Neuropilin-2/Semaphorin-3F-mediated repulsion promotes inner hair cell innervation by spiral ganglion neurons
Figures
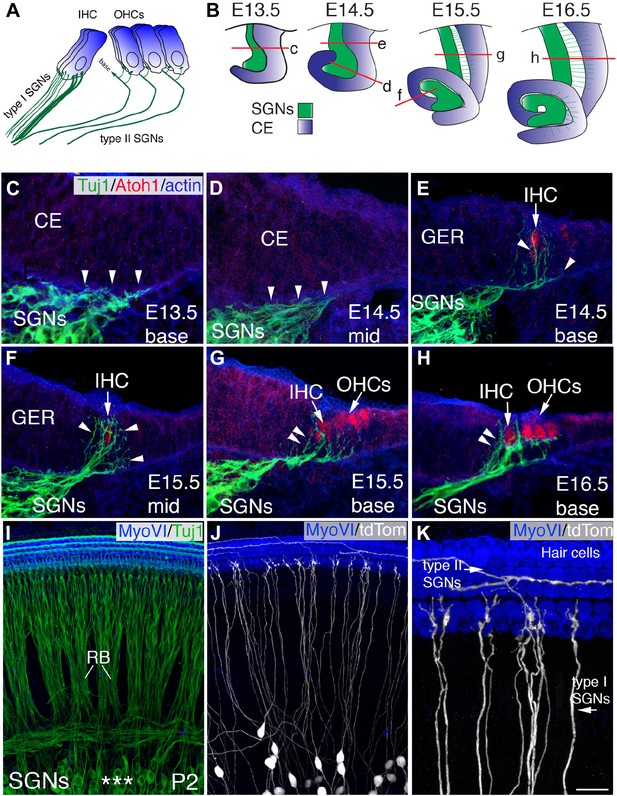
Development of SGN innervation patterns.
(A) Illustration of the innervation pattern of inner hair cells (IHCs) and outer hair cells (OHCs) by type I and II spiral ganglion neurons (SGNs). (B) Illustrations showing the morphology of the cochlear duct and spiral ganglion (SG) for the indicated cross-sections. Letters on each illustration correspond to the position of the section in the adjacent panels. ce, cochlear epithelium. (C–H) Cross-sections of the cochlea and associated SG at the indicated time points and positions. SGN processes are labeled with Tuj1 (green), hair cells (HCs) with anti-Atoh1 (red), and actin with phalloidin (blue). SGN processes do not enter the epithelium until HCs being to differentiate (E14.5 base) but then rapidly extend towards the developing IHCs (C–E). As OHCs begin to develop at E15.5, some processes extend past the IHCs to form contacts with OHCs (F–H). (I) Confocal image of flat-mounted cochleae from a Neurog1CreERT2; R26R-tdTomato mouse at P2. HCs are labeled with anti-myosin VI (blue) and all SGNs are labeled with Tuj1 (green). Asterisks mark the SGN somata, which extend SGN fibers that form into radial bundles (RB) prior to forming synapses with HCs. (J) The same preparation as in panel I, but illustrating expression of tdTomato (anti-dsRed) to visualize sparsely labeled individual fibers. (K) High-magnification view from panel J illustrating type I SGNs innervating IHCs and type II SGNs passing IHCs to innervate OHCs. Scale bar in K: 20 μm, C–H; 45 μm, I and J; 15 μm, K.
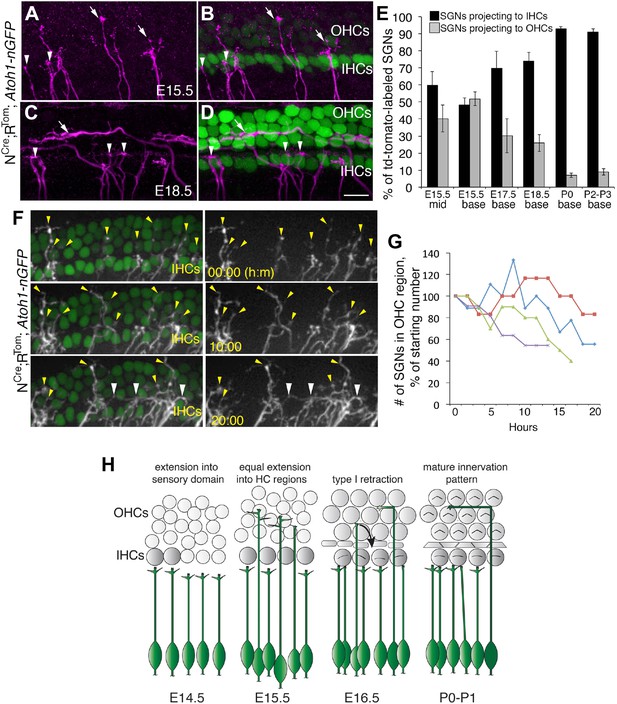
Sparse SGN labeling reveals retraction of processes from the OHC region during embryonic development.
(A–D) Fixed whole-mount cochleae from Neurog1CreERT2;R26RtdTomato; Atoh1nGFP mice at E15.5 (A, B) and E18.5 (C, D). Panels A and C show sparse-labeling of SGN fibers (tdTomato in magenta), while panels B and D show the same regions but include HCs in green. Note that at E15.5, the number of projections terminating in the IHC (arrowheads) and OHC (arrows) regions appears roughly equivalent. In contrast, at E18.5, the majority of SGNs terminate in the IHC region. (E) Comparison of the percentage of labeled SGNs that terminate in the IHC (black bars) or OHC region (gray bars) at different developmental time points (minimum of 6 cochleae per time point). Error bars are standard error of the mean (sem). (F) Sequential images at the indicated times from a single time-lapse experiment showing sparsely labeled SGNs with HCs (left) or alone (right) in an E16.5 cochlear explant. Yellow arrowheads indicate SGN fiber terminals in the OHC region. White arrowheads point to SGNs that are clustered around IHCs that had originally been in the OHC region. (G) Data from four imaging experiments illustrate a decrease in the number of fibers positioned in the OHC region over the course of experimental time. Since there was variability in the number of labeled SGNs between experiments, the data for each experiment were normalized to the number of labeled fibers present in the OHC region at time 0. (H) Illustration of the time-line of cochlear innervation. Fibers initially arrive at the IHC region around E14.5. At E15.5, when both IHCs and OHCs are present, SGN fiber distribution is roughly equal between the IHC and OHC regions. In contrast, by E16.5, many SGN fibers have withdrawn from the OHC region. At P0, the vast majority of SGNs terminate at the IHC region. These events occur before the final stages of synapse formation and subsequent fiber pruning. Scale bar in D: 15 μm, A–D; 20 μm, F.
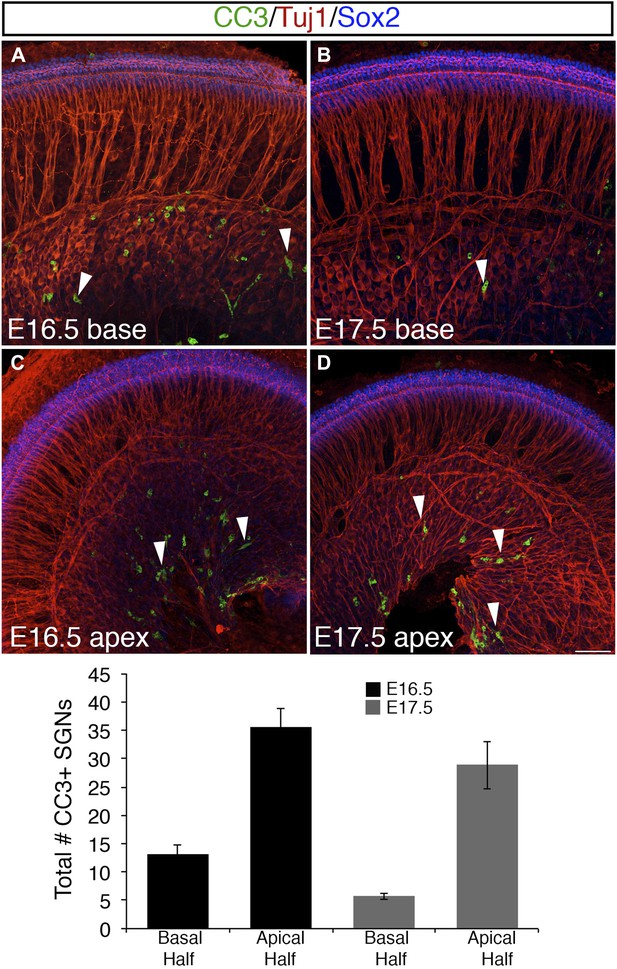
Programmed cell death (apoptosis) is limited in SGNs at E16.5 and E17.5.
(A–D) Confocal images from whole-mount preparations marked with anti-cleaved caspase-3 (CC3) antibodies, Tuj1 antibodies (to show SGN somata), and anti-Sox2 antibodies (to show the position of the cochlear sensory epithelium). The arrowheads in these panels point to SGNs that are CC3 positive. Scale bar in D, 40 μm. Histogram showing average numbers of CC3-positive SGN in the base and apex of cochleae at each stage. n = 7 cochleae per group.
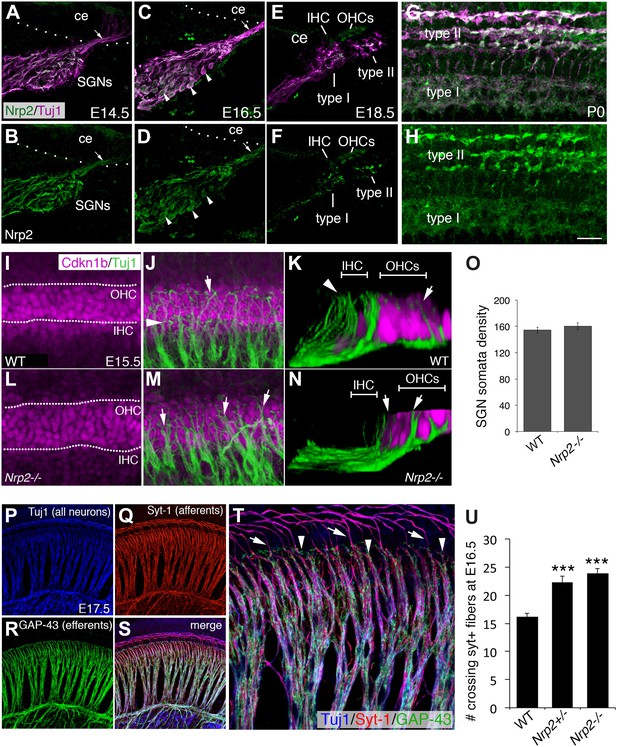
Nrp2 is expressed in SGNs and is required for normal HC innervation.
(A–H) Cross-sections of the cochlear duct and associated SGNs from the indicated ages. Upper panels show both SGNs (Tuj1 in magenta) and anti-Nrp2 (green). Lower panels show anti-Nrp2 alone. (A, B) At E14.5, Nrp2 is present in SGN somata and in peripheral axons (arrow) projecting into the cochlear epithelium (ce). (C, D) At E16.5, Nrp2 is detectable in virtually all SGN somata (arrowheads). (E, F) At E18.5, Nrp2 is detectable in putative type I and type II processes. (G, H) Whole-mount preparation at P0 indicates expression of Nrp2 in both type I and type II SGNs. (I–N) Comparison of SGN process distribution at E15.5 in WT and Nrp2−/− cochleae. Anti-Cdkn1b (magenta) labels the developing cochlear prosensory cells prior to HC formation (I) while Tuj1 (green) marks the SGNs (J). The dotted lines in I delineate the IHC and OHC regions. (K) Three-dimensional Y-Z view of the processes in J. Note that roughly equal number of processes terminate in both the IHC and OHC regions. In contrast, in a cochlea from an Nrp2−/− mutant, more processes appear to extend into the OHC region (arrowheads in M, N). (O) Quantification of cell bodies indicates no difference in number of SGNs between WT and Nrp2−/−. (P–T) E17.5 whole-mount cochlea stained with markers for all neurons (Tuj1; blue), SGN afferents (anti-Syt-1; red), and olivocochlear efferents (anti-GAP-43; green). The high-magnification view in T is from the same preparation shown in P–S. (U) Quantification of number of processes crossing into the OHC region in the base of the cochlea at E16.5. Both Nrp2−/− and Nrp2+/− mice show significantly higher numbers of SGN processes in the OHC region as compared to WT. ***p ≤ 0.001. n = 9, WT; 6, Nrp2−/−; 4, Nrp2+/−. Error bars, sem. Scale bar in H: 35 μm, A–H; 20 μm, F, J, L, M; 10 μm, K, N; 55 μm, P–S; 20 μm, F.
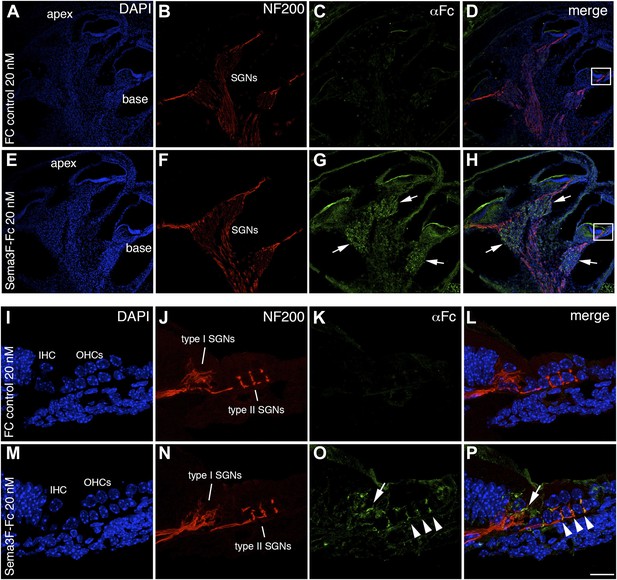
Semaphorin3F-Fc recognizes binding partners in vivo.
Representative images from wild-type P0 cochleae stained with Fc (A–H) or Sema3F-Fc (I–P) fusion proteins. Samples were counterstained with DAPI (blue) to reveal overall morphology and anti-Neurofilament 200 antibodies (red) to reveal SGN positions. (A–D) A low-magnification view shows very little binding by control Fc. (E, F) However, binding by Sema3F-Fc in a similar preparation is clearly evident in the SGNs (arrows). (I–L) A high-magnification view from the boxed region in D. Fc control does not show binding to type I or II SGNs. (M–P) A high-magnification view from the boxed region in H. Sema3F-Fc binds to type I (arrow) and type II (arrowheads) SGNs. Scale bar in P: approximately 450 μm for A–H and 20 μm for I–P.
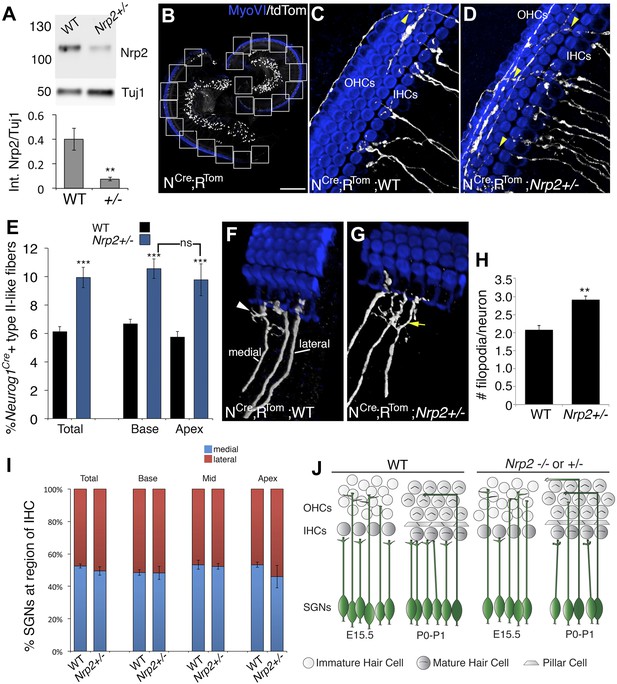
Nrp2 regulates SGN axonal complexity.
(A) Western blot demonstrating significantly reduced Nrp2 protein in cochleae from Nrp2+/− mice as compared to WT. **p ≤ 0.01; n = 9 WT; 7 Nrp2+/−. (B) Low-magnification view of a Neurog1CreERT2; R26R-tdTomato cochlea with boxed regions showing the locations where confocal z-stacks were acquired. In all of the micrographs in this figure, white represents anti-tdTomato and blue represents anti-myosin VI. (C, D) Compared to WT, Nrp2+/− cochleae with sparse SGN labeling show higher numbers of fibers in the OHC region (yellow arrowheads). (E) Quantification of the percentage of sparsely labeled type II-like fibers (defined as SGNs that had crossed into the OHC region and turned toward the base) in each genotype. ***p ≤ 0.001; n = 9 WT; 6 Nrp2+/−; ns, not significant. (F, G) 3D reconstructions of type I SGNs (relative positions are indicated in F). In Nrp2+/− cochleae, there were many type I SGNs with increased branching (yellow arrow in G), suggesting absence of an inhibitory cue. (H) Quantification of number of filopodia per individual type I SGN in WT and Nrp2+/− cochleae indicates significantly higher numbers of small branches in Nrp2+/− heterozygotes. **p ≤ 0.01; n = 4 WT; 6 Nrp2+/−. (I) The number of medially and laterally positioned type I SGNs does not change in Nrp2+/− cochleae. n = 4 cochleae each genotype. (J) Model illustrating phenotypic changes in cochlear innervation as a result of a decrease or absence of Nrp2. Error bars, sem. Scale bar in B: 150 μm, B; 20 μm, C, D, F, G.
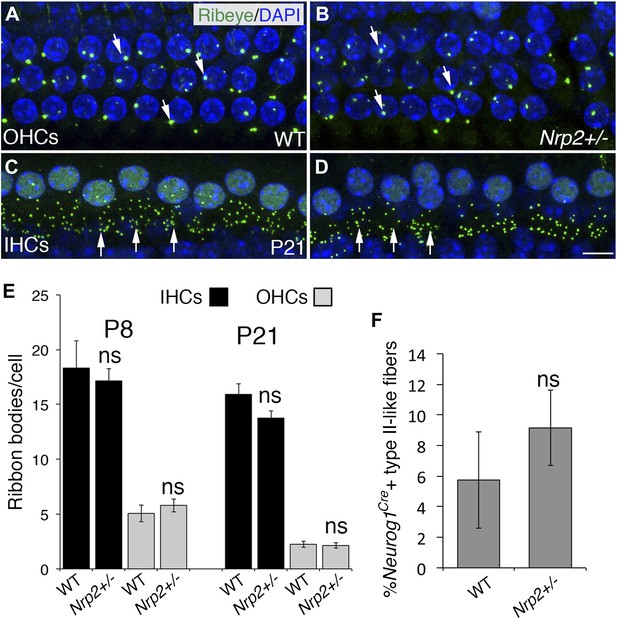
The number of HC ribbon synapses is unchanged in cochleae with reduced Nrp2.
(A–D) Visualization of ribbon synapses (anti-Ribeye in green; white arrows) on IHCs and OHCs at P21. Nuclei are labeled with DAPI (blue). Overall no differences in ribbon synapse numbers were observed between genotypes. (E) Quantification of ribbon bodies per IHCs and OHCs in P8 and P21 cochleae. All differences were not significant (ns). n = 6 cochleae per genotype per age. (F) Percentage of labeled type II-like fibers (defined as SGNs that had crossed into the OHC region and turned toward the base) in each genotype at P21. n = 6 WT; 6 het. Although the data trended toward more fibers in the Nrp2+/− cochleae, the difference compared to the control group was not significant (ns). Error bars, sem. Scale bar, 8 μm.
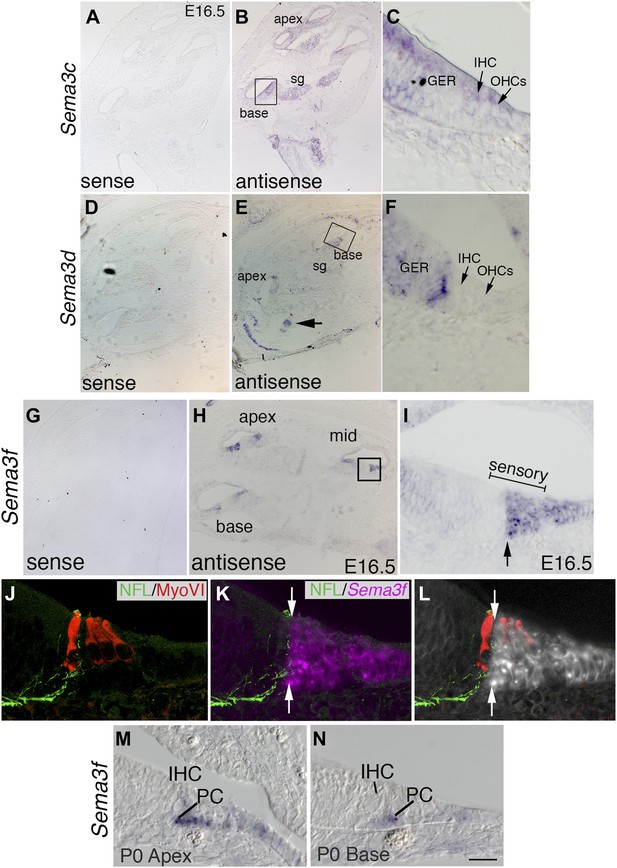
The expression pattern of Sema3f suggests a role in Nrp2 signaling during cochlear innervation.
In situ hybridizations for Sema3c, Sema3d, and Sema3f. (A–J) Low-magnification sense controls (A, D, G) demonstrate specificity for each probe. Low (B, E, H) and high-magnification views (C, F, J) from the boxed regions in B, E, and H for antisense probes indicate unique patterns of expression. (B, C) Sema3c transcripts are present in the SG, greater epithelial ridge (GER), and faintly in the IHCs and OHCs. (E, F) Sema3d transcripts are weakly present in the GER and in a striped pattern that includes non-sensory cells within the medial portion of the cochlear epithelium. (H, I) Sema3f transcripts are present in the lateral portion of the cochlear sensory domain (bracketed region of I), which includes OHCs, outer pillar cells (PCs), Dieters' cells, and Henson's cells. (J–L) To better resolve the expression pattern for Sema3f, the preparation in panel I was counterstained with anti-neurofilament light chain and anti-myosin VI. Results indicate a sharp medial boundary of Sema3f expression that correlates with the lateral edge of type I SGN innervation. (M, N) Sema3f expression in the apex and base of the cochlea at P0. IHC, inner HC; PC, pillar cell. Sema3f levels are diminished at this stage. By P5, Sema3f transcripts are not detectable (not shown). Scale bar in N: 350 μm, A, B, D, E, G, H; 20 μm, C, F, I, M, N; 15 μm, J–L.
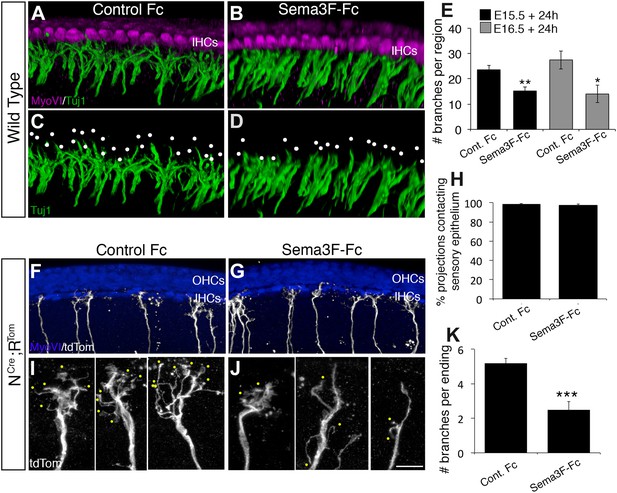
Exogenous Sema3f inhibits SGN outgrowth in vitro.
(A–D) Three-dimensional renderings of confocal z-stacks from cultured cochleae treated with either control IgG-Fc or Sema3F-Fc at 20 nM. The cultures were stained with anti-myosin VI to show the position of the HCs (magenta) and Tuj1 to mark SGNs (green). By comparison with an Fc-treated control (A, C) in which the SGNs show extensive branching, Sema3F-Fc-treatment significantly inhibited branching (B, D). The white dots in C and D mark individual branch points. (E) Quantification of SGN branch numbers normalized to the length of the cochlea within the photographed region. For this normalization, we measured the length of the cochlea along the IHC border. Addition of Sema3F-Fc led to significantly fewer SGN branches in explants initiated at E15.5 (black bars) or E16.5 (gray bars). **p ≤ 0.01; *p ≤ 0.05. n = 6 cochleae per treatment group. (F–K) To determine whether Sema3F-Fc reduced the number of SGNs in the cochlear epithelium or reduced the number of secondary branches per SGN, the Sema3F-Fc experiments were repeated using cochleae from Neurog1CreERT2; R26R-tdTomato mice. (F, G) Low-magnification micrographs from cochleae stained with anti-myosin VI (HCs, blue) and tdTomato to mark individual SGNs (white). (H) There were no differences in the percentages of labeled SGNs contacting the cochlear sensory epithelium in either group. (I, J) High-magnification images of representative individually labeled SGN terminals. Yellow dots mark small secondary branches extending from each SGN peripheral process. Note the reduced number of secondary branches in Sema3F-Fc-treated explants. (K) Quantification of the number of secondary branches per SGN ending showing the inhibitory effect of Sema3F-Fc. ***p ≤ 0.001. n = 6 cochleae per treatment group. Error bars, sem. Scale bar in J: 15 μm, A–D; 30 μm, F and G; 7 μm, I and J.
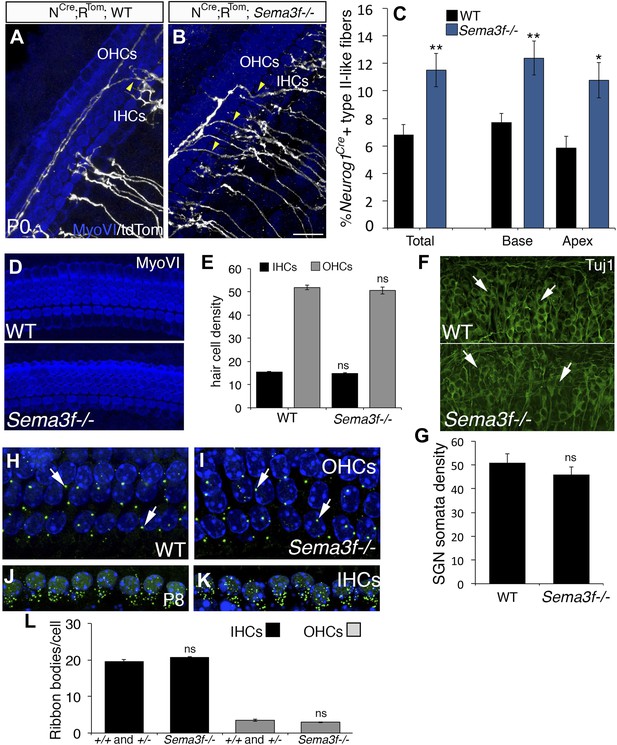
Deletion of Sema3f leads to increased SGNs in the OHC region.
(A, B) Representative images from Neurog1CreERT2;R26RtdTom; WT or Sema3f−/− cochleae at P0 stained with anti-myosin VI (HCs, blue) and tdTomato to mark individual SGNs (white). Increased numbers of crossing fibers (yellow arrowheads) projecting into the OHC region are present in the absence of Sema3f. IHCs, inner HCs; OHCs, outer HCs. (C) Absence of Sema3f leads to a significant increase in the percentage of labeled type II-like fibers (defined as SGNs that had crossed into the OHC region and turned toward the base). Note that the change in percentage in the absence of Sema3f is comparable to the change observed in Nrp2+/− heterozygotes (Figure 4E). **p ≤ 0.01; *p ≤ 0.05. n = 6 WT; 6 Sema3f−/−; error bars, sem. (D) Whole-mount images of HCs in WT and Sema3f−/− cochleae after myosin-VI immunostaining. (E) Quantification of number of IHCs and OHCs indicates no difference between WT and Sema3f−/−. n = 7 each genotype. (F) Whole-mount images of SGNs in WT and Sema3f−/− cochleae after Tuj1 immunostaining. The white arrows point to individually identifiable SGN somata. (G) Quantification indicates no difference in SGN density between WT and Sema3f−/−. n = 5 each genotype (H–K) Visualization of ribbon synapses (anti-Ribeye in green; white arrows) on IHCs and OHCs at P8. Nuclei are labeled with DAPI (blue). Overall, no differences in ribbon synapse numbers were observed between genotypes. (L) Quantification of ribbon bodies per IHCs and OHCs in P8. All differences were not significant (ns). Sample sizes: n = 6 for the control group, which is a combination of heterozygous and WT littermates; 4 Sema3f−/−. The scale bar in B = 15 μm for A and B; 30 μm, D; 40 μm, F and 12 μm, H–K.
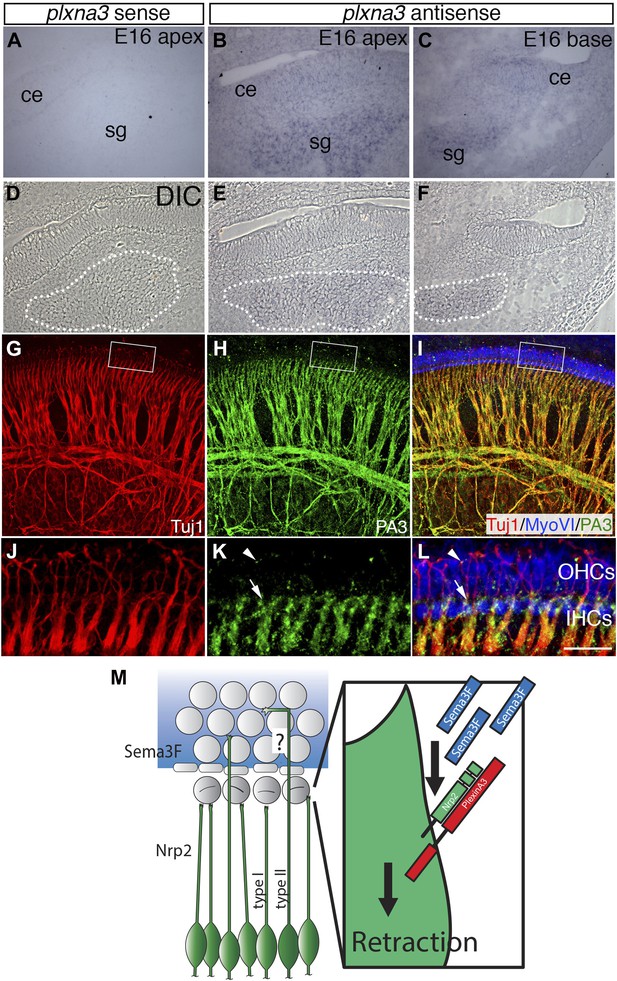
Differential expression of Plexin-A3 in SGNs and a proposed model.
(A–F) In situ hybridization for Plxna3 at E16.5. (A) A Plxna3 sense control probe does not show any reactivity. (B, C) Plxna3 transcripts are present in the SG, and possibly faintly in cells throughout the cochlear epithelium (ce). (D–F) Differential interference contrast (DIC) images for A–C were used to identify SGNs by morphology. The white dotted lines encircle the SGNs in each panel. (G–I) Plexin-A3 antibody staining in an E16.5 whole-mount cochlea. (G–I) Expression of Plexin-A3 (green) in SGNs appears to overlap with a neuronal marker (Tuj1 in red) except in the HC region (MyoVI staining in blue). (J–L) High-magnification views from the boxed regions in G, H, and I. Plexin-A3 is strongly expressed in SGN peripheral processes located adjacent to the IHC region (arrows) but appears markedly reduced in fibers that cross into the OHC region. Limited Plexin-A3 is detectable on sparse numbers of SGN processes in the OHC region (arrowhead). Scale bar in L: 100 μm, A–I, 25 μm, J–L. (M) Proposed model: Sema3F (blue shading and blue rectangles) is expressed by cells in the OHC region of the cochlea with a sharp boundary along the PCs. Binding of Sema3F to Nrp2 (green SGNs and green rectangle) induces retraction events that inhibit the growth of the type I SGN peripheral processes into the OHC domain. PlexinA3 co-receptors, which likely serve a role in this process, are shown in red. ‘?’ illustrates an outstanding question of how type II SGNs, which express Nrp2 and are confronted by Sema3F, are able to extend into the OHC domain.

Plxna3 expression in the inner ear at E11.5-E15.5.
In situ hybridization using Plxna3 antisense probe in C57BL/6J mice from the Allen Brian Atlas (http://developingmouse.brain-map.org/gene/show/18610) shows that Plxna3 is broadly expressed in the cochleovestibular ganglion neurons (cvg) at E11.5 and in the SGNs (sg) at E13.5 and E15.5 Specific images can found at: http://developingmouse.brain-map.org/experiment/show/100047035 for E11.5, http://developingmouse.brain-map.org/experiment/show/100049528 for E13.5, and http://developingmouse.brain-map.org/experiment/show/100049508 for E15.5.
Videos
A cochlea from a mouse carrying the Neurog1CreERT2; Rosa-tdTomato; Atoh1-nGFP alleles that was imaged by spinning disk confocal microscopy for approximately 20 hr.
The orange dots mark SGN processes that can be seen projecting into the OHC region and then falling back to the IHC region.





