The molecular mechanism of nuclear transport revealed by atomic-scale measurements
Figures
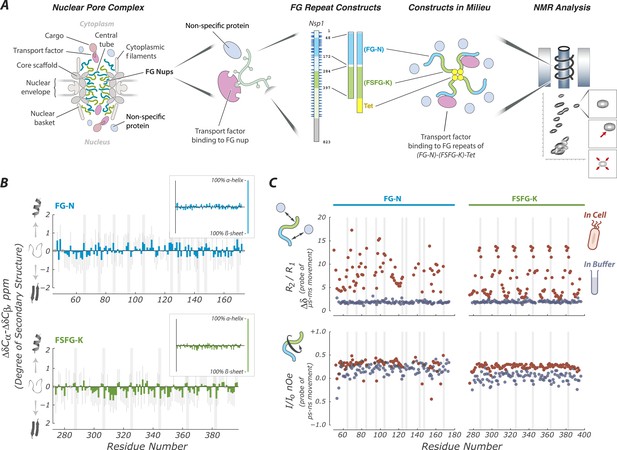
FG Nups are normally in a fully disordered and highly dynamic fluid state.
(A) Our experimental approach includes key features of the NPC; a mixture of FG flavors, attachment at one end, and both specific (TF) and non-specific interactions with the cellular milieu. For example, our largest construct (FG-N-FSFG-K-tet) contains two fragments from Nsp1 (FG-N, turquoise; FSFG-K, green; full-length Nsp1 also shown with residue numbering), a separator (white) and the tetramerization domain of p53 (yellow). NMR analysis is performed on this construct and its variants, in milieu of various types; changes in position or intensity of peaks (bottom right) indicate changes in structure or interactions of the FG motifs. (B) Deviations of chemical shift values in cell (Escherichia coli) from predicted (colored bars) showing that FG-N and FSFG-K fragments are fully disordered, with no propensity for secondary structure. Also shown are standard errors of the mean (gray bars) and positions of FG motifs in the sequence (gray columns). Chemical shift values expected for an α-helix or β-sheet are approximately −4 and +4 ppm, respectively, as shown in the small insets. (C) The FG-N and FSFG-K constructs show significant interactions with the cellular milieu and exhibit very rapid motions. The upper panels show the ratio of R2/R1 indicative of overall motion and effects of multiple environments (chemical exchange) in cell (E. coli), compared to buffer A. The heteronuclear nuclear overhauser effect (nOe) is shown in lower panels, indicative of backbone motions. Gray columns indicate the locations of the FG motifs. Full experimental details and interpretations are available in ‘Materials and methods’ and Figure 1—figure supplements 1–4.
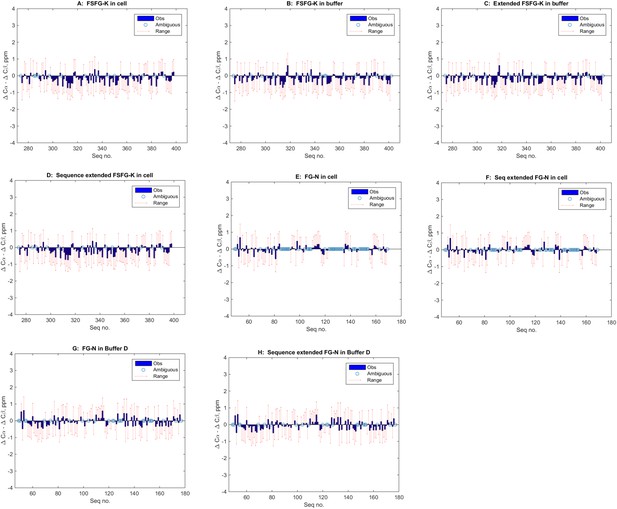
Lack of indicated secondary structure in FG Nup constructs (panels A–H, text).
The most generally accepted indicator of propensity for secondary structure is the differences in deviations of 13C shifts of Cα and Cβ's (e.g., Spera and Bax, 1991; Wishart and Sykes, 1994; Wishart and Case, 2001, illustrated in e.g., Fushman et al., 1998; McDonnell et al., 1999; Mittag and Forman-Kay, 2007), with α-helical segments showing a value of ∼ +4, and β sheets a value of ∼ −4. However, the standard deviations of the underlying reference values have not been widely available until recently (Tamiola et al., 2010). In the above panels, the observed values of the difference of deviations for each residues is shown in blue (-●) while the range of the calculated standard errors is shown in red ( -- ). Values which are ambiguous or missing because of overlaps are indicated by an open circle ( O ). Panels are (A) FSFG-K in cell; (B) FSFG-K in buffer A; (C) same as B but those ‘degenerate’ sequences otherwise unassigned, are duplicated at the additional positions; for example, in this case the sequence surrounding P397 was most similar to that of P282 (ATSKPAFSF and ATSKPALEH) and the P282 values are used at position P397 for the 13Cα and 13Cβ assignments; (D) assignments of B extended; (E) FG-N in cell; (F) sequence duplicated values of E; (G) FG-N in buffer D; (H) Sequence extended values of set G. The data presented indicate no significant runs of deviation of the Δδ shift value, and none exceeding the standard deviations of the measurements. All conclusions in the paper are based on specific assignment, or grouped assignments in the case of overlaps and extended assignment, and do not rely on specific assignment in ambiguous/overlapped cases. When in cell assignments were ambiguous, and in buffer assignments were consistent with in cell observation, the in buffer assignments for specific residues were used.
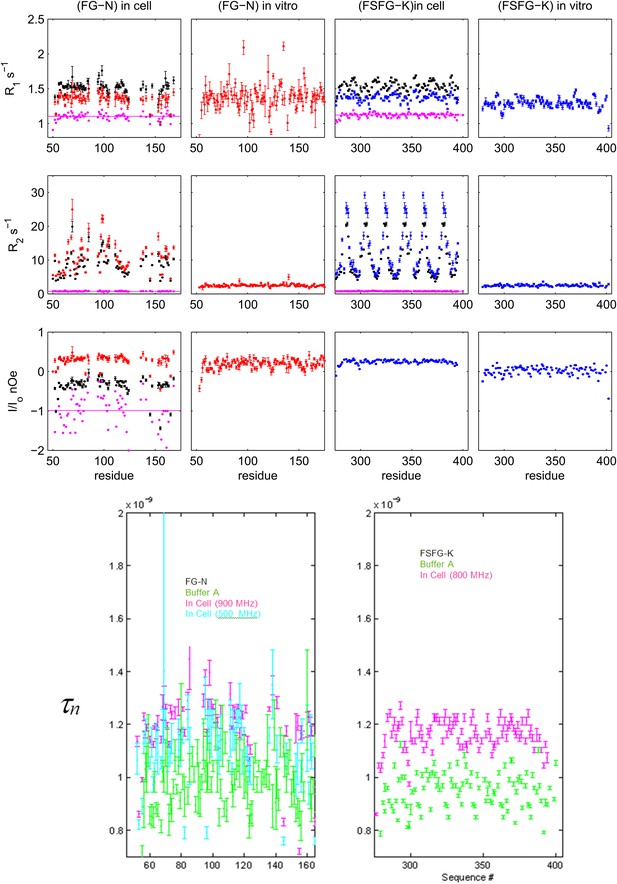
Details of NMR relaxation data and derived correlation times.
Upper: 15N[1H] relaxation data for FG-N and FSFG-K in respectively E. coli cell, and in buffer A (‘in vitro’). Measurements were at 900 MHz (red), 800 MHz (blue), and 500 MHz (black). For those with dual field measurements, the ratio at the two fields is shown in purple. The nOe is used to analyze rapid backbone motion in the panel below, and data will be further examined in future work. Lower: derived rotational correlation times, τn, for the N-H bond vector from the heteronuclear nOe. Data indicate that the field dependence of the observed nOe for FG-N in cell (red and black in low left above) is almost solely from the expected field dependence of the expected correlation, since the derived τn's are very similar (purple and cyan in left below). The in buffer average values, ∼0.9 ns, are similar for the two constructs, consistent with their similar chemical shift and dynamic light scattering properties (Figure 1, Figure 4G). The measured errors for the FG-N are greater than those of the FSFG-K set, predominantly because accurate estimates of peak heights are more difficult in the presence of overlaps in the spectra. Although there appears to be some significant sequence dependence in the data for FSFG-K in-buffer (green, right), the S/N in the FG-N case obscures whether this is present there also. Sequence dependence of the τn may arise from local restriction by immediately adjacent side chains. Values of τn in cell are increased modestly (to ∼1.17 ns) for both constructs and are assumed to reflect partial restriction of fast internal motions (Neuhaus and Williamson, 2000).

HSQC spectra of FSFG-K construct in buffer A vary with pH.
HSQC spectra of FSFG-K in buffer A adjusted to the indicated pH's. In buffer, fast solvent exchange of significant number of peaks is apparent as expected (Croke et al., 2008, 2011; Burz et al., 2012). The His6 tag signal titrates with the pH change and is circled at the lower right of each panel.
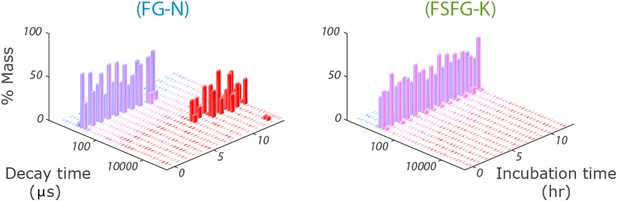
Stability of FG Nup constructs by DLS.
Stability of size for FG-N (left) and FSFG-K (right) in buffer A by dynamic light scattering. The raw decay times are used so no interpretive models are applied. FG-N shows a sharp transition to a gel-like state after about 7 hr. Protein concentrations 8 mg/ml. For detailed methods see ‘Dynamic Light Scattering’.
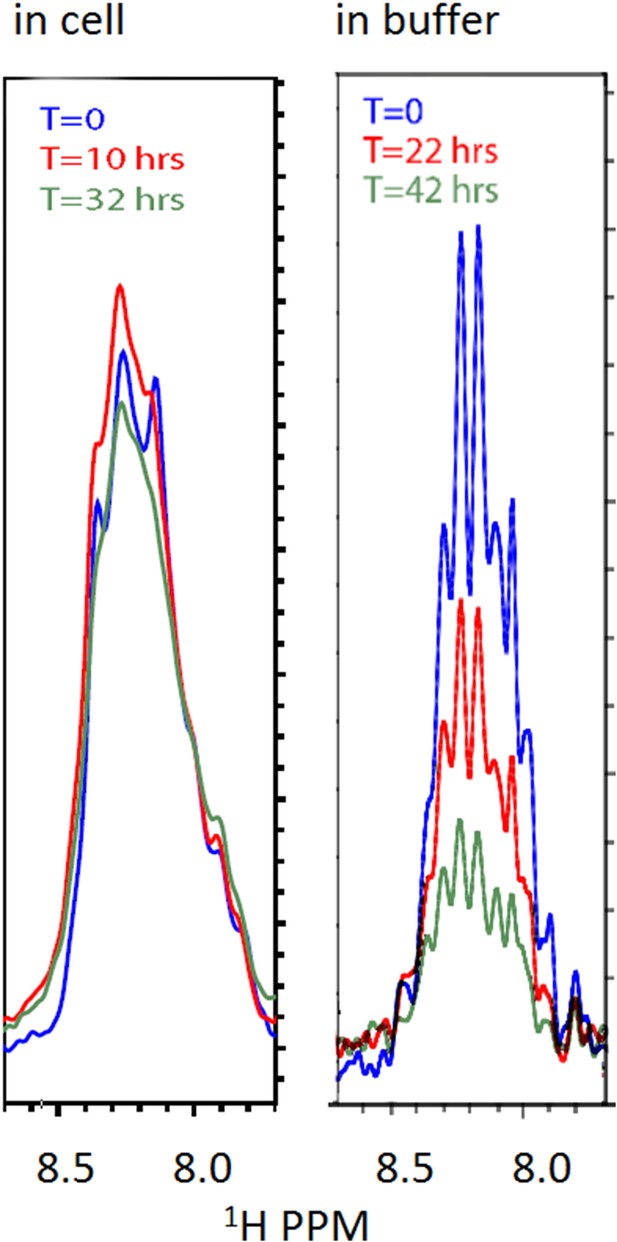
Stability of the FG-N construct in cell (left) and in buffer (right) by NMR.
Left panel: freshly made cells (grown as ‘Materials and methods’); induced at OD 0.8 for 200 m were spun and resuspended in buffer E. Right panel: purified and concentrated FG-N was buffer exchanged directly from 8 M urea to buffer A. All spectrometer conditions remained constant between scans. Samples were contained in 5 mm Shigemi NMR tubes. The spectra are 1-D traces from the first block of HSQC spectra initiated at the listed times.
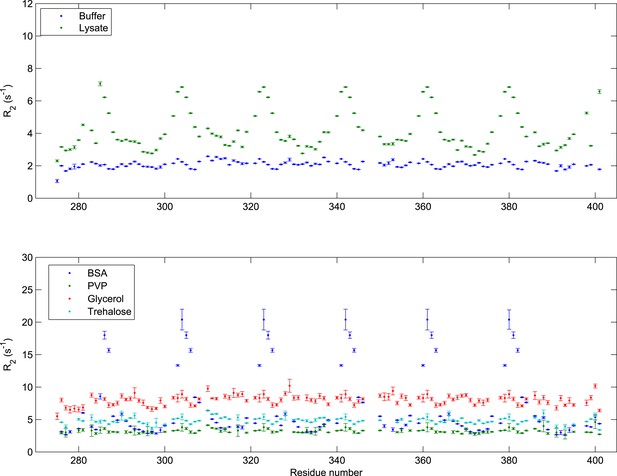
Transverse relaxation of FSFG-K in multiple environments.
All R2 data were collected at 25°C with 140 μM 15N-labeled FSFG-K in buffer A with the following delays: 0, 32.64, 65.28, 97.92, 130.56, 163.20, 195.84, 228.48, 261.12, and 326.40 ms. The concentrations of the different additives are as follows: E. coli lysate (Wang et al., 2011; Latham and Kay, 2014) 230 mg/ml; BSA 205 mg/ml Glycerol (Li et al., 2009; Wang et al., 2011) 60% wt/wt; PVP-10 (Li et al., 2008, 2009) 111 mg/ml; Trehalose (Lins et al., 2004; Chakrabortee et al., 2010) 154 mg/ml. Sequence-dependent fluctuation is seen only with lysate or with BSA, indicating non-specific rapid interactions, in contrast to changes of viscosity (Serber et al., 2005; Li et al., 2009; Wang et al., 2011), solvation (Lim and Deng, 2009; Xue and Skrynnikov, 2011; Kim et al., 2013), or packing (Li et al., 2009; Uversky, 2009; Milles et al., 2013). The E. coli lysate concentration is comparable to the estimated in vivo total concentration (Cayley et al., 1991). Interaction with BSA is consistent with previous observations (Parks et al., 1983; Higuchi et al., 1994; Clarkson et al., 1996; Fielding et al., 2005; Simard et al., 2006).
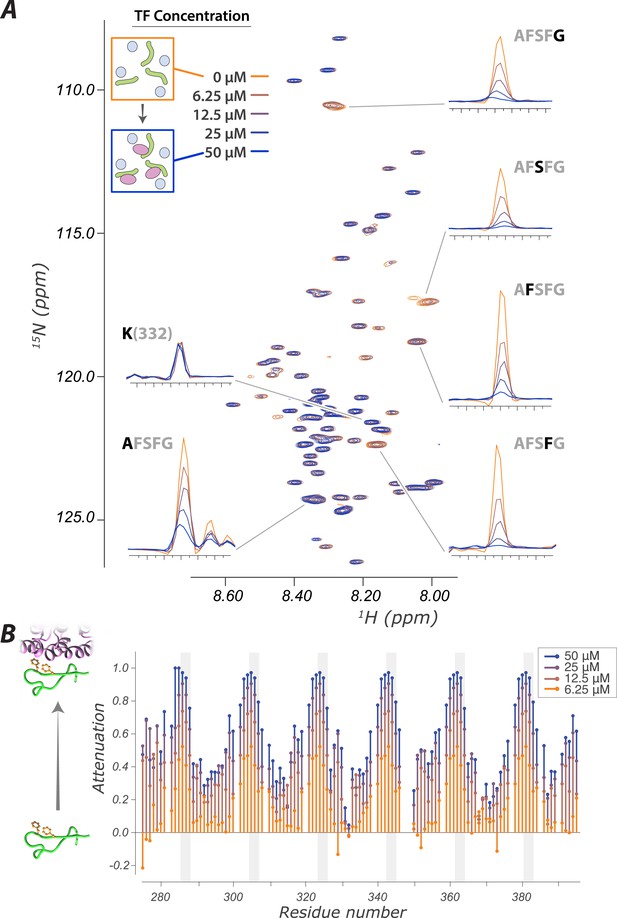
The interaction of FG Nups with the transport factor Kap95 is specific to the FG sites and leaves the spacers highly mobile
(A) Overlay of spectra with varying concentrations of Kap95 (0, 6.125, 12.5, 25, 50 μM) in the presence of [U-15N] FSFG-K (25 μM) in Xenopus egg extract, showing only the FG motifs strongly interact with the TF. Several peaks represent overlapping similar sequences with indistinguishable attenuation. (B) Superimposed values of attenuation ((1 − I)/Io) on addition of Kap95 across the sequence; increased attenuation indicates a stronger interaction propensity for that residue (positions of FG motifs in the sequence are indicated by gray columns).
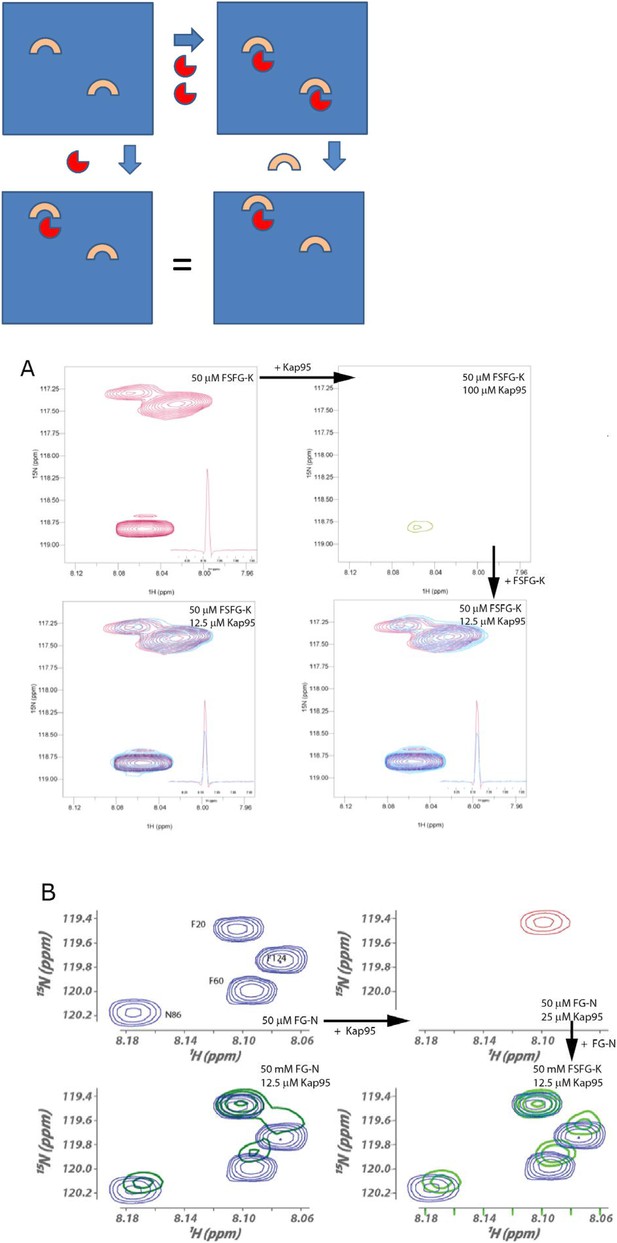
Reversibility of TF/FG Nup interactions.
For each of FSFG-GK and FG-N's interactions with Kap95, we demonstrated reversibility as follows. The spectra below can be cartooned as arising from the following, where the upper left panel represents the solution of an FG Nup. Addition of TFs (red) at high concentration forms a substantial amount of complex (upper right). Addition of more FG Nup at the same concentration as upper left results in reversibility of a portion of the complex formed, and spectra identical to those formed by direct addition of TF in smaller amounts (lower panels). If the complex was irreversible, then the dilution from the right upper complex, would lead to a summation of the upper two figures for the right lower, and would not show the intermediate chemical shifts seen. (A) HSQC spectra of complexes of FSFG-K and Kap95, illustrating the formation of the same complex (lower spectra) by either admixture of the components (lower left) or dilution of the TF concentration from a higher value to the final by addition of FSFG-K (lower right). At upper left, the region of the S, F residues of FSFG-K 50 μM is shown in the absence of Kap95 in buffer C. On makeup to 100 μM Kap95, the resulting spectrum is at the upper right. Dilution with additional FSFG-K produces the spectra at the lower right. The lower spectra superimpose the starting spectrum (upper left, red) and the complex spectrum (azure). (B) HSQC spectra of complexes of FG-N and Kap95 components (lower left) or dilution of the TF concentration from a higher value to the final by addition of FG-N (lower right) in buffer A. At upper left, the region of the N and F residues of FG-N 50 μM is shown in the absence of Kap95. On addition of Kap95 to 0.5 molar equivalence (25 μM), the resulting spectrum is at the upper right. Dilution with additional FG-N produces the spectra at the lower right. The lower right spectra superimpose the starting spectrum (upper left, blue) and the complex spectrum (green).

15N R2 titration of Kap95 and [15N]FSFG-K.
Observed values of 15N R2 for FSFG-K at various concentrations, in the presence of 20 μM Kap95. Experimental details in ‘NMR analysis’ in ‘Materials and methods’. The 15N R2 values and their standard errors of the mean are plotted vs the log10 of the 15N FGFG-K concentration all in the presence of 20 μM Kap95 for the residue types of the FSFG motif. The values of 15N R2 are shown in green at the extreme right axis for free FSFG-K. The derived value fitted for the equation of Section S2.3 is shown as the dotted line with ‘X’ marks. The fitted values of Kd and Rb for the derived line are 36.1 μM and 25.5 s−1. The apparent Kd is then equivalent to 216 μM for each FSFG site in FSFG-K. Lines connect the observed or calculated values.
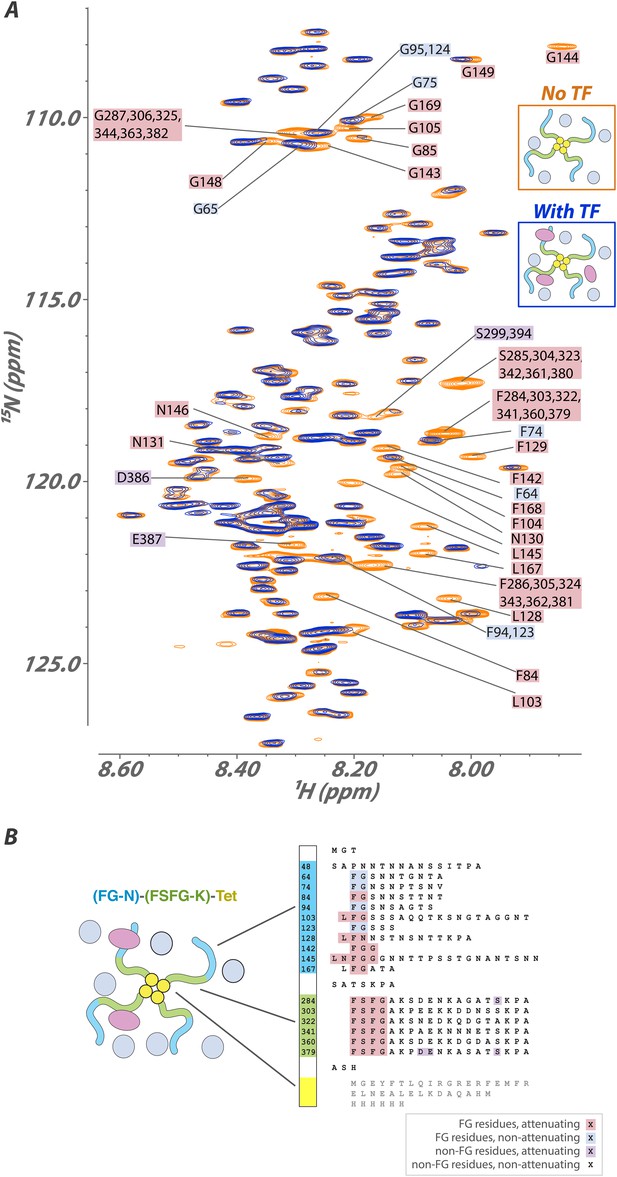
Interaction of a high molecular-weight tetramerized hybrid construct indicates that the binding mechanism is robust to changes in FG type, packing, and environment.
(A) 1H[15N] Heteronuclear Single Quantum Coherence (HSQC) spectra of the FG-N-FSFG-K-tet sequence (Tables 1 and 2) (MW 120 kDa) in the unbound (orange) and Kap95-bound (blue) states with Xenopus egg extract as the milieu. Peaks that undergo signal attenuation in the Kap95-bound spectrum indicate binding of those residues to Kap95. Typically, we observe that it is the FG residues (pink boxes), but not intervening spacer residues (no boxes), that attenuate upon binding to the transport factor Kap95; however, there are four FG repeats that are minimally attenuated (blue boxes), and a small number of spacer residues that are attenuated (purple boxes). (B) Schematic of the construct used, labeled as in (A). Very small chemical shift changes are observable for a few sites, consistent with rapid exchange into multiple inhomogeneous sites.
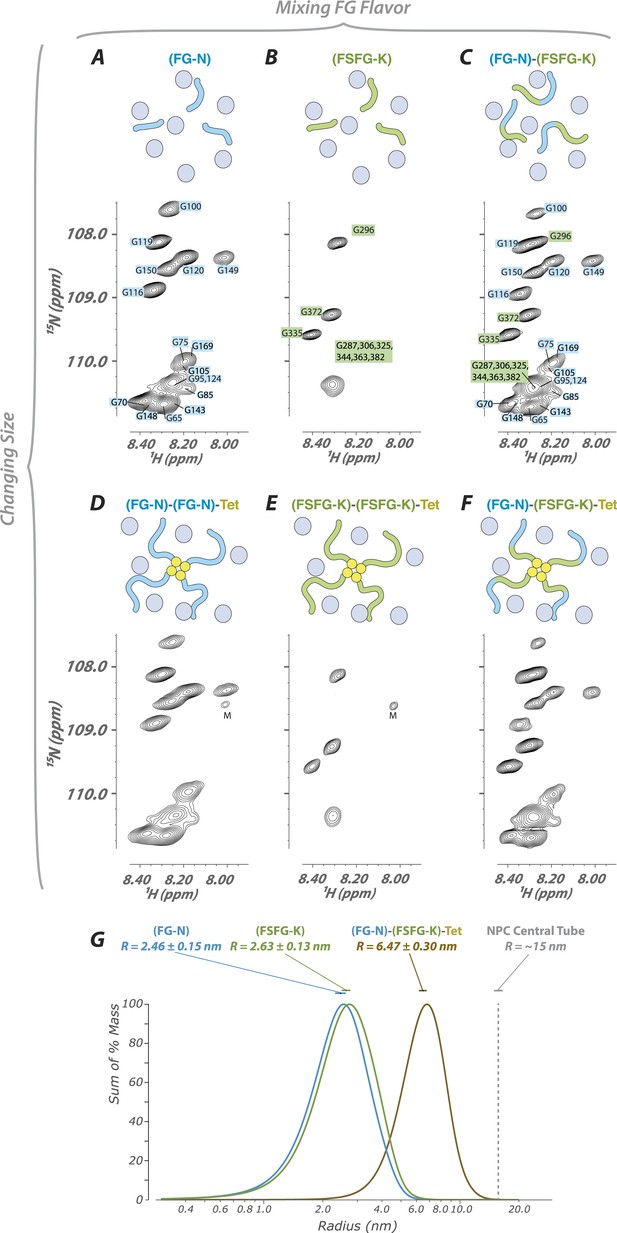
FG constructs show no significant FG–FG interactions, and remain fully disordered even when tethered at one end and packed at high density.
(A–F) We developed a wide range of constructs containing fragments from Nsp1 (turquoise and green, numbering in Figure 1) and the small tetramerization domain from p53 (yellow). Schematics illustrate the different constructs as studied within the cellular milieu (blue circles) which vary both in total molecular weight, crowding of the FG repeats via tetramerization, and composition of FG fragments (homotypic or heterotypic). Below each schematic is the HSQC spectrum of the glycine region of the represented construct observed in cell (E. coli). All constructs are tabulated in Table 1. ‘M’ identifies apparent 15N-labeled metabolites which are variable in intensity from preparation to preparation. (G) Dynamic light scattering of three representative constructs in buffer, showing homogeneity and consistency with expected size of constructs.
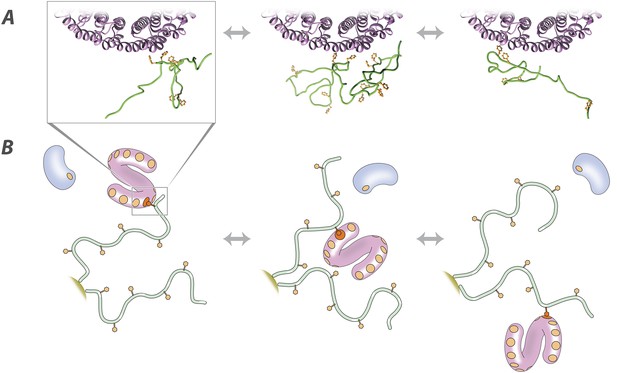
Molecular description of the nuclear transport mechanism.
Specific conclusions from our results, as discussed in the main text, are illustrated with (A) details from a docked molecular simulation of the TF Kap95 (purple) and the FG repeat region FSFG-K, rendered in Chimera (see ‘Materials and methods’), and (B) a diagrammatic view of the molecules involved. FG repeat (green; an ensemble of disordered conformers is illustrated in A). Phe (orange), Phe-binding pockets (B, orange circles), TF (pink), non-specific macromolecule (B, blue).
Tables
FG constructs prepared
| Abbreviation used in text | Simple structure | Sequence | Number of residues / monomer | MW | Nsp 1 ref start number | Data Figure |
|---|---|---|---|---|---|---|
| FSFG-K | M-Nsp1(274–397) | MDNKTTNTTPSFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPFSFGAKPDENKASATSKPA | 125 | 13,070 | 274 | 1, 1S1,1S2 |
| FSFG-K | M-Nsp1(274–397)-LEHHHHHH | MDNKTTNTTPSFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPALEHHHHHH | 133 | 14,135 | 274 | 1, 2, 1S1-6, 2S1-2 |
| FG-N | MGT-Nsp1(48–172)-SHMHHHHHH | MGTSAPNNTNNANSSITPAFGSNNTGNTAFGNSNPTSNVFGSNNSTTNTFGSNSAGTSLFGSSSAQQTKSNGTAGGNTFGSSSLFNNSTNSNTTKPAFGGLNFGGGNNTTPSSTGNANTSNNLFGATASHMHHHHHH | 137 | 13,649 | 48 | 1, 1S1-2,1S4-5, 2S1 |
| (FG-N)-(FSFG-K)-Tet-6His | MGT-Nsp1(48–172)-ASATSKPA-Nsp1(284–397)-SHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | MGTSAPNNTNNANSSITPAFGSNNTGNTAFGNSNPTSNVFGSNNSTTNTFGSNSAGTSLFGSSSAQQTKSNGTAGGNTFGSSSLFNNSTNSNTTKPAFGGLNFGGGNNTTPSSTGNANTSNNLFGATAASATSKPAFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPASHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | 292 | 30,235 | 48, 284 | 3, 4 |
| FSFG-K | MGTSATSKPA-Nsp1(284–397)-SHHHHHH | MGTSATSKPAFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPASHHHHHH | 131 | 13,721 | 284 | 4 |
| (FSFG-K)-(FSFG-K)-Tet-6His | MGTSATSKPA-Nsp1(284–397)-ATSKPA-Nsp1(284–397)-SHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | MGTSATSKPAFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPASATSKPAFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPASHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | 287 | 30,504 | 284 | 4 |
| (FG-N)-(FG-N)-Tet-6His | MGCT-Nsp1(48–172)-Nsp1(48–172)-SHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | MGCTSAPNNTNNANSSITPAFGSNNTGNTAFGNSNPTSNVFGSNNSTTNTFGSNSAGTSLFGSSSAQQTKSNGTAGGNTFGSSSLFNNSTNSNTTKPAFGGLNFGGGNNTTPSSTGNANTSNNLFGATASAPNNTNNANSSITPAFGSNNTGNTAFGNSNPTSNVFGSNNSTTNTFGSNSAGTSLFGSSSAQQTKSNGTAGGNTFGSSSLFNNSTNSNTTKPAFGGLNFGGGNNTTPSSTGNANTSNNLFGATASHMGEYFTLQIRGRERFEMFRELNEALELKDAQAHMHHHHHH | 296 | 29,927 | 48 | 4 |
| (FG-N)-(FSFG-K)-6His | MGT-Nsp1(48–172)-ASATSKPA-Nsp1(284–397)-SHHHHHH | MGTSAPNNTNNANSSITPAFGSNNTGNTAFGNSNPTSNVFGSNNSTTNTFGSNSAGTSLFGSSSAQQTKSNGTAGGNTFGSSSLFNNSTNSNTTKPAFGGLNFGGGNNTTPSSTGNANTSNNLFGATAASATSKPAFSFGAKSDENKAGATSKPAFSFGAKPEEKKDDNSSKPAFSFGAKSNEDKQDGTAKPAFSFGAKPAEKNNNETSKPAFSFGAKSDEKKDGDASKPAFSFGAKPDENKASATSKPASHHHHHH | 257 | 25,955 | 48, 284 | 4 |
Sequence assignments filed with BRMB (Ulrich et al., 2008)
| FGFG-K in cell E. coli expression | 25,182 |
| FSFG-K in buffer | 25,183 |
| FG-N in cell E. coli expression | 25,184 |
| FG-N in buffer | 25,185 |





