TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst
Figures
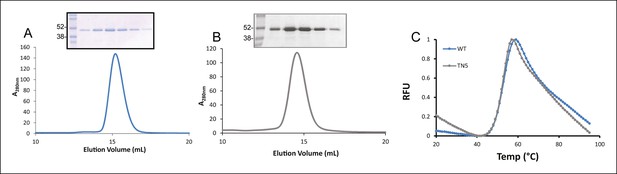
Expression of TAPBPR.
Size exclusion chromatograms of (A) TAPBPR and (B) TAPBPRTN5 purified from cell culture supernatants. The protein peaks were analysed by SDS-PAGE followed by Coomassie staining. (C) Differential scanning fluorimetry of TAPBPR and TAPBPRTN5 demonstrate equivalent thermal denaturation profiles. The data is representative of two independent experiments.
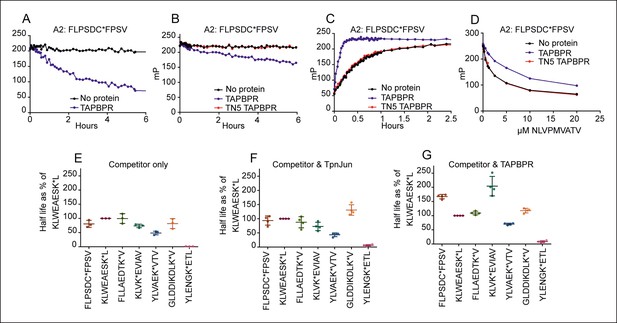
TAPBPR functions as a peptide loading catalyst and peptide editor for HLA-A2.
(A,B) Dissociation, (C) association and (D) peptide competition of fluorescent peptide FLPSDC*FPSV on HLA-A*02:01 in the absence or presence of TAPBPR or TAPBPRTN5 as measured by fluorescence polarisation. (A) 0.15 µM HLA-A*02:01fos molecules were mixed with 1.2 µM human β2m and loaded with 0.1 μM FLPSDC*FPSV and (B) 0.5 µM HLA-A*02:01 molecules were loaded with 0.125 μM FLPSDC*FPSV. The complexes were then split and incubated with 1000 fold molar excess of (A) FLPSDCFPSV or (B) NLVPMVATV with either A) buffer (No protein) or supplemented with 0.75 μM TAPBPR, or (B) buffer (No protein) or supplemented with 0.25 μM TAPBPR or TAPBPRTN5. Note, the slight difference in dissociation rate observed in A & B is likely to be related to the concentration of TAPBPR used in each experiment and not to the sequence of the competing peptide used. The data shown in Figure 2A,B is representative of 13 independent experiments, which all produced similar results. (C) 0.5 µM HLA-A*02:01 molecules were made peptide receptive and then the binding of 0.125 µM FLPSDC*FPSV was followed in the presence or absence of 0.05 µM TAPBPR or TAPBPRTN5. One representative association experiment of 13 experiments is shown. (D) 0.5 µM HLA-A*02:01 molecules were made peptide-receptive and incubated with 0.125 μM high affinity peptide FLPSDC*FPSV and various concentrations of the lower affinity competing peptide NLVPMVATV (0–20 μM) in presence or absence of 0.25 μM TAPBPR or TAPBPRTN5. One of two experiments is shown. (E–G) Comparison of the dissociation of seven peptides from HLA-A*02:01fos in the presence or absence of TAPBPR or tapasin-Jun as performed in Figure 2—figure supplement 1. The results of three (E) and four (F,G) independent experiments were combined and are shown. Data from each experiment was processed in GraphPad Prism using one-phase exponential decay non-linear regression. The half-lives that were calculated for the dissociation of the indicated peptide in the presence of either (E) Competitor only, (F) Competitor and Tapasin-Jun or (G) Competitor and TAPBPR were plotted as a percentage of the half-life calculated for the dissociation of KLWEAESK*L in the equivalent condition. Error bars show the standard deviation of the relative half-lives measured in the different experiments. While there were slight modifications of experimental conditions between replicate experiments, results consistent with the presented results were observed.
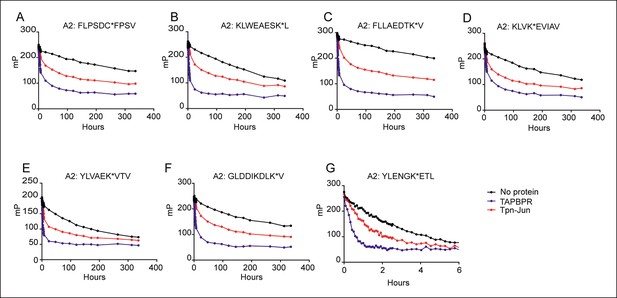
Dissociation of seven different peptides from HLA-A*02:01fos in the presence or absence of TAPBPR or Tapasin-Jun.
0.15 µM HLA-A*02:01fos molecules were mixed with 1.5 µM human β2m and made peptide receptive by UV exposure and loaded with 0.1 µM of the indicated labelled peptide overnight at 4°C. Next day the sample was split and either: 1000 excess unlabelled FLPSDCFPSV in buffer (No protein); 1000 excess unlabelled FLPSDCFPSV supplemented with 0.35 µM TAPBPR; or 1000 excess unlabelled FLPSDCFPSV supplemented with 0.1 μM Tapasin-Jun was added. One experiment from a total of four experiments is shown.
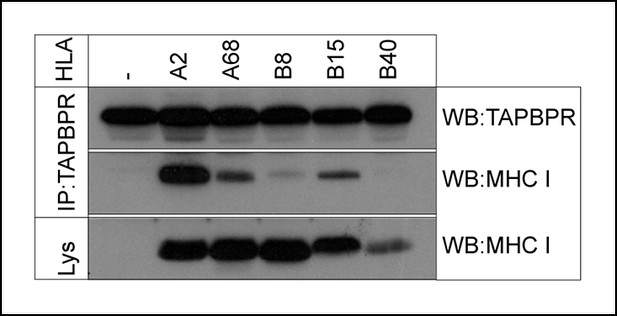
TAPBPR associates with HLA-A and HLA-B molecules.
TAPBPR was isolated by immunoprecipitation (using R014) from the MHC class I negative cell line 721.221 and 721.221 stably transduced with HLA-A2, A68, B8, B15 or B40. Western blot analysis was performed for TAPBPR and the MHC class I heavy chain (using HCA2 and HC10) on lysates (labelled lys) and TAPBPR immunoprecipitates as indicated. The data is representative of three independent experiments.
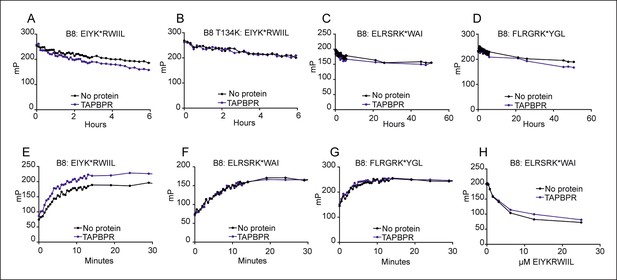
TAPBPR can function as a peptide loading catalyst and peptide editor for HLA-B8.
(A–D) Dissociation, (E–G) association and (H) peptide competition of fluorescent peptides on HLA-B*08:01 in the absence or presence of TAPBPR. 2 μM HLA-B*08:01fos or HLA-B*08:01fos T134K molecules were mixed with 20 μM human β2m and made peptide receptive then loaded with 1 μM (A,B) EIYK*RWIIL (C) ELRSRK*WAI or (D) FLRGRK*YGL. Dissociation was subsequently followed after the addition of a 250 molar excess of (A,B) EIYKRWIIL (C) ELRSRKWAI, or (D) FLRGRKYGL in the absence or presence of 0.75 µM TAPBPR. A total of eight dissociation experiments have been conducted for wild-type HLA B*08:01fos, and the T134K mutant was included in six of these experiments. (E–G) 0.6 μM HLA B*08:01fos molecules were mixed with 6 µM human β2m and made peptide-receptive, then the binding of 1 µM of (E) EIYK*RWIIL, (F) ELRSRK*WAI or (G) FLRGRK*YGL was followed in the absence or presence of 0.175 µM TAPBPR. One of three experiments is shown. (H) 0.6 μM HLA-B*08:01fos molecules were mixed with 0.6 µM human β2m and were made peptide-receptive, then incubated with 0.1 μM high affinity peptide ELRSRK*WAI and various concentrations of the lower affinity competing peptide EIYKRWIIL (0–25 μM) in presence or absence of 0.3 μM TAPBPR. One of six experiments is shown. Fluorescence polarisations measurements were taken after being left at room temperature overnight. While there were slight modifications of experimental conditions between replicate experiments, results consistent with the presented results were observed.
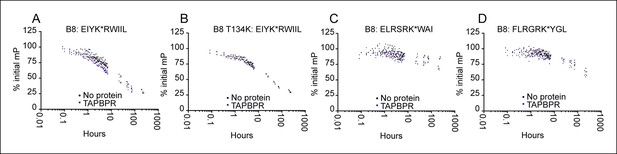
Dissociation of three different peptides from HLA B*08:01fos in the presence or absence of TAPBPR.
2 µM HLA-B*08:01fos molecules or T134K mutant were mixed with 20 µM human β2m and made peptide receptive by UV exposure and then loaded with 1 μM of the indicated peptide overnight at 4°C. Dissociation was subsequently followed at room temperature after the addition of a 250 molar excess of (A B) EIYKRWIIL (C) ELRSRKWAI, or (D) FLRGRKYGL in the absence (black dots) or presence of 0.75 µM TAPBPR (blue dots) for up to 240 hr. The results of three independent experiments are combined, with each polarisation measurement plotted as a percentage of the initial polarisation measurement taken in the absence of competing peptide from the same experiment.
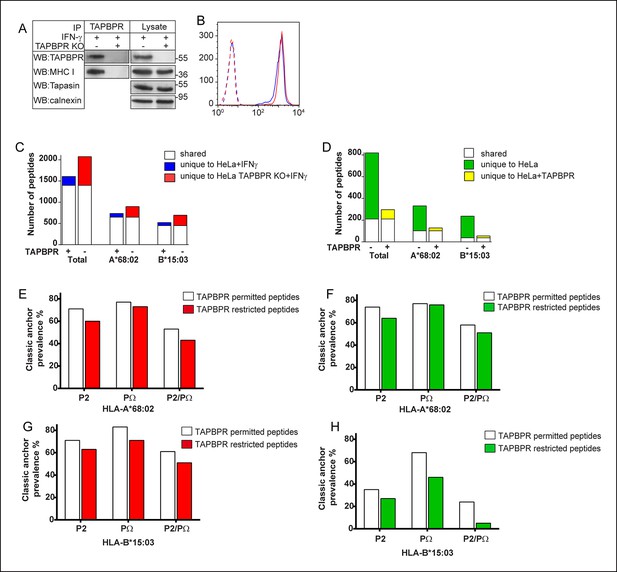
TAPBPR expression alters the peptide repertoire presented by MHC class I on cells.
(A) The TAPBPR:MHC class I complex was immunoprecipitated from IFN-γ treated HeLa and HeLa-TAPBPR KO cells. Western blot analysis was performed for TAPBPR (mouse anti-TAPBPR), tapasin (Rgp48N), MHC class I (HC10) and calnexin on lysates and TAPBPR immunoprecipitates as indicated. The data is representative of three independent experiments. (B) Cytofluorometric analysis of MHC class I detected with W6/32 on IFN-γ treated HeLa (Blue line) and HeLa-TAPBPR KO cells (Red line). Staining with an isotype control on both cell lines (dashed lines) is included as a control. (C,D) Peptide-MHC class I complexes were isolated by affinity chromatography using W6/32 from (C) IFN-γ treated HeLa and HeLa-TAPBPR KO cells or (D) HeLa and HeLa overexpressing WT-TAPBPR. Eluted peptides were analysed using LC-MS/MS. Graphs show the total number of peptides and also the number of peptides assigned as HLA-A*68:02 and HLA-B*15:03 binders based on their peptide motifs using the online programme NetMHC. The number of peptides shared between the cell lines (white bar) and the number of peptides unique to each cell line (coloured bars) is shown. The data was generated from tandem MS analysis performed five times on one immunoprecipitate. Two independent biological repeats have been performed in two different cell lines (HeLa-S cells shown in Figure 5—figure supplement 1, and KBM-7 cells shown in Figure 6) in which a similar pattern of increased peptide diversity in TAPBPR depleted cells was observed. (E–H) Conservation of P2 and C-terminal anchor residues (PΩ). Plots show prevalence of classic peptide anchors on (E,F) HLA-A*68:02 or (G, H) HLA-B*15:03. Bars show classic anchor conservation at P2, PΩ and P2/PΩ combined for (E,G) shared peptides found in both IFN-γ treated HeLa and HeLa TAPBPR KO cells (white bar) (presumably permitted expression in the presence of TAPBPR) and peptides unique to HeLa TAPBPR KO + IFNγ cells (red bar) (presumably restricted in the presence of TAPBPR), (F,H) shared peptides found in both HeLa and HeLa over-expressing TAPBPR (white bar) (presumably permitted expression in the presence of TAPBPR) and peptides unique to HeLa (green bar) (presumably restricted in the presence of TAPBPR).
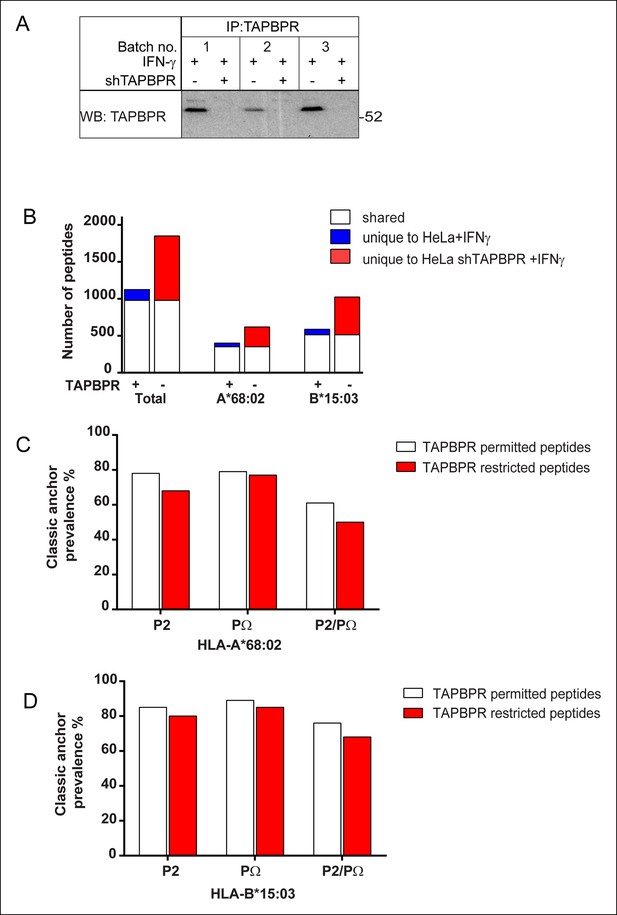
Increased peptide diversity on MHC class I in the absence of TAPBPR in IFN-γ treated HeLa-S cells.
Approximately 2 × 108 HeLa-S and HeLa-S depleted of TAPBPR expression using shRNA specific for TAPBPR (Boyle et al., 2013) were induced with IFN-γ treated for 72 hr. This was repeated three time (Batch no. 1–3) in order to harvest >5 × 108 cells of each line. (A) To confirm the efficient induction of TAPBPR by IFN-γ and the depletion of TAPBPR in shTAPBPR treated cells, a small fraction of the cells were lysed in 1% digitonin-TBS, TAPBPR was immunoprecipitated using the rabbit anti-TAPBPR polyclonal R014 and western blot analysis was performed for TAPBPR (using mouse anti-TAPBPR). (B) Peptide-MHC class I complexes were isolated using affinity chromatography using W6/32 from IFN-γ treated HeLa-S and shTAPBPR depleted HeLa-S cells. Eluted peptides were analysed using LC-MS/MS. The Graph show the total number of shared and unique peptides, and also the number of peptides assigned as HLA-A*68:02 and HLA-B*15:03 binders based on their peptide motifs. Note: surface expression of MHC class I detected by W6/32 is similar in both cells as demonstrated in Boyle et al., 2013. The number of peptides shared between the cell lines (white bar) and the number of peptides unique to each cell line (coloured bars) is shown. (C&D) Conservation of P2 and C-terminal anchor residues (PΩ). Plots show prevalence of classic peptide anchors on (C) HLA-A*68:02 or (D) HLA-B*15:03. Bars show classic anchor conservation at P2, PΩ and P2/PΩ combined for shared peptides found in both IFN-γ treated HeLa and HeLa shTAPBPR cells (white bar) (presumably permitted expression in the presence of TAPBPR) and peptides unique to HeLa shTAPBPR IFNγ cells (red bar) (presumably restricted in the presence of TAPBPR).
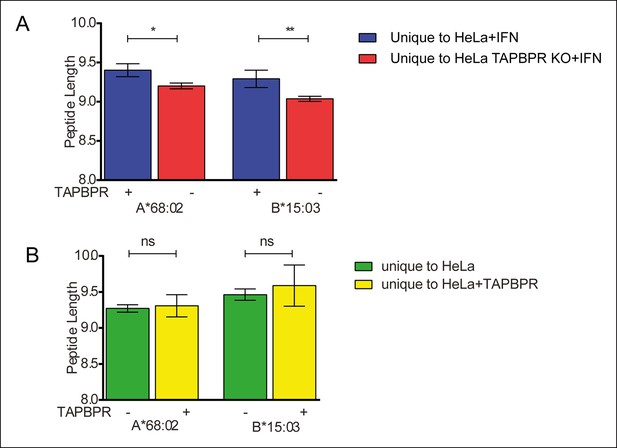
TAPBPR expression does not influence peptide length.
The average length of peptide eluted from HLA-A*68:02 and HLA-B*15:03 was calculated using GraphPad Prism and compared between (A) IFN-γ treated HeLa and HeLa-TAPBPR KO cells and (B) HeLa cells and HeLa overexpressing TAPBPR. Error bars show SEM of peptide length. The p value was calculated by two-tailed t-test.
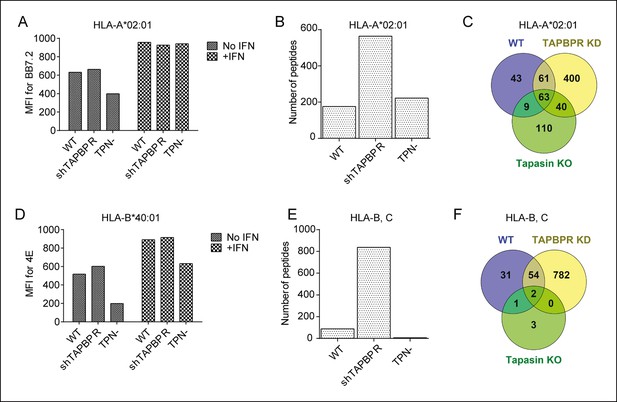
Distinct alterations to the peptide repertoire expressed on MHC class I molecules in IFN-γ treated KBM-7 cells upon TAPBPR depletion or tapasin deficiency.
Cytofluorometric analysis of (A) HLA-A2 expression (detected with BB7.2) and (D) HLA-B expression (detected with 4E) on the cell surface of WT, TAPBPR stably depleted (shTAPBPR) and tapasin deficient (TPN-) KBM-7 cells treated with and without IFN-γ for 48 hr. The data is representative of three independent experiments. (B,E) Peptide-MHC class I complexes were isolated using affinity chromatography using (B) BB7.2 (HLA-A2) and (E) B1.23.2 (HLA-B,-C) from IFN-γ treated WT, TAPBPR stably depleted (shTAPBPR) and tapasin deficient (TPN-) KBM-7 cells. Eluted peptide were analysed using LC-MS/MS. Graphs show the total number of peptides. The data was generated from tandem MS analysis performed five times on one immunoprecipitate. Two independent biological repeats comparing the peptide repertoire expressed on MHC class I under conditions of TAPBPR competency and TAPBPR deficiency in two other cell lines have been performed (HeLa-M cells shown in Figure 5 and HeLa-S cell shown in Figure 5—figure supplement 1) in which a similar pattern of increased peptide diversity upon TAPBPR depletion was observed. (C,F) Venn diagrams show the overlap of peptides on (C) HLA-A2 and (F) HLA-B,-C eluted from IFN-γ treated WT, TAPBPR stably depleted (TAPBPR KD) and tapasin deficient (Tapasin KO) KBM-7 cells.
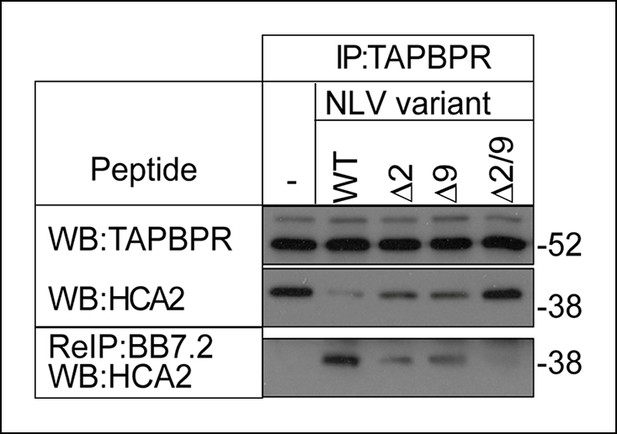
The MHC I bound to TAPBPR is peptide receptive.
The TAPBPR:MHC I complex was immunoprecipitated from IFN-γ induced KBM-7 cells. Equal aliquots were divided, then incubated -/+ 100 μM (20 μg) of the indicated peptide (WT:NLVPMVATV, Δ2: NAVPMVATV, Δ9: NLVPMVATM, Δ2/9:NAVPMVATM) for 30 min at 4°C. Subsequently all eluates (- or + peptide) were re-immunoprecipitated with BB7.2. Extensive washing was performed to remove any released MHC I before denaturation. Western blot analysis was performed for TAPBPR, and HLA-A2 (using HCA2) under reducing conditions. Data shown are representative of three independent experiments.
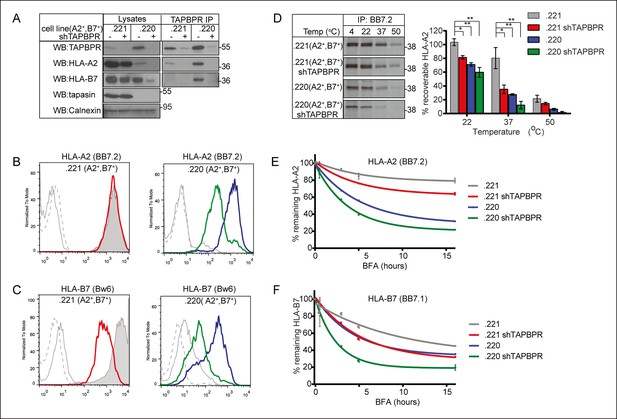
TAPBPR depletion reduces MHC class I stability.
(A) The MHC class I negative cell line 721.221 and the tapasin and HLA-A and -B negative cell line 721.220 were transduced with HLA-A2 and –B7. shRNA specific for TAPBPR (shTAPBPR) was used to produce TAPBPR depleted versions of these cell lines. TAPBPR was isolated by immunoprecipitation from these four cell lines. Western blot analysis was performed for TAPBPR, tapasin, HLA-B (3B10.7), HLA-A2 (HCA2), and calnexin on lysates and TAPBPR immunoprecipitates as indicated. This data is representative of three independent repeats. Cytofluorometric analysis of (B) HLA-A2 (detected with BB7.2) or (C) HLA-B7 (detected with anti-Bw6 antibody) on .221 (A2+ , B7+ ) (grey filled histogram), .221 (A2+ ,B7+) shTAPBPR (red line histogram), .220 (A2+ ,B7+ ) (blue line histogram), .220 (A2+ ,B7+ ) shTAPBPR (green line histogram). Staining on the non-transduced 721.221 and 721.220 cells with BB7.2 and Bw6 (grey solid line) or with an isotype control (grey dashed line) are included as controls. The data is representative of three independent experiments. (D) Thermal stability of HLA-A2 expressed in .221 (A2+ ,B7+ ) and .220 (A2+ ,B7+) -/+ stable depletion of TAPBPR (shTAPBPR). Cells were radiolabelled for 60 min with [35S] cysteine/methionine, lysed, then equal aliquots of cleared lysates were either kept at 4°C or heated at 22°C, 37°C or 50°C for 12 min. Peptide loaded HLA-A2 was immunoprecipitated using BB7.2 post-preclear. After separation by SDS-PAGE, the signal intensity of the radiolabelled HLA-A2 band was determined by phosphorimaging. The results are representative of four independent experiments. The graph shows the percentage of peptide loaded HLA-A2 recoverable at each temperature as a percentage of the signal intensity at 4°C. Error bars show SEM from four independent experiments. Surface expression of (E) HLA-A2 (detected with BB7.2) and (F) HLA-B7 (detected with BB7.1) on cells treated with 5 µg/ml brefeldin A (BFA), which inhibits egress of newly assembled molecules from the ER, for 0, 0.5, 3, 6 and 16 hr. The level of remaining HLA-A2 and HLA-B7 at each time point is expressed as percentage of the mean fluorescence at time 0. Error bars represent SEM of duplicate samples and the data is representative of three independent experiments.
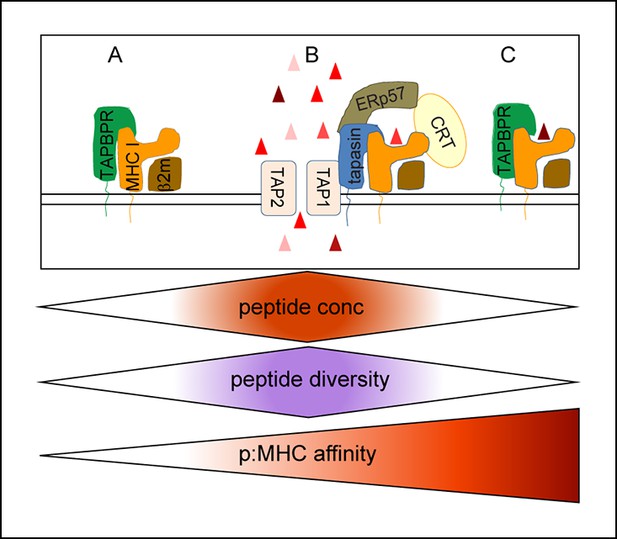
Model of the relationship between tapasin and TAPBPR in shaping the peptide repertoire expressed on MHC class I.
(A) Upstream of the PLC, in an environment in which peptides suitable for MHC class I binding are likely to be at a low concentration, TAPBPR might help stabilise a peptide-receptive conformation of MHC class I, but would not directly assist in peptide loading, peptide exchange or directly increase the abundance of peptide:MHC complexes due to a shortage of peptide. (B) The optimal peptide loading environment for MHC class I is presumably within the PLC where the concentration of peptides suitable for MHC class I is the highest and where MHC I are held in a peptide-receptive conformation by tapasin. (C) Once peptide:MHC I complexes are released from the PLC, TAPBPR-mediated dissociation would attempt to remove peptide from MHC class I. If a high affinity peptide was bound to the MHC I molecules, TAPBPR would not be able to remove it. If TAPBPR can cause the dissociation of peptide, peptide exchange could ensue if suitable replacement peptides were available. However, the further away from the PLC and/or the further down the MHC class I peptide gradient TAPBPR works there may be a shortage of suitable replacement peptides. In this situation TAPBPR is unlikely to load the peptide-receptive MHC class I and may assist in the recycling of MHC I molecules. By functioning as a peptide exchange catalyst in a relatively peptide deficient environment, TAPBPR could increase the affinity of the peptide:MHC I complexes or restrict the diversity of peptide:MHC I complexes which are presented to the cell surface.





