H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast
Figures
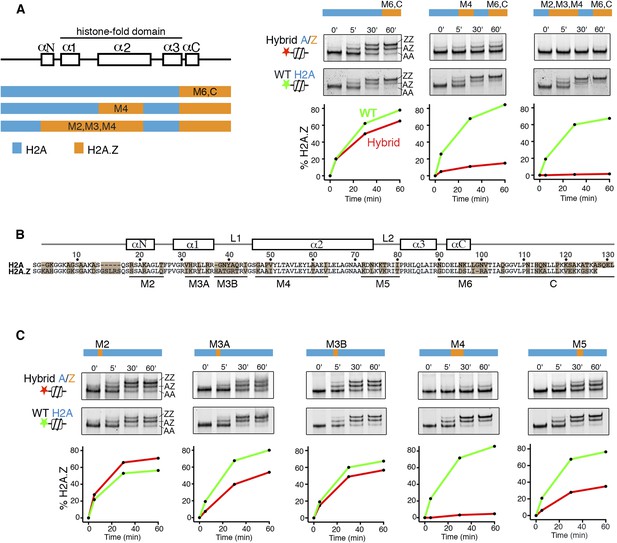
H2A histone regions in the canonical nucleosome that activate H2A.Z replacement by SWR1.
(A) Left: H2A/H2A.Z hybrid histones used for reconstituting nucleosomes. Right: Histone H2A.Z replacement assay. Hybrid (red) and WT (green) nucleosomes (2.5 nM) were incubated with SWR1 (2 nM), H2A.Z-3F-H2B (22 nM), and ATP (1 mM) for the indicated times, and nucleosomes containing zero (AA), one (AZ), or two copies (ZZ) of H2A.Z-3F were resolved by 6% native PAGE. Top: EMSA (electrophoretic mobility shift assay) and fluorescence imaging. Bottom: H2A.Z incorporation curves. (B) Sequence alignment of histone H2A and H2A.Z from budding yeast. (C) Histone replacement assay as above, with hybrid nucleosomes containing fine H2A/H2A.Z interchanges. Bottom: H2A.Z incorporation curves.
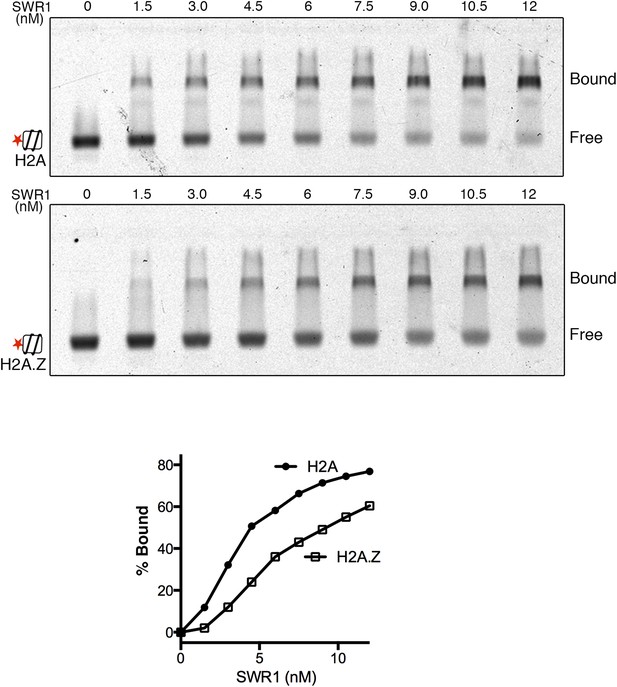
SWR1 binding to nucleosome core particles containing H2A or H2A.Z histone.
EMSA shows SWR1 binding to Alexa 647-labeled H2A- and H2A.Z-nucleosome core particles (1 nM). Free and bound complexes are resolved on 1.3% agarose gel. Bottom: binding curves for H2A- and H2A.Z-nucleosome core particles.
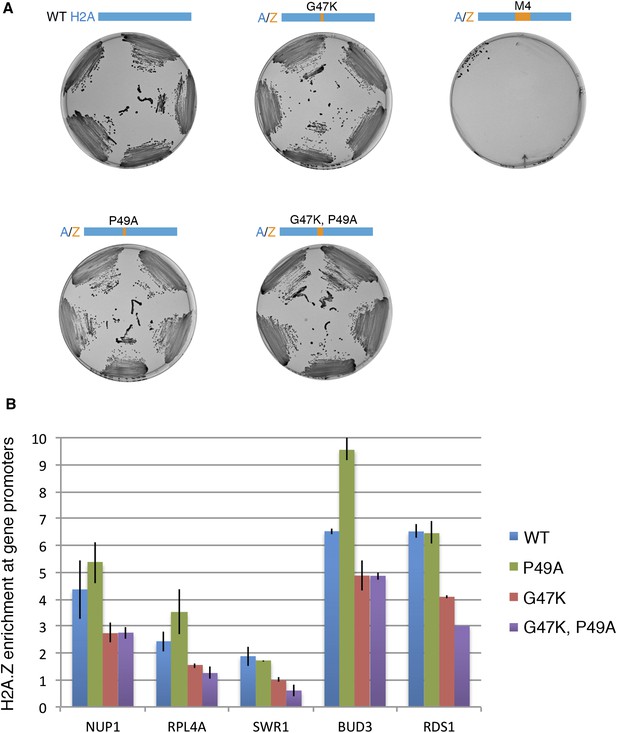
Effect of H2A M4 on H2A.Z enrichment at gene promoters.
(A) Viability of H2A mutants. In the plasmid shuffle experiment, yeast cells containing episomal copies of WT HTA1/HTB1 (under URA selection) and a second plasmid of WT or indicated mutants (under HIS selection) were plated on CSM-His/5-FOA plates. (B) ChIP-PCR for H2A.Z-HA for WT and mutant cells. The signal from gene promoters was normalized to a sub-telomeric region on chromosome 6. Error bars are standard deviations from technical repeat.
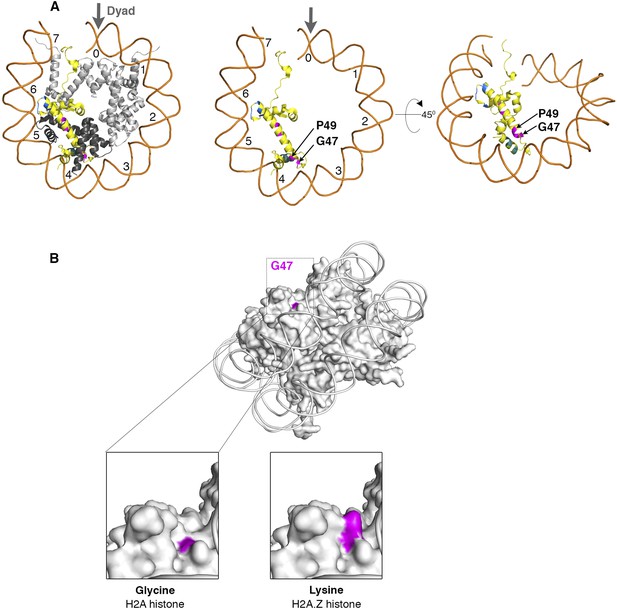
Nucleosome structure showing critical H2A residues that effect SWR1 activity.
(A) Left: The yeast nucleosome crystal structure 1ID3 in Protein Data Bank was modeled to show histones on one face of nucleosome. Histone H2A is yellow, H2B is black and H3, H4 are gray. The domains of H2A that affect SWR1 activity-M3A (cyan), M4 (magenta), and M5 (blue) are marked. Center and right: Buried residues of histone H2A are shown by removing other histones and rotating on X-axis by 45°. (B) The H2A surface residue G47 in 1ID3 is shown in magenta. Bottom left: Zoom-in view shows that G47 is at the bottom of a cleft. Bottom right: Replacing Glycine for Lysine in H2A.Z histone shows the long side-chain of Lysine filling the cleft.
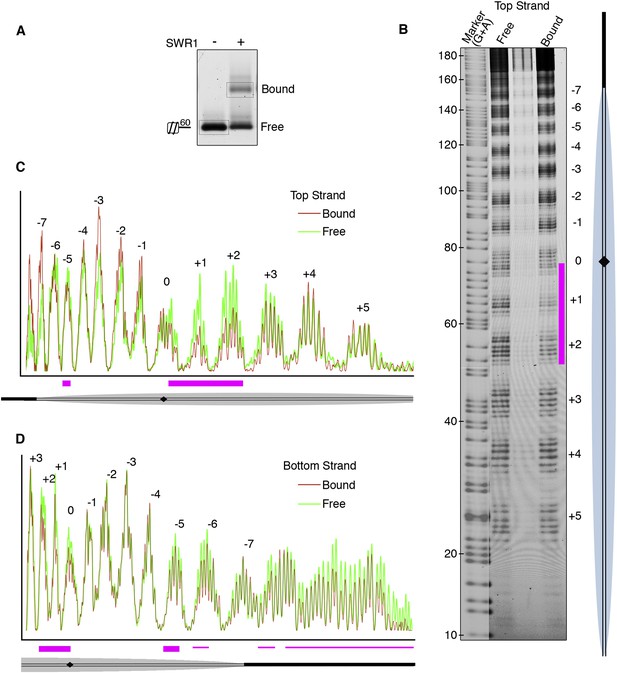
Hydroxyl radical footprinting of SWR1 on nucleosomal DNA.
(A) EMSA (1.3% agarose gel) shows SWR1 (12 pmole; 240 nM) binding to a fluorescent end-labeled asymmetric 60 bp long linker nucleosome (7.4 pmole; 150 nM) after reaction with hydroxyl radical. (B) DNA samples resolved on 8% sequencing gel. Top strand is fluorescently labeled. The strongest protected area is shown as magenta bar. (C) Top strand intensity plots of free (green) and bound (red) nucleosome corresponding to B. (D) Bottom strand intensity plots for free and bound samples were normalized to signals at +2 and +3 SHL from dyad axis.
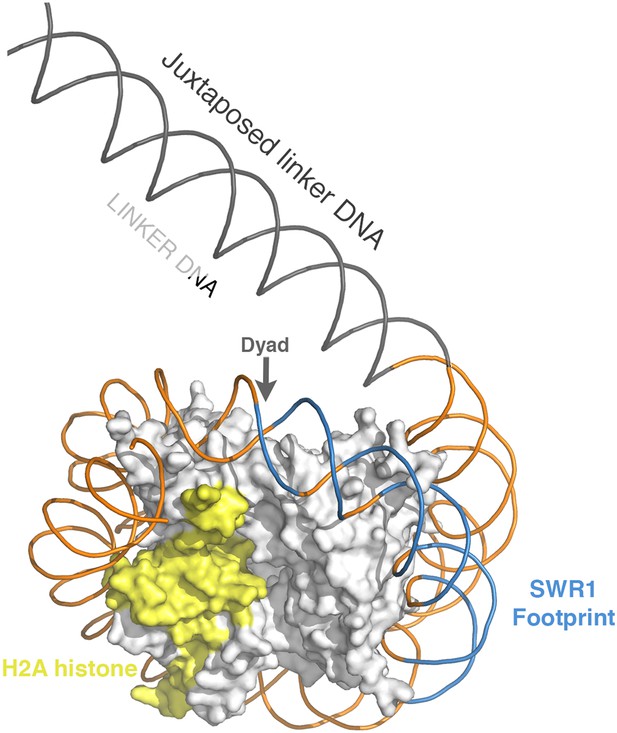
Position of SWR1 footprint on linker-distal face of nucleosome.
The 601 DNA-containing nucleosome structure PDB 3MVD was modeled to highlight the position of the SWR1 footprint in blue on the linker-distal side of the dyad axis. The H2A on the linker-distal face is in yellow.
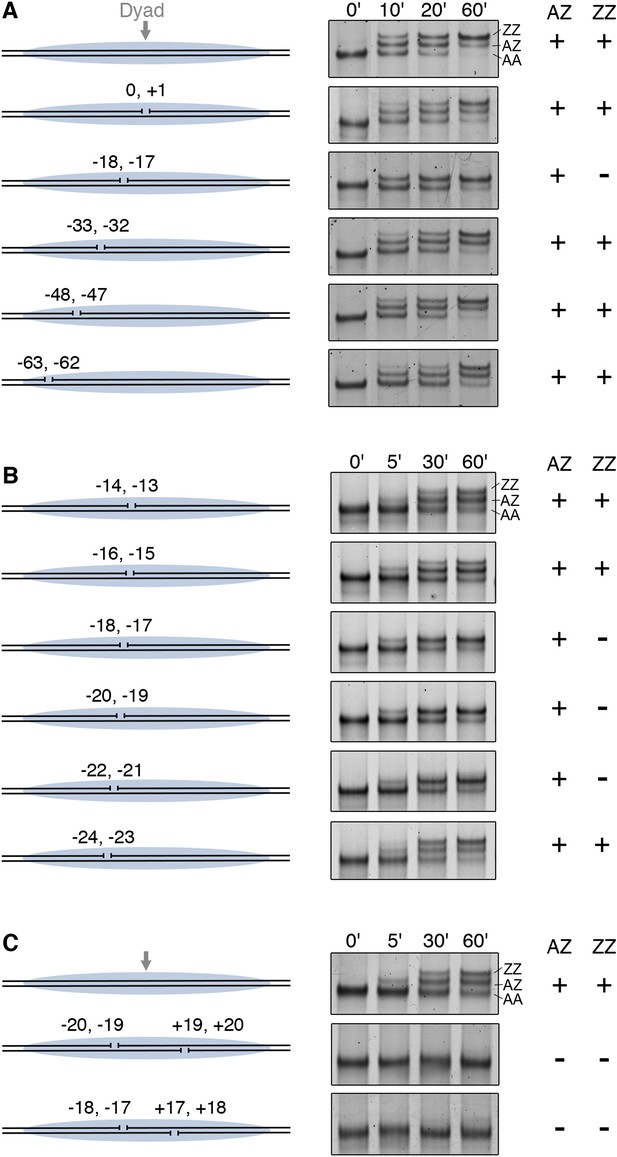
DNA gaps block SWR1 activity when positioned 17–22 bp on either side from dyad.
All nucleosomes have a 20 bp linker DNA at both ends, and a two-nucleotide gap introduced at indicated positions. EMSA (6% native PAGE) shows the H2A.Z replacement reaction, terminated at the indicated times, using fluorescently labeled nucleosomes (4 nM), SWR1 (2 nM), and H2A.Z-3F-H2B dimer (10 nM). Nucleosome products with 0, 1, and 2 H2A.Z-3FLAG molecules are resolved (AA, AZ, ZZ). (A) Mapping of gap sites that block SWR1 activity. Left: Design of WT and gap nucleosomes. Right: (+) and (−) denote presence and absence of the AZ or ZZ species. (B) Fine mapping of the gap-sensitive region near two turns from nucleosome dyad. (C) Gaps within the sensitive region on both sides of nucleosome completely block SWR1 activity.
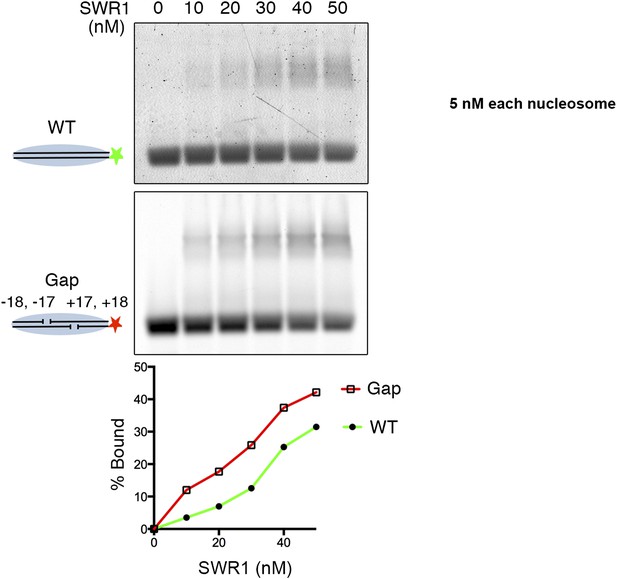
SWR1 binding to nucleosome core particle with gaps on both sides of dyad.
Fluorescently labeled WT (green) and Gap (red) nucleosome core particles (5 nM) were mixed with indicated amounts of SWR1. Free and SWR1-bound nucleosome core particles were resolved on a 1.3% agarose gel. Bottom: binding curves for WT and Gap particles.
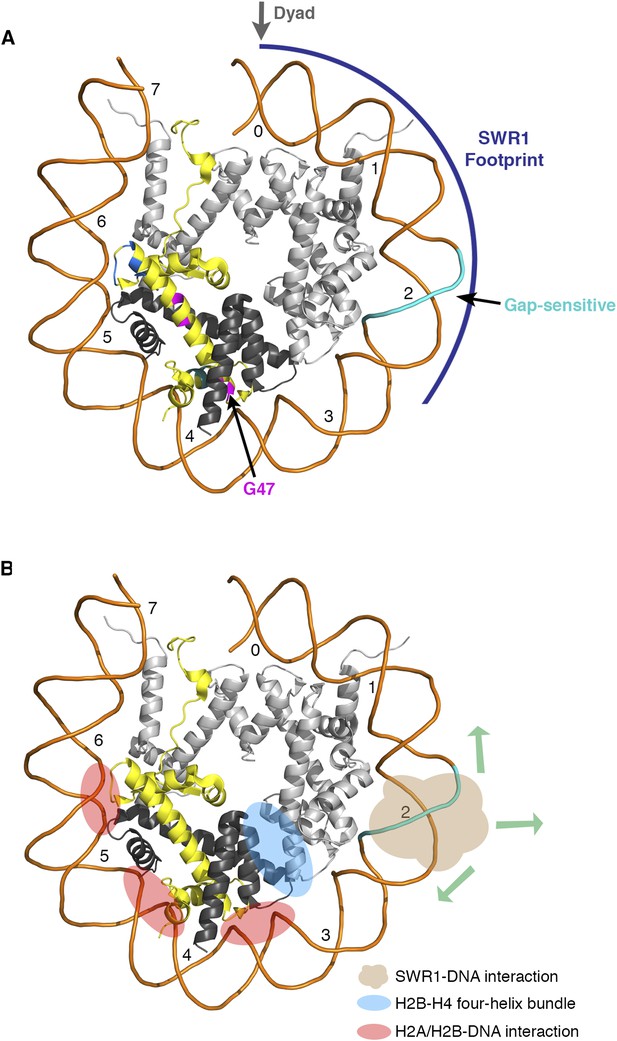
Nucleosomal histone and DNA elements critical for SWR1 activity and model for SWR1-mediated H2A-H2B displacement.
(A) Yeast nucleosome structure PDB 1ID3 was modeled to show one face of the nucleosome and the histone-fold elements that are critical for SWR1 activation. The SWR1 footprint is shown in blue. The gap-sensitive region, 17–22 nt from dyad, is shown in cyan. Residues of H2A that affect SWR1 activity are shown in magenta. (B) Nucleosome model showing histone-DNA and histone–histone interactions that hold H2A-H2B within the nucleosome. Also shown is the gap-sensitive region, where SWR1 interacts with nucleosome DNA leading to eviction of H2A/H2B and concomitant deposition of H2A.Z/H2B.
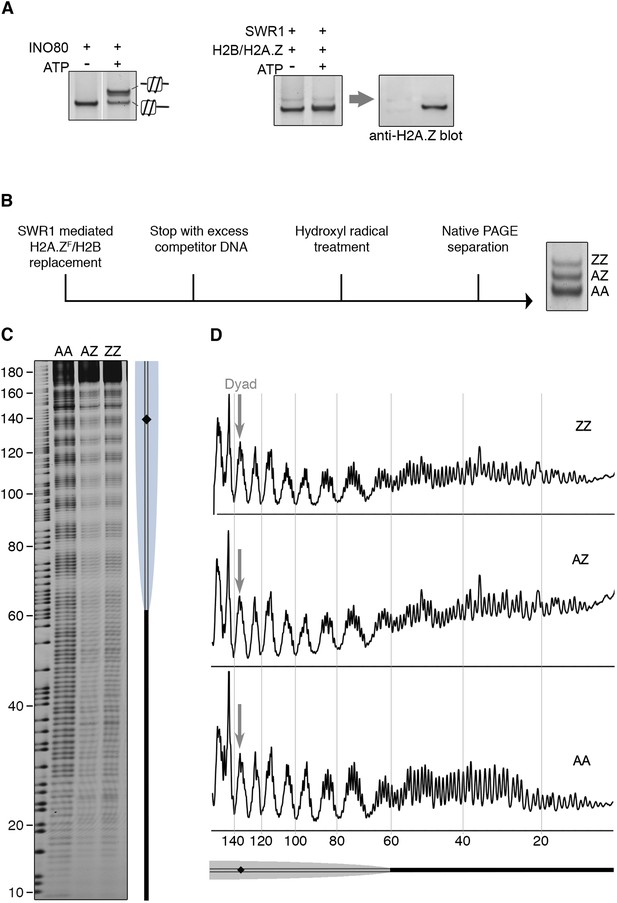
SWR1 mediates histone exchange without net change of nucleosome position.
(A) Left: EMSA (6% native PAGE) shows INO80-mediated nucleosome sliding. An asymmetrically positioned 601 nucleosome with a 43 bp and 0 bp DNA linker was used for the sliding assay. Right: SWR1-mediated incorporation of H2A.Z-H2B dimer (without 3FLAG epitope tag). Incorporation of H2A.Z in nucleosome was confirmed by immunoblotting with anti-H2A.Z antibody. (B) Hydroxyl radical footprinting strategy. A canonical nucleosome with 60 bp and 0 bp linker DNA and fluorescence end-label (bottom strand) was used as substrate for histone replacement, followed by hydroxyl radical treatment and separation by 6% native PAGE. (C) Recovered DNA from gel slices containing AA, AZ, and ZZ states was analyzed on DNA sequencing gels. (D) Intensity plots for AA, AZ, and ZZ nucleosomes.
Additional files
-
Supplementary file 1
Genotype of strains used in this study, related to Figure 1.
- https://doi.org/10.7554/eLife.06845.013





