The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix
Figures
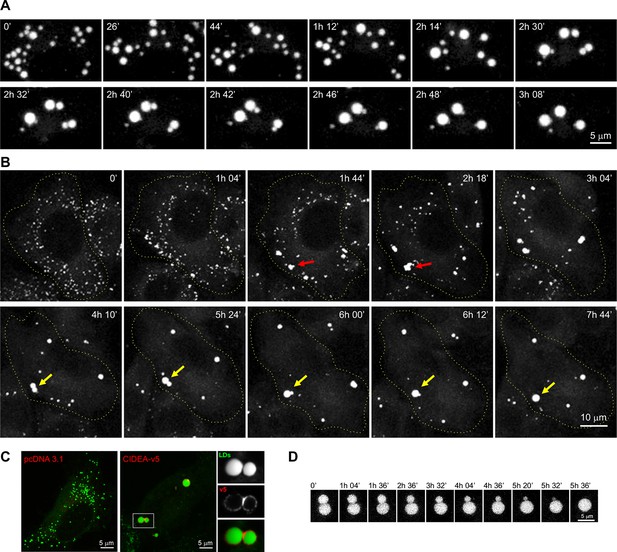
CIDEA promotes lipid droplet (LD) enlargement by transference of lipids.
(A) Live imaging of the LD dynamics during the differentiation of a brown pre-adipocyte, showing the characteristic CIDE-triggered LD enlargement, characterized by the progressive transference of lipids from a donor to an acceptor LD until it is completely absorbed. (B) Live imaging of the LD dynamics in an undifferentiated 3T3-L1 cell 6 hr after infection with adenoviral particles carrying the Cidea gene. Red arrows highlight the transient formation of irregularly shaped LD clusters, while yellow arrows mark the fusion of two droplets by transference of lipids. (C) CIDEA-v5 expression in Hela cells induces LD enlargement. An enrichment in CIDEA-v5 (red) can be observed in the contact site between two LDs (green). (D) Detail of LD fusion by slow transference of lipids in a 3T3-L1 cell stably expressing CIDEA-v5.
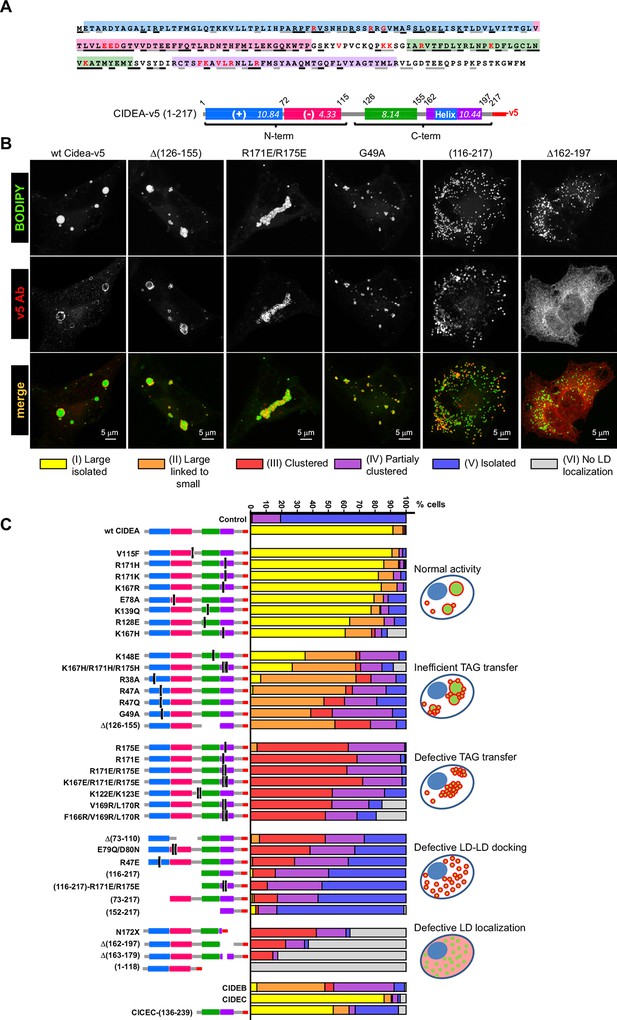
Mapping the functional domains of CIDEA.
(A) Amino acid (aa) sequence of murine CIDEA highlighting the residues conserved in either CIDEB or CIDEC (grey underline), or in both proteins (black underline). The substituted aa in mutant constructs appear in red, and a positively charged sequence necessary for the TAG transfer step is encircled in orange. Four highly conserved regions are defined and symbolized by colour boxes in a linear representation of CIDEA-v5. The theoretical isoelectric point of each fragment is indicated inside the boxes. (B) Representative images of the different phenotypes observed in Hela cells overexpressing mutated forms of CIDEA-v5 24 hr after transfection cells were treated with oleic acid and incubated for further 18 hr prior to fixation. Cells were classified into six major phenotypes. Cells expressing fully active forms of CIDEA had few and large LDs (Type I). In some mutants, the large LDs remained attached to many small LDs, indicating that lipid transfer was inefficient or inactive for some LDs (Type II). When CIDEA alterations blocked the lipid transfer process, the LDs remained small and grouped in a few large clusters (Type III). If this was accompanied by inefficient LD–LD docking, the cells contained a number of small LD clusters combined with isolated LDs (Type IV). The CIDEA forms that could not stabilize LD–LD interactions displayed a phenotype similar to the mock transfected cells, with most of the LDs dispersed through the cytoplasm (Type V). Finally, some CIDEA constructs were unable to target the LDs, indicating an alteration of the LD-binding domain (Type VI). (C) Morphologic distribution of cells expressing each of the studied CIDE constructs. The phenotypic distribution was performed in a minimum of three independent experiments for every construct (n>50 cells).

Alignment of amino acid sequences of CIDEA, CIDEB, and CIDEC.
Clustal format alignment of murine CIDEA, CIDEB, and CIDEC, performed using T-COFFEE (www.tcoffee.org). Amino acid sequences were obtained from http://ensembl.org. "*" means residues are identical in all sequences in the alignment; ":" means conserved substitutions have been observed, "." means that semi-conserved substitutions are observed. Four highly conserved regions are defined by colour boxes.
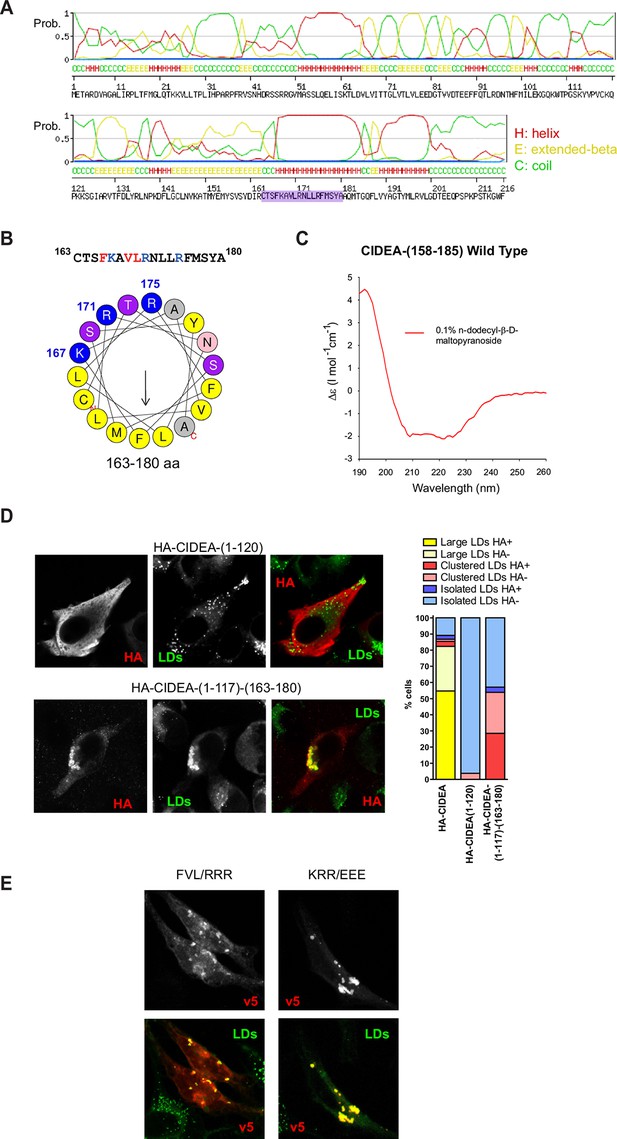
CIDEA targets the LD monolayer through a cationic amphipathic helix.
(A) Secondary structure of CIDEA predicted by SWISS-MODEL server. (B) Helical wheel representation of the putative amphipathic α-helix (163–180) generated at http://heliquest.ipmc.cnrs.fr/. (C) Circular dichroism (CD) spectra of a 28-aa peptide corresponding to the 158–185 sequence of CIDEA (41 μM) solubilized in 50 mM potassium phosphate, pH 6.2 plus 0.1% n-dodecyl-β-D-maltopyranoside. (D) A Hela cell expressing HA-CIDEA-(1–120)-v5 (red) or HA-CIDEA-(1–117)-(163–180) (red), showing the inclusion of aas 163–180 enhances LD localization and the ability to promote LD docking. The phenotypic distribution was performed in a minimum of three independent experiments for every construct (n>50 cells). HA signal in LDs was only detected in a proportion of the cells where HA-CIDEA constructs had induced LD enlargement or clustering, possibly due to the formation of CIDEA complexes reducing antibody accessibility to the HA epitope at the N-term. (E) A Hela cell expressing CIDEA-(F166R/V169R/L170R)-v5 (red) showing aa substitutions to compromise amphipathicity of the helix disrupt LD targeting, and a Hela cell expressing CIDEA-(K167E/R171E/R175/E)-v5 (red) showing amino acid (aa) substitutions to invert the charge of the helix but maintaining amphipathicity retains predominantly LD localization.
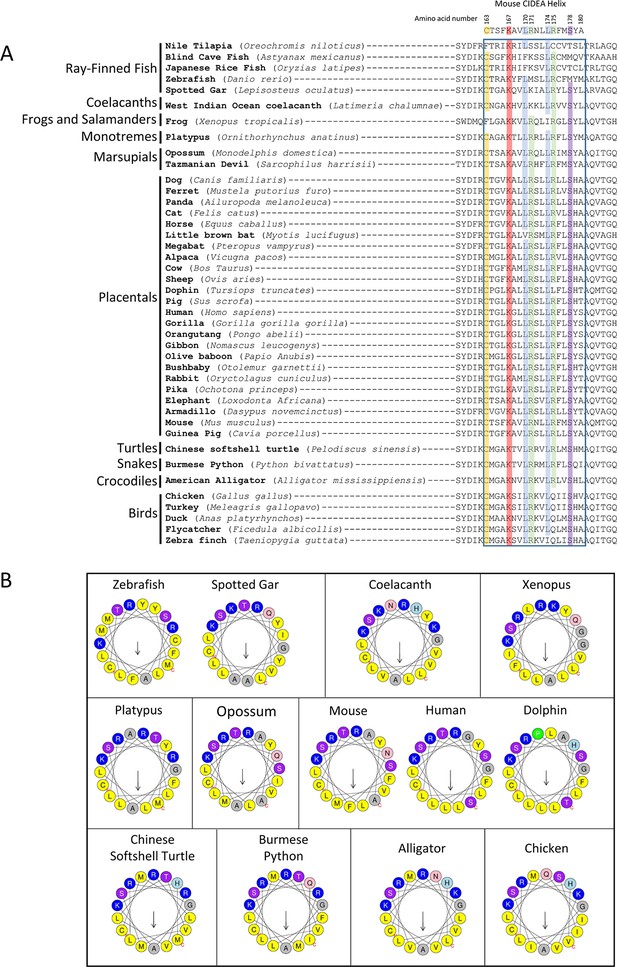
Conservation of amino acids (aas) for CIDEA amphipathic helix across vertebrate species.
(A) Alignment of aa sequences for CIDEA over a range of vertebrate species corresponding to the amphipathic α-helix (murine aas 163–180). Amino acid sequences were obtained from http://ensembl.org and initially aligned using WebPRANK at http://ebi.ac.uk. Sequences were then grouped based on species phylogeny. (B) Helical wheel representation of the CIDEA putative amphipathic α-helix for indicated species (corresponding to murine aas 163–180) generated at http://heliquest.ipmc.cnrs.fr/.
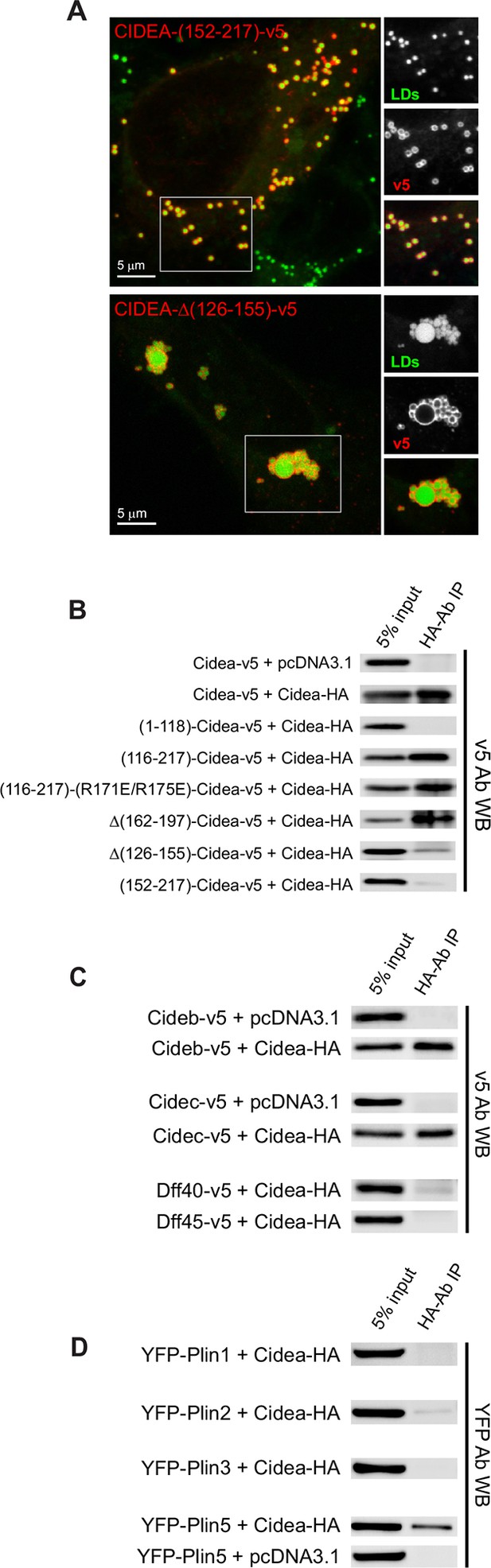
Liquid droplet (LD)–LD docking and CIDEA interactions.
(A) A Hela cell expressing CIDEA-(152–217)-v5 showing normal recruitment to LDs, but no LD docking. A Hela cell expressing CIDEA-△(126–155)-v5 showing normal LD–LD docking but inefficient LD enlargement as revealed by the presence of clusters of small and large LDs. Representative images are shown of experiments performed in a minimum of three independent experiments for every construct (n>50 cells). (B) Co-immunoprecipitation (co-IP) assays between CIDEA-HA and different CIDEA-v5 constructs. The observed CIDEA–CIDEA interaction was driven by the C-term domain and required the presence of the 126–155 aa sequence. (C, D) Co-IP assays showing CIDEA interactions with CIDEB, CIDEC, and PLIN5. Each co-IP assay was performed at least in triplicate, producing similar results in each experiment.
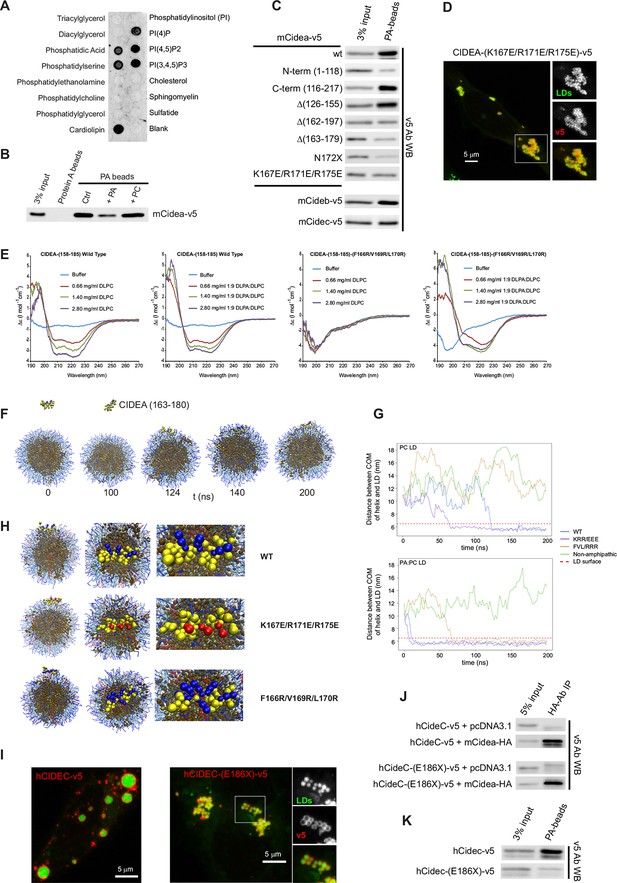
CIDEA is a phosphatidic acid (PA)-binding protein.
(A) Lipid strip assay showing the affinity of CIDEA-v5 for certain anionic phospholipids. (B) Interaction of CIDEA-v5 with PA beads. Binding was reduced by pre-incubation of the lysate with soluble PA, but not phosphatidylcholine (PC). (C) The affinity for PA beads was highly reduced in CIDEA-v5 constructs with alterations in its C-term hydrophobic and basic region (162–197). (D) CIDEA-(K167E/R171E/R175E)-v5 localizes to LDs and induces their clustering but cannot promote their enlargement by lipid transfer. Representative images are shown of experiments performed in a minimum of three independent experiments for every construct (n>50 cells). (E) Circular dichroism spectra of the synthetic wild type (wt) or mutant (F166R/V169R/L170R) CIDEA peptides encompassing aas 158–185 solubilized in 25 mM sodium phosphate (pH 7.2) at concentrations of 70 μM (wt) and 47 μM (mutant). Peptide samples were prepared in the absence and presence of increasing amounts of DPLC or DLPC:DLPA (9:1 molar ratio). (F–H) Coarse-grained molecular dynamics (CG-MD) simulations of peptide interactions with LDs (PC: hydrophobic chains, transparent blue, polar heads, opaque blue; TAG: hydrophobic chains, dark brown, glycerol chain, light brown; PA: hydrophobic chains, transparent red, polar heads, opaque red; peptides: yellow, with cationic aa in blue and anionic in red). (F) Selected time points of the wt helix simulation with PC-LDs. At 124 ns the helix initiates the contact through its hydrophobic face, being rapidly embedded in the phospholipid monolayer. TAG molecules can abandon the neutral lipid core and are integrated in the hydrophobic region of the phospholipid monolayer. (G) Distance between the peptide and LD centre of mass (COM) versus time for the different helices with a PC-LD and a PC:PA-LD. The dashed line represents the approximate location of LD phospholipid head groups. (H) Different views of the configuration of the LD helix at the end of the simulations. Interaction between the polar head of PA and the helix can be observed for the wt and F166R/V169R/L170R but not K167E/R171E/R175E. (I–K) Comparison of full-length hCIDEC-v5 and the lipodystrophy-associated truncation hCIDEC-(E186X)-v5, including LD localization and morphology (I), co-IP with CIDEA-HA (J), and affinity for PA beads (K). Each co-IP, PA-binding assay, and lipid strip assay was performed at least in triplicate, producing similar results in each experiment.
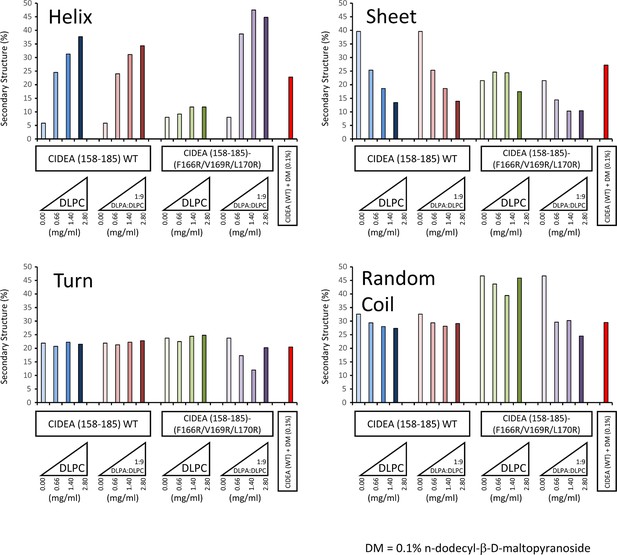
Secondary structure determination of CIDEA amino acids 158–185 (wild type and F166R/V169R/L170R) by CDPro DATABASE 4 (43 soluble proteins) using the CONTINLL program.
https://doi.org/10.7554/eLife.07485.012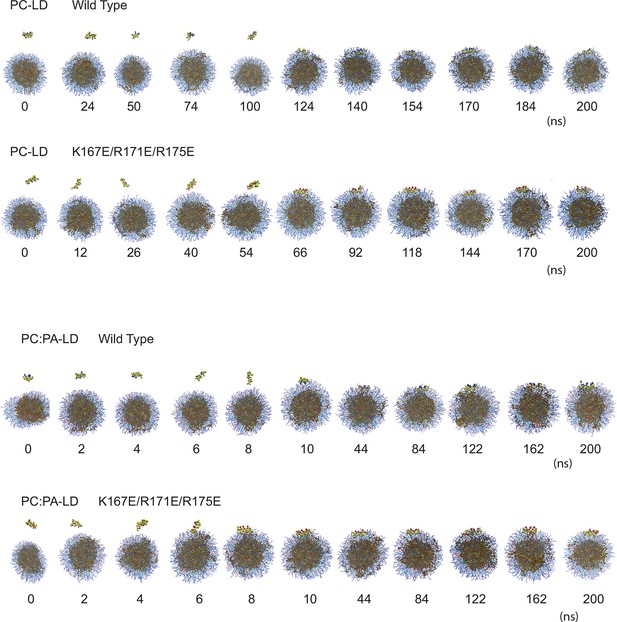
Computational prediction of the amphipathic helix and LD interactions.
CG-MD simulations of peptide interactions with LDs (PC: hydrophobic chains, transparent blue, polar heads, opaque blue; TAG: hydrophobic chains, dark brown, glycerol chain, light brown; PA: hydrophobic chains, transparent red, polar heads, opaque red; peptides: yellow, with cationic aa in blue and anionic in red). Selected time points of the wt helix and K167E/R171E/R175E helix simulation with PC-LDs or PC:PA-LDs are indicated.
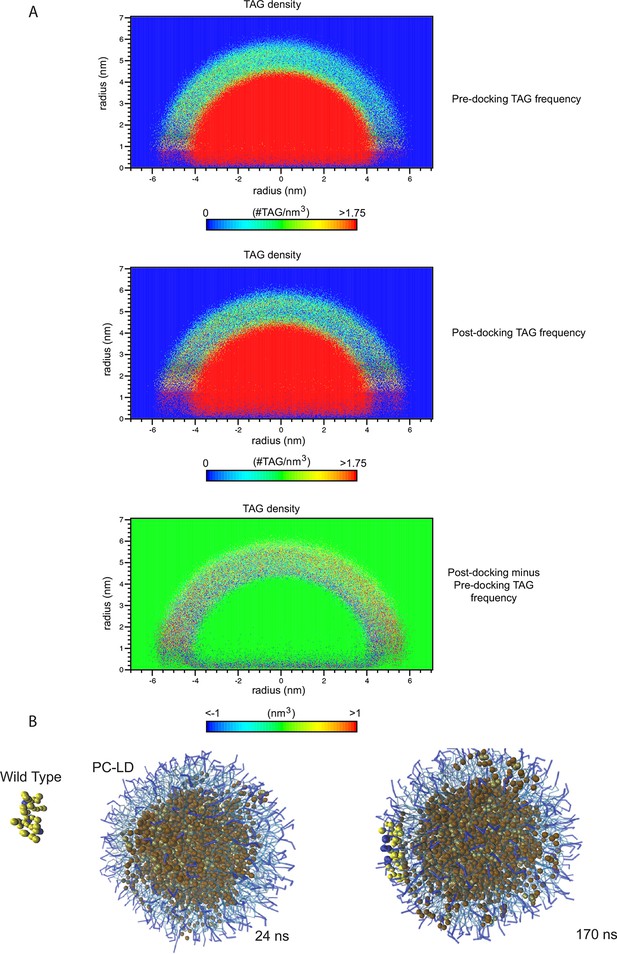
TAG infiltration into the phospholipid monolayer.
(A) 2D number density maps of TAG molecules along the z direction from the CG-MD simulations. TAG frequency was measured over 76 ns before and after the docking of the wt helix to the membrane. The differential distribution before and after docking reveals an increase in the presence of TAG molecules in the membrane, which was revealed by the red spots in the outer arch and blue in the inner. (B) CG-MD simulations of peptide interactions with LDs (PC: hydrophobic chains, transparent blue, polar heads, opaque blue; TAG: hydrophobic chains, dark brown, glycerol chain, light brown; PA: hydrophobic chains, transparent red, polar heads, opaque red; peptides: yellow, with cationic aa in blue and anionic in red). Selected time points of the wt helix with PC-LD show TAG molecules in the LD membrane were increased after CIDEA peptide docking (170 vs 24 ns).
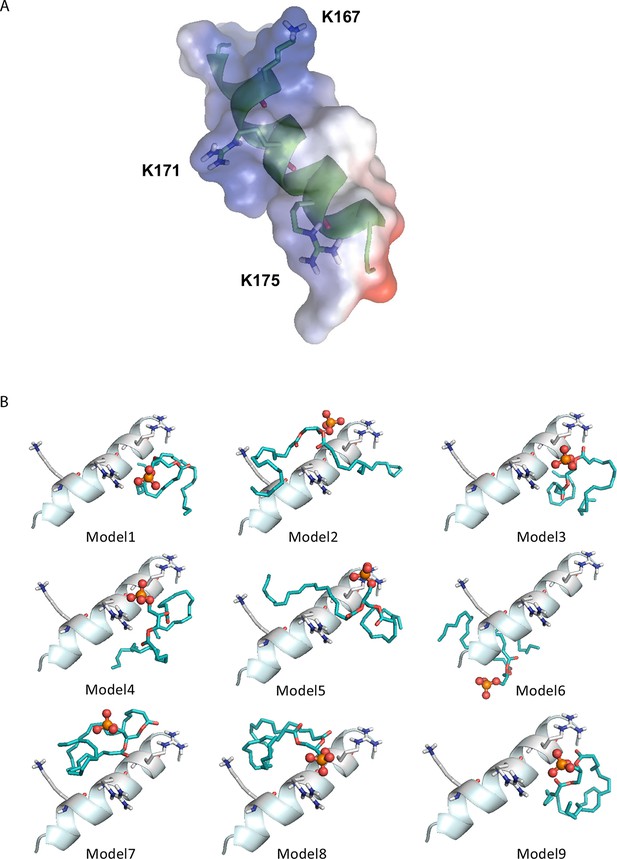
Computational prediction of PA docking to the amphipathic helix structure of CIDEA.
(A) Charge-smoothed potential of the predicted putative amphipathic helix of murine CIDEA (aas 163–180). Stick representation has been employed to highlight the positively charged residues K167, R171, and R175. (B) The nine top-ranked models from molecular docking of PA and the amphipathic helix using Autodock Vina. Interaction of the PA phosphate group with R171 and R175 is observed in eight of them, while the hydrophobic chains of the phospholipid can adopt multiple configurations.
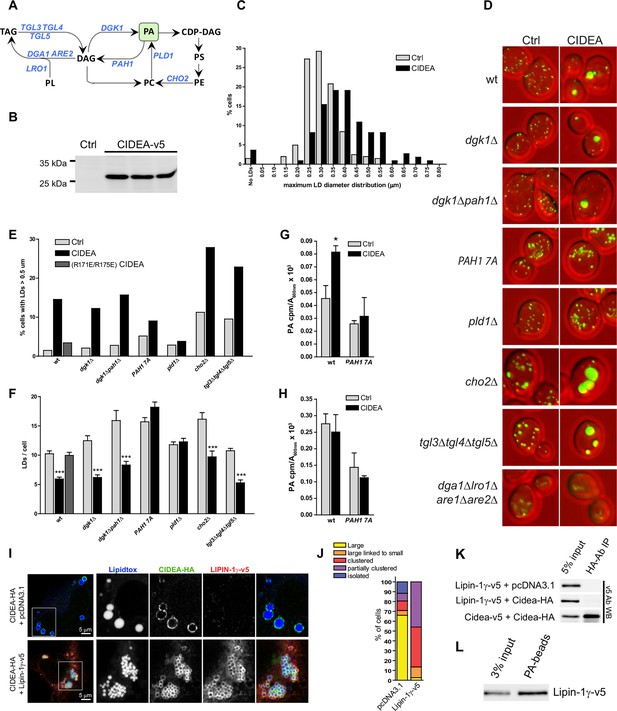
CIDEA is functional in yeast and requires PA.
(A) Pathway showing the reactions catalyzed by the enzymes altered in the studied yeast strains. (B) Stable expression of mCIDEA-v5 in three transformed yeast clones. (C) Frequency distribution of the diameter of the largest LD per cell. (D) LD staining in the studied yeast strains transformed with pRS316-CYC1p-Cidea or the empty vector. (E, F) Quantification of LD size and number per cell in randomly acquired images (100–200 cells/condition). CIDEA activity in yeast was measured by its ability to increase the percentage of cells with supersized LDs (E) and reduce the total number of LDs per cell (F). (G, H) Effect of CIDEA and PAH1-7A expression in the cellular levels of PA (G) and its synthesis rate (H). Three different yeast clones per condition were analysed, and results are shown as the mean ± SEM. One-way ANOVA with Bonferroni post-test was performed to determine significant differences due to the presence of CIDEA (*p<0.05; ***p<0.001). (I–L) Coexpression of hLIPIN-1γ-v5 and CIDEA-HA in Hela cells. (I) Representative immunofluorescence images showing LD staining (blue) in Hela cells expressing CIDEA-HA (green) in the presence or absence of hLIPIN-1γ-v5 (red). Twenty-four hours after overexpression of hLIPIN-1γ-v5, cells were transfected with pcDNA3.1/Cidea-HA and incubated for a further 24 hr. (J) Phenotypic distribution in randomly selected cells (n>50) showing the average values for three independent experiments. (K) Co-IP assay in lysates of transfected Hela cells. (L) PA beads binding assay for hLIPIN-1γ-v5. Each co-IP and PA-binding assay was performed at least in triplicate, producing similar results in each experiment.
-
Figure 6—source data 1
List of yeast strains used in this study.
- https://doi.org/10.7554/eLife.07485.017
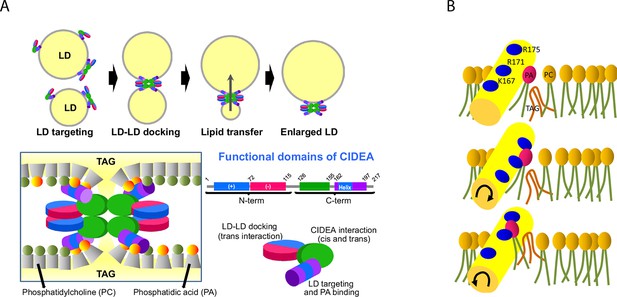
Proposed molecular mechanism.
(A) CIDEA targets the LD through its C-term amphipathic helix and once diffused to the LD surface, it forms cis CIDEA complexes by interacting through the C-term (126–155) region. When two CIDEA-containing LDs make contact, trans interactions between CIDEA molecules in each droplet can be established, which will facilitate the docking of the LDs. Both the N-term and the C-term would contribute by dimerizing with their counterparts of the neighbour LD. This trans interaction will anchor the CIDEA complexes in the LD–LD contact site, promoting a local enrichment of CIDEA. The monolayers of the two LDs will be maintained at short distance by the CIDEA complex. The amphipathic helices, embedded in the hydrophobic region of the membrane, will interact with the cone-shaped PA, creating a local perturbation in the phospholipid barrier that will increase its permeability to TAG. (B) The docking of the amphipathic helix to the membrane could facilitate the integration of TAG molecules within the phospholipid hydrophobic tails. Although the helix will be stabilized with its cationic residues pointing outwards, it will interact with PA molecules in its vicinity, which could be pulled out of the monolayer by the helix molecular dynamics. This could create a transitory discontinuity in the polar barrier that will reduce the energy required to transfer the TAG molecules present in the membrane. This alteration, together with the microenvironment created by the CIDEA complex, will reduce the energy barrier necessary to transfer TAG molecules between LDs, allowing the LD growth by lipid transference.
Videos
Lipid droplet (LD) enlargement in differentiating immortalized brown adipose tissue (imBAT) cells.
Immortalized brown pre-adipocytes were induced to differentiate by incubation for 48 hr + 6 hr with the described differentiation cocktails. The cell displays the characteristic LD enlargement pattern triggered by CIDE proteins, defined by the progressive fusion by lipid transference of the pre-existing LDs.
Lipid droplet (LD) enlargement induced by CIDEA.
LD dynamics in undifferentiated 3T3-L1 cells 6 hr after infection with adenoviral particles carrying the mouse Cidea gene. After CIDEA induction, the initial individual LDs form stable contacts reflected by small irregularly shaped clusters of LDs. These interacting LDs undergo an enlargement process by lipid transference, characterized by the progressive enlargement of the acceptor LD and shrinkage of the donor LD until only a few large LDs remain in the cell.





