with asterisks. (C) pH-sensitive residues from panel A mapped onto the PhoQ PD structure (PDB 1YAX); pH-sensitive residues (magenta), pH-insensitive residues (teal), and ambiguous or non-assigned residues (black). pH-sensitive secondary structural features are labeled with yellow circles (NT, N-termini; CT, C-termini). (D) Continuous surface representation (1.4 Å probe) of pH-sensitive (magenta) and pH-insensitive (teal) residues from panel C mapped onto the PhoQ PD crystal structure.
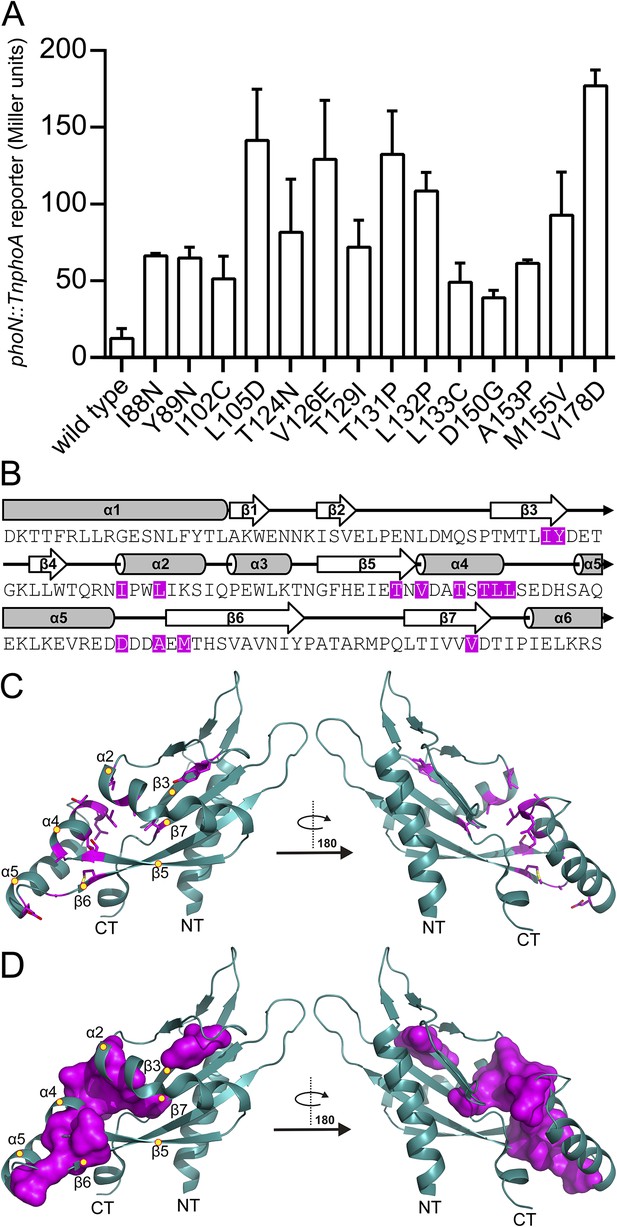
Residues involved in PhoQ activation and repression form a buried network connecting α4 and α5 to the α/β-core.
(A) Mutations identified by random and site-directed mutagenesis confer increased PhoQ-dependent phoN::TnphoA alkaline phosphatase activity when grown in N-mm supplemented with 10 mM MgCl2. The data shown are representatives from at least three independent experiments performed in duplicate and presented as the mean ± SD. (B) Activating mutations from panel A (magenta) mapped onto the S. enterica Typhimurium PhoQ PD primary and secondary structures (residues 45–188). (C) The locations of activating mutations from panel A (magenta sticks) mapped onto the PhoQ PD structure (PDB 1YAX). Secondary structural features with activating mutations are labeled with yellow circles (NT, N-termini; CT, C-termini). (D) Continuous surface representation (1.4 Å probe) of activating mutations from panel C mapped onto the PhoQ PD crystal structure.
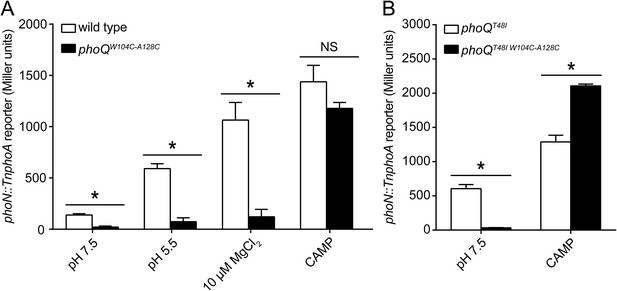
A disulfide bond between α-helices 2 and 4 inhibits PhoQ activation by acidic pH and divalent cation limitation, but does not inhibit activation by CAMP.
PhoQ-dependent phoN::TnphoA alkaline phosphatase activity of (A) wild-type and phoQW104C-A128C or (B) phoQT48I and phoQT48I W104C-A128C S. enterica Typhimurium strains grown in basal (pH 7.5) or activating (pH 5.5, 10 µM MgCl2, or CAMP) N-mm. (A and B) The data shown are representatives from at least three independent experiments performed in duplicate and presented as the mean ± SD. Unpaired Student's t-test was performed between wild type and phoQW104C-A128C or phoQT48I and phoQT48I W104C-A128C for all conditions; (*) p ≤ 0.05, (NS) not significantly different.
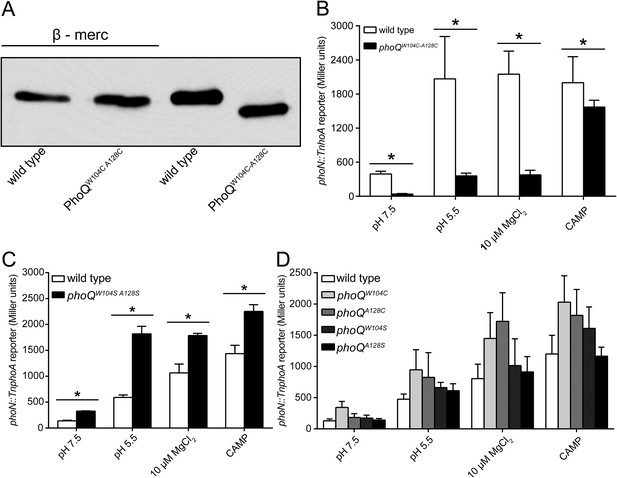
The PhoQW104C-A128C disulfide forms in the Salmonella periplasm and individual mutations at W104 or A128 do not inhibit activation by acidic pH or divalent cation limitation.
(A) Non-reducing SDS-PAGE and Western blotting of wild-type and phoQW104C-A128C membranes with an anti-PhoQ PD antibody. Membranes were treated with or without sample buffer containing β-mercaptoethanol to show the effect of disulfide reduction on migration rate. (B, C, and D) PhoQ-dependent phoN::TnphoA alkaline phosphatase activity of S. enterica Typhimurium strains grown in basal (pH 7.5) or activating (pH 5.5, 10 µM MgCl2, CAMP) N-mm. (B) Chromosomal wild-type and phoQW104C-A128C S. enterica Typhimurium. (C) Wild-type and phoQW104S-A128S S. enterica Typhimurium. (B and C) Unpaired Student's t-test was performed between wild type and phoQW104C-A128C or phoQW104S-A128S for all conditions; (*) p ≤ 0.05. (D) Single cysteine or serine mutations at position W104 and A128 in PhoQ. The data shown are representatives from at least three independent experiments performed in duplicate and presented as the mean ± SD.
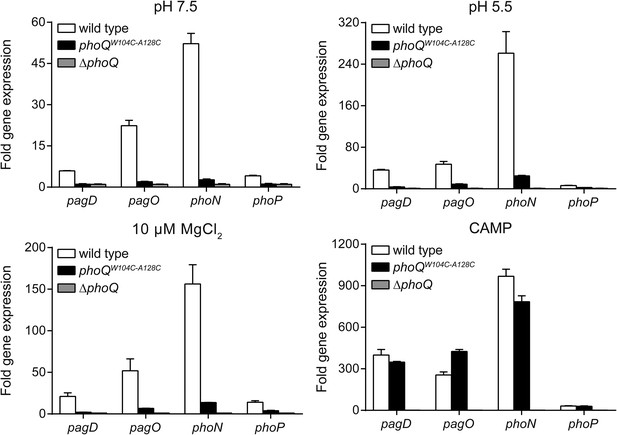
Multiple PhoQ-dependent genes in phoQW104C-A128C Salmonella are induced by CAMP, but not by acidic pH or divalent cation limitation.
PhoQ-dependent gene expression from S. enterica Typhimurium strains grown in basal (pH 7.5) or activating (pH 5.5, 10 µM MgCl2, or CAMP) N-mm. Gene expression was normalized to rpoD and represented as fold-induction relative to ΔphoQ. The data shown are representatives from at least three independent experiments performed in duplicate and presented as the mean ± SD.
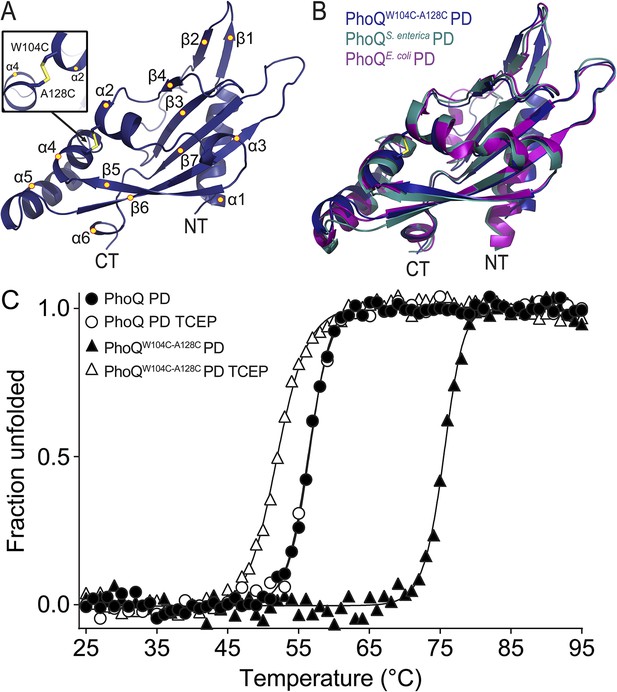
The PhoQW104C-A128C PD is structurally similar to wild type and has increased stability.
(A) 1.9 Å crystal structure of the S. enterica Typhimurium PhoQW104C-A128C PD (PDB 4UEY). The W104C-A128C disulfide bond (inset) is located between α2 and α4. Secondary structural features are annotated with yellow circles (NT, N-termini; CT, C-termini). (B) Structural comparison of the PhoQW104C-A128C PD (blue), the wild-type S. enterica Typhimurium PhoQ PD (PDB 1YAX, teal), and the wild-type E. coli PhoQ PD (PDB 3BQ8, purple). (C) Thermal denaturation of wild-type S. enterica Typhimurium PhoQ PD and PhoQW104C-A128C PD treated with or without TCEP reducing agent monitored by CD spectroscopy at 212 nm. Raw data were normalized to give the fraction unfolded protein assuming a two-state denaturation process. A sigmoidal curve was fit to the processed data. The data shown are representatives from three independent experiments.
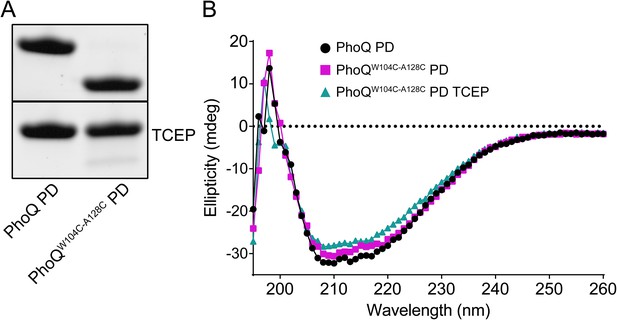
Wild-type and PhoQW104C-A128C PD have similar secondary structure.
(A) Non-reducing SDS-PAGE of purified wild-type S. enterica Typhimurium PhoQ PD and PhoQW104C-A128C PD treated with or without TCEP reducing agent. (B) CD wavelength scan of the wild-type PhoQ PD, PhoQW104C-A128C PD, and PhoQW104C-A128C PD treated with TCEP at 25°C buffered to pH 5.5.
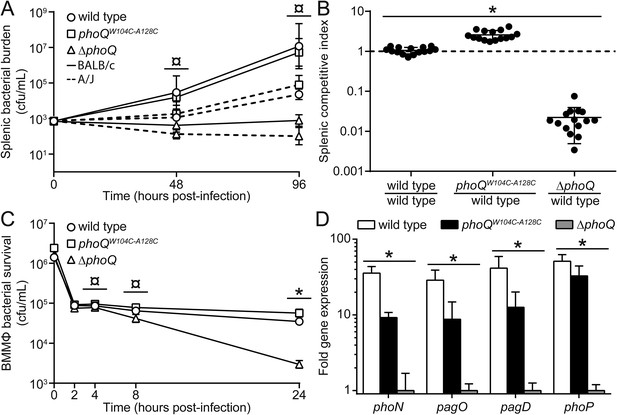
phoQW104C-A128C Salmonella survive within host organisms and exhibits PhoQ-dependent gene expression within macrophage.
(A) Individual S. enterica Typhimurium strains administered IP to BALB/c (solid lines) or A/J (dotted lines) mice. The inoculum is shown at T = 0 hpi. Spleens were harvested and bacterial burden quantified. (B) Competition between S. enterica Typhimurium strains administered IP to BALB/c mice. Spleens were harvested, bacteria quantified 48-hpi and CI determined. (A and B) The data shown are representatives from at least three independent experiments performed in quintuplet and presented as the mean ± SD. (C) BALB/c BMMΦ infected with strains of S. enterica Typhimurium. Bacteria were harvested and quantified at the indicated time-points. The inoculum is shown at T = 0 hpi. The data shown are representatives from at least three independent experiments performed in triplicate and presented as the mean ± SD. (D) PhoQ-dependent gene expression from S. enterica Typhimurium strains within BALB/c BMMΦ 4-hpi. Gene expression was normalized to rpoD and presented as fold-induction relative to ΔphoQ. The data shown are representatives from at least three independent experiments and presented as the mean ± SD. (A, B, C, and D) Unpaired Student's t-test was performed between all strains (bar) for each time-point or gene. Symbols for significant difference; (¤) wild type and phoQW104C-A128C are not significantly different from each other (p ≥ 0.05), but are significantly different from ΔphoQ (p ≤ 0.05), (*) all strains are significantly different from each other (p ≤ 0.05).
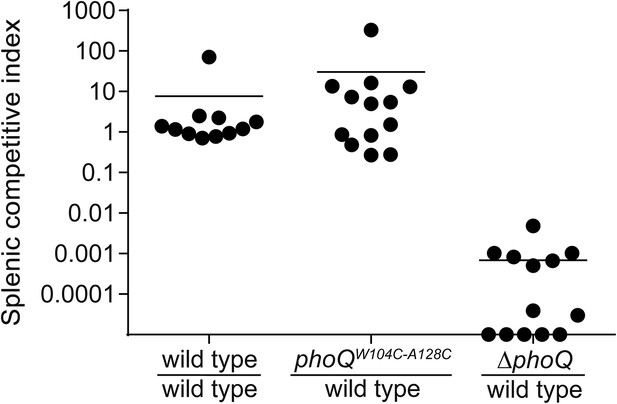
Acidic pH and divalent cation sensing by PhoQ are dispensable for PO systemic competition of S. enterica Typhimurium.
Competition between S. enterica Typhimurium strains administered PO to BALB/c mice. Spleens were harvested, bacteria quantified 96-hpi and CI determined. The data shown are from three independent experiments and presented as the mean ± SD. Data points on the x-axis represent samples with a CI of zero.
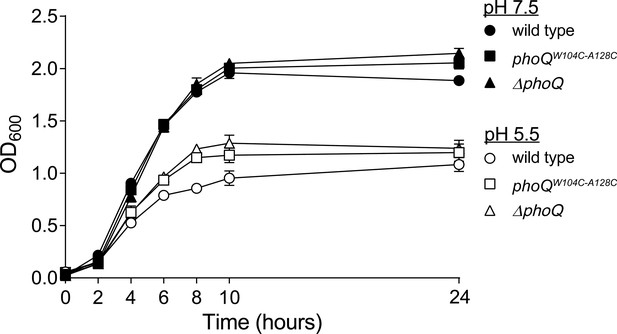
The in vitro growth rate of wild-type Salmonella is decreased relative to phoQW104C-A128C and ΔphoQ when grown at pH 5.5.
S. enterica Typhimurium strains were grown in N-mm buffered to pH 7.5 (closed symbols) or pH 5.5 (open symbols) supplemented with 1 mM MgCl2. Bacterial growth was monitored by OD600 at the indicated time-points. The data shown are representatives from at least three independent experiments performed in duplicate and presented as the mean ± SD.
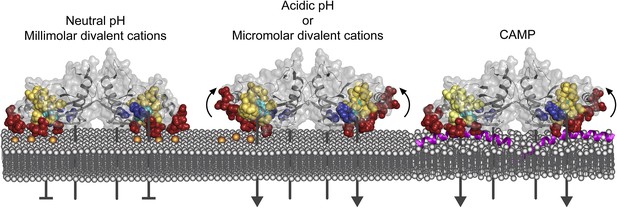
Model of PhoQ activation and repression.
(Left) At neutral pH and millimolar divalent cation concentration, the PhoQ PD is maintained in a repressed conformation due to rigidified interactions between the α/β-core (yellow spheres), α4 and α5, and salt-bridges (bronze spheres) formed between the acidic patch (red spheres) and inner membrane. (Middle) Transition to a mildly acidic (left protomer) or divalent cation limited (right protomer) environment promotes flexibility in α4 and α5 (bent arrows) and conformational dynamics in the α/β-core surrounding H157 (teal spheres). Movement in α4 and α5 due to acidic pH or divalent cation limitation destabilizes salt-bridges between the acidic patch and inner membrane perturbing the TDK network (blue spheres) resulting in activation. (Right) CAMP (magenta helices) intercalates into the inner membrane and promotes PhoQ activation by directly interacting with the PhoQ transmembrane domains and/or by disrupting local phospholipid packing (left protomer) and/or by overcoming constraints in α4 and α5 (right monomer, bent arrow).
Tables
Crystallographic data collection and refinement
| PhoQW104C-A128C PD | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 128.04, 45.37, 81.37 |
| α, β, γ (°) | 90, 102.53, 90 |
| Resolution (Å) | 31.3–1.9 (2.01–1.90)* |
| Rsym or Rmerge | 0.05 (0.51) |
| I/σI | 12.9 (1.6) |
| Completeness (%) | 97.8 (93.8) |
| Redundancy | 3.6 (3.2) |
| Refinement | |
| Resolution (Å) | 31.3–1.90 (1.95–1.90)* |
| No. reflections | 35,633 |
| Rwork/Rfree | 0.23/0.26 (0.38/0.44) |
| No. atoms (all) | |
| Protein | 3391 |
| Water | 138 |
| Ca2+ | – |
| B-factors | |
| Protein | 44.8 |
| Water | 40.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.2 |
| Ramachandran statistics | |
| Residues in favored region no (%) | 409 (98.3) |
| Residues in allowed region no (%) | 7 (1.7) |
| Residues in outlier region no (%) | 0 (0) |
| PDB-entry | 4UEY |
| Crystallization conditions | 0.1 M Bis-Tris pH 6.5, 200 mM MgCl2, 25% Peg3350 |
-
*
Values in parentheses are for highest-resolution shell.
Strains and plasmids used in this study
| Strain | Description | Source |
|---|---|---|
| CS093 | 14028s wild type S. enterica Typhimurium | ATCC |
| CS1081 | CS093 phoQ::TPOP phoN::TnphoA | Bader et al., 2005 |
| CS1083 | CS1081 pBAD24 | Bader et al., 2005 |
| CS1084 | CS1081 pBAD24-phoQ | Bader et al., 2005 |
| CS1399 | CS1081 pBAD24-phoQI88N | This work |
| CS1400 | CS1081 pBAD24-phoQY89N | This work |
| KH45 | CS1081 pBAD24-phoQI102C | This work |
| KH140 | CS1081 pBAD24-phoQL105D | This work |
| CS1402 | CS1081 pBAD24-phoQT124N | This work |
| CS1403 | CS1081 pBAD24-phoQV126E | This work |
| CS1404 | CS1081 pBAD24-phoQT129I | This work |
| CS1405 | CS1081 pBAD24-phoQT131P | This work |
| CS1406 | CS1081 pBAD24-phoQL132P | This work |
| KH28 | CS1081 pBAD24-phoQL133C | This work |
| CS1407 | CS1081 pBAD24-phoQD150G | This work |
| CS1408 | CS1081 pBAD24-phoQA153P | This work |
| CS1409 | CS1081 pBAD24-phoQM155V | This work |
| CS1410 | CS1081 pBAD24-phoQV178D | This work |
| CS1374 | CS1081 pBAD24-phoQW104C | This work |
| CS1386 | CS1081 pBAD24-phoQA128C | This work |
| CS1382 | CS1081 pBAD24-phoQW104C-A128C | This work |
| KH48 | CS1081 pBAD24-phoQW104S | This work |
| KH49 | CS1081 pBAD24-phoQA128S | This work |
| KH50 | CS1081 pBAD24-phoQW104S A128S | This work |
| CS1101 | BL21 pET11a-phoQ 45-190-(His)6 | Bader et al., 2005 |
| KH85 | NEB SHuffle T7 express pET11a-phoQW104C-A128C 45-190-(His)6 | This work |
| KH23 | phoQ::tetRA | This work |
| KH163 | phoQW104C-A128C | This work |
| CS1350 | ΔphoQ | Prost et al., 2008 |
| KH127 | phoQ phoN105::TnphoA | This work |
| KH130 | phoQW104C-A128C phoN105::TnphoA | This work |
| KH111 | CS093 pWSK129Kan | This work |
| KH112 | CS093 pWSK29Amp | This work |
| KH113 | phoQW104C-A129C pWSK29Amp | This work |
| KH114 | ΔphoQ pWSK29Amp | This work |
Primer sequences used in this study
| Primer # (name) | Sequence (5′–3′) |
|---|---|
| LP135 (RM_Fwd) | CTGGTCGGCTATAGCGTAAGTTTTG |
| LP136 (RM_Rev) | CACGTATACGAACCAGCTCCACAC |
| LP178 (I88N_Fwd) | CGACCATGACGCTGAATTACGATGAAACGG |
| LP179 (I88N_Rev) | CCGTTTCATCGTAATTCAGCGTCATGGTCG |
| LP180 (Y89N_Fwd) | CCATGACGCTGATTAACGATGAAACGGGC |
| LP181 (Y89N_Rev) | GCCCGTTTCATCGTTAATCAGCGTCATGG |
| KH81 (I102C_Fwd) | GACGCAGCGCAACTGTCCCTGGCTGATTAAAAG |
| KH82 (I102C_Rev) | CTTTTAATCAGCCAGGGACAGTTGCGCTGCGTC |
| LP184 (T124N_Fwd) | CTTCCATGAAATTGAAAACAACGTAGACGCCACC |
| LP185 (T124N_Rev) | GGTGGCGTCTACGTTGTTTTCAATTTCATGGAAG |
| LP186 (V126E_Fwd) | GAAATTGAAACCAACGAAGACGCCACCAGCAC |
| LP187 (V126E_Rev) | GTGCTGGTGGCGTCTTCGTTGGTTTCAATTTC |
| LP188 (T129I_Fwd) | CAACGTAGACGCCATCAGCACGCTGTTG |
| LP189 (T129I_Rev) | CAACAGCGTGCTGATGGCGTCTACGTTG |
| KH192 (L105D_Fwd) | GCGCAACATTCCCTGGGATATTAAAAGCATTCAAC |
| KH193 (L105D_Rev) | GTTGAATGCTTTTAATATCCCAGGGAATGTTGCGC |
| LP190 (L131P_Fwd) | CAACGTAGACGCCACCAGCCCACTGTTGAGCGAAGACCATTC |
| LP191 (L131P_Rev) | GAATGGTCTTCGCTCAACAGTGGGCTGGTGGCGTCTACGTTG |
| LP192 (L132P_Fwd) | GACGCCACCAGCACGCCATTGAGCGAAGACCATTC |
| LP193 (L132P_Rev) | GAATGGTCTTCGCTCAATGGCGTGCTGGTGGCGTC |
| KH85 (L133C_Fwd) | CACCAGCACGCTGTGTAGCGAAGACCATTC |
| KH86 (L133C_Rev) | GAATGGTCTTCGCTACACAGCGTGCTGGTG |
| LP194 (D150G_Fwd) | GTACGTGAAGATGGCGATGATGCCGAG |
| LP195 (D150G_Rev) | CTCGGCATCATCGCCATCTTCACGTAC |
| LP196 (A153P_Fwd) | GAAGATGACGATGATCCCGAGATGACCCAC |
| LP197 (A153_Rev) | GTGGGTCATCTCGGGATCATCGTCATCTTC |
| LP198 (M155V_Fwd) | GACGATGATGCCGAGGTAACCCACTCGGTAGC |
| LP199 (M155V_Rev) | GCTACCGAGTGGGTTACCTCGGCATCATCGTC |
| LP200 (V178D_Fwd) | CCATCGTGGTGGACGATACCATTCCG |
| LP201 (V178D_Rev) | CGGAATGGTATCGTCCACCACGATGG |
| LP141 (W104C_Fwd) | GCGCAACATTCCCTGCCTGATTAAAAGCATTC |
| LP142 (W104C_Rev) | GAATGCTTTTAATCAGGCAGGGAATGTTGCGC |
| LP145 (A128C_Fwd) | GAAACCAACGTAGACTGCACCAGCACGCTGTTG |
| LP146 (A128C_Rev) | CAACAGCGTGCTGGTGCAGTCTACGTTGGTTTC |
| KH61 (W104S_Fwd) | CAGCGCAACATTCCCAGCCTGATTAAAAGCATTC |
| KH62 (W104S_Rev) | GAATGCTTTTAATCAGGCTGGGAATGTTGCGCTG |
| KH63 (A128S_Fwd) | GAAACCAACGTAGACAGCACCAGCACGCTGTTG |
| KH64 (A128S_Rev) | CAACAGCGTGCTGGTGCTGTCTACGTTGGTTTC |
| LP164 (T48C_Fwd) | GTAAGTTTTGATAAAACCTGCTTTCGTTTGCTGCGCG |
| LP165 (T48C_Rev) | CGCGCAGCAAACGAAAGCAGGTTTTATCAAAACTTAC |
| LP168 (K186C_Fwd) | CCATTCCGATAGAACTATGCCGCTCCTATATGGTGTG |
| LP169 (K186C_Rev) | CACACCATATAGGAGCGGCATAGTTCTATCGGAATGG |
| KH35 (T48S_Fwd) | GTTTTGATAAAACCAGCTTTCGGCTGCG |
| KH36 (T48S_Rev) | CGCAGCAAACGAAAGCTGGTTTTATCAAAA |
| KH39 (K186S_Fwd) | CATTCCGATAGAACTAAGTCGCTCCTATATGGTG |
| KH40 (K186S_Rev) | CACCATATAGGAGCGACTTAGTTCTATCGGAATG |
| KH45 (PhoQ_tetRA_knock-in_Fwd) | GAATAAATTTGCTCGCCATTTTCTGCCGCTGTCGCTGCGGTTAAGACCCACTTTCACA |
| KH46 (PhoQ_tetRA_knock-in_Rev) | CCTCTTTCTGTGTGGGATGCTGTCGGCCAAAAACGACCTCCTAAGCACTTGTCTCCTG |
| KH93 (ST-PhoQ_N-term_Fwd) | ATGAATAAATTTGCTCGCCATTTTC |
| KH94 (ST-PhoQ_N-term_Rev) | TTATTCCTCTTTCTGTGTGGG |
| KH265 (ST-rpoD_Fwd_qRT) | GGGATCAACCAGGTTCAATG |
| KH266 (ST-rpoD_Rev_qRT) | GGACAAACGAGCCTCTTCAG |
| KH269 (ST-pagD_Fwd_qRT) | GTTCAGGCCATTGTTCTGGT |
| KH270 (ST-pagD_Rev_qRT) | TAATCTGCCTGGCTTGCTTT |
| KH273 (ST-pagO_Fwd_qRT) | CGGGCTTAACTATCGCAATC |
| KH274 (ST-pagO_Rev_qRT) | CAGCAGAAATAAGCGCAGTG |
| KH275 (ST-phoP_Fwd_qRT) | TGCCAGGGAAGCTGATTACT |
| KH276 (ST-phoP_Rev_qRT) | CAGCGGCGTATTAAGGAAAG |
| KH277 (ST-phoN_Fwd_qRT) | CCGGCTTACCGCTATGATAA |
| KH278 (ST-phoN_Rev_qRT) | CGCTTACATCTGCATCCTCA |





