Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha
Figures
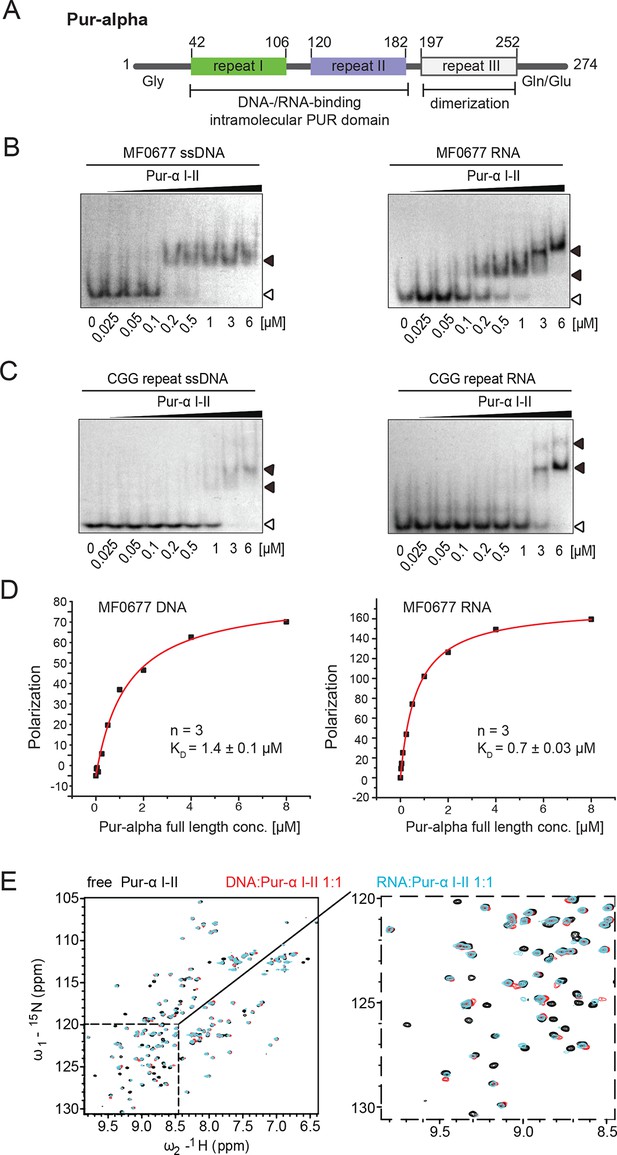
Pur-alpha uses similar binding modes for DNA and RNA.
(A) Schematic representation of the Drosophila Pur-alpha protein, comprising 274 amino acids. Cartoon shows PUR repeat I (green) and II (blue), forming the intramolecular DNA-/RNA-binding PUR domain, and PUR repeat III (grey) mediating dimerization. The N-terminal unstructured, glycine-rich region and the C-terminal glutamine-/glutamate-rich region are indicated by Gly and Gln/Glu, respectively. Numbers indicate amino-acid positions of domain boundaries. (B, C) Radioactive EMSA with Drosophila Pur-alpha repeat I-II. (B) Pur-alpha repeat I-II binds to MF0677 ssDNA (left) and ssRNA (right) with similar affinities. (C) Pur-alpha repeat I-II binds to CGG-repeat ssDNA (left) and RNA (right) also with similar affinity, but less strong than to the MF0677 sequence. Open arrowheads indicate free and filled arrowheads indicate protein-bound DNA/RNA oligonucleotides. (D) Fluorescence-polarization experiments with full-length Pur-alpha and nucleic acids. The full-length protein shows a twofold stronger binding to MF0677 ssRNA when compared to MF0677 ssDNA. (E) Binding of unlabeled GCGGA ssDNA and ssRNA to 15N-labeled Pur-alpha repeat I-II (50 µM) followed by NMR spectroscopy. (Left) Overlay of 1H,15N HSQC NMR spectra of free (black), DNA-bound (red, 1:1 ratio) and RNA-bound (cyan, 1:1 ratio) protein, respectively. (Right) Close-up on the dashed area with the same color code.
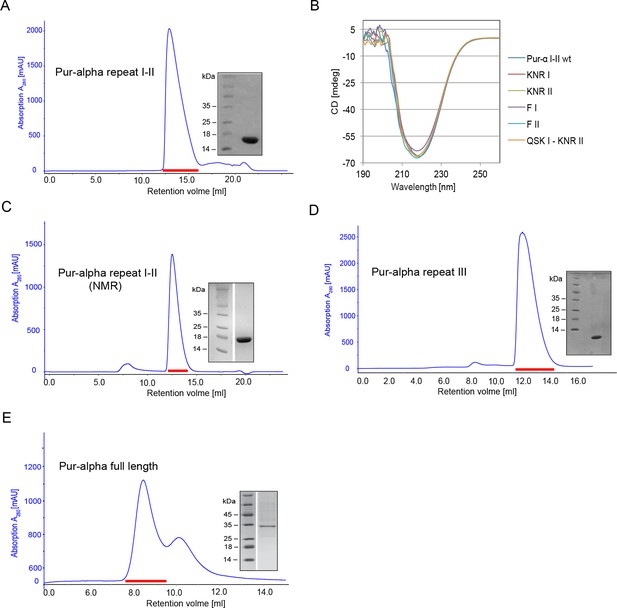
Purification and quality control of Drosophila Pur-alpha protein derivatives used in this study.
(A, C–E) Size exclusion chromatogram (blue) of the final purification step with the Superdex 75 10/300 GL column. Peak fractions (red dash) were pooled, concentrated and analyzed on SDS–PAGE. (A) Pur-alpha repeat I-II (17 kDa), (C) Pur-alpha repeat I-II (17 kDa) in NMR buffer, (D) Pur-alpha repeat III (10 kDa), (E) Pur-alpha full length. (B) Overlay of CD spectra of Pur-alpha wild-type and mutant protein (color code as indicated in the figure).
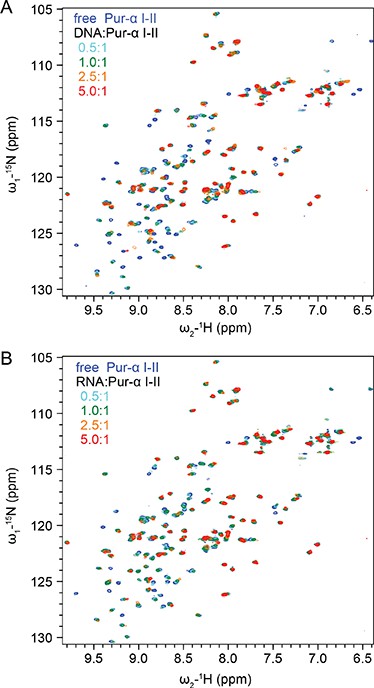
1H,15N HSQC NMR spectra showing NMR titrations of 15N-labeled Pur-alpha repeat I-II (50 µM) with increasing amounts of unlabeled GCGGA ssDNA and RNA, respectively.
(A) Titration with DNA. Peaks corresponding to the free and DNA-bound protein states (protein:DNA 1:0, 1:0.5, 1:1, 1:2.5 and 1:5 ratio) are represented in blue, cyan, green, orange and red, respectively. (B) Titration with RNA. Peaks corresponding to the free and RNA-bound protein states (protein:RNA 1:0, 1:0.5, 1:1, 1:2.5, and 1:5 ratio) are represented with the same color code as in (A).
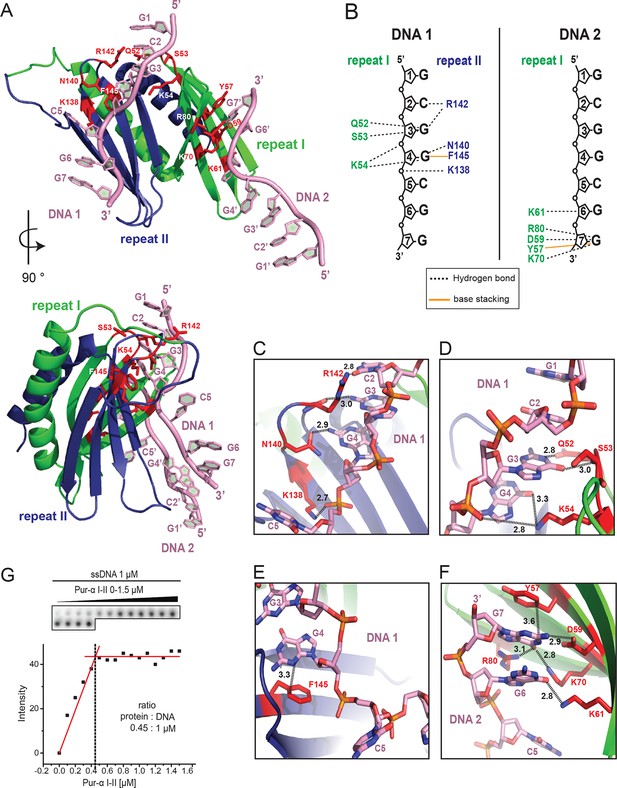
Crystal structure of Drosophila Pur-alpha repeat I-II in complex with the GCGGCGG ssDNA reveals that one molecule of Pur-alpha repeat I-II can bind two molecules of ssDNA.
(A) Cartoon representation of the backbone model of the DNA-/RNA-binding domain formed by PUR repeat I (green) and II (blue) in complex with two DNA molecules (pink). Important protein residues involved in DNA interactions are depicted in red with side chains. (B) Schematic representation of Pur-alpha interaction with DNA molecules 1 and 2. Both PUR repeats are involved in DNA binding. Pur-alpha mainly binds to guanine bases, but also to one cytosine and the sugar phosphate backbone. (C–F) Detail of the protein-DNA interaction sites. (G) Nitrocellulose filter (top) from binding assays showing the titration of Pur-alpha repeat I-II to a constant amount of MF0677 ssDNA. The measured intensities from the filters were quantified. The graph (bottom) shows intensities from one representative binding assay, with the concentration of saturation marked with a dashed line. Three independent filter-binding assays yielded a mean saturation of 1 : 0.58 ± 0.1 µM (ssDNA : Pur-alpha repeat I-II).
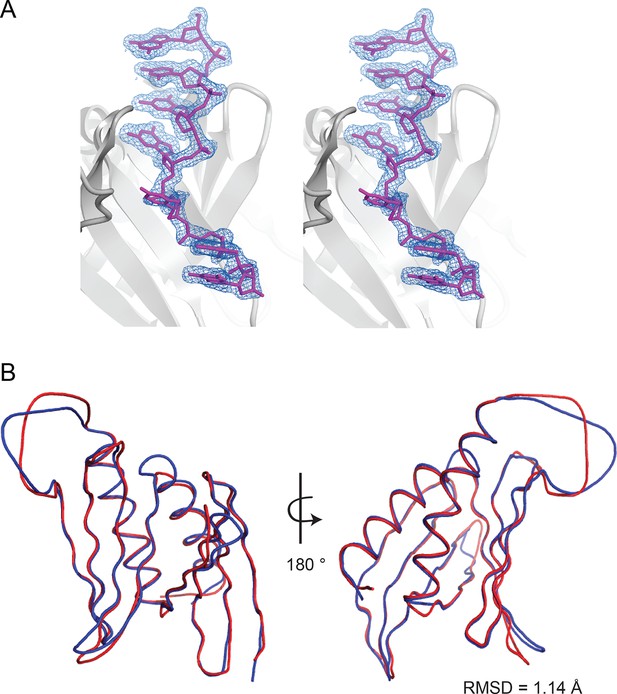
Analysis of the structural model of Drosophila Pur-alpha repeat I-II in complex with DNA.
(A) Stereo view of the Pur-alpha/DNA structure model showing the protein (grey), the DNA molecule 1 (magenta), and the (2Fo-Fc) electron density map (blue). (B) Structure alignment of the protein backbones of Pur-alpha repeat I-II apo-structure (PDB ID 3K44; depicted in red) and in complex with ssDNA (this study; depicted in blue) reveals no major conformational changes upon DNA binding. RMSD value is indicated in the figure.
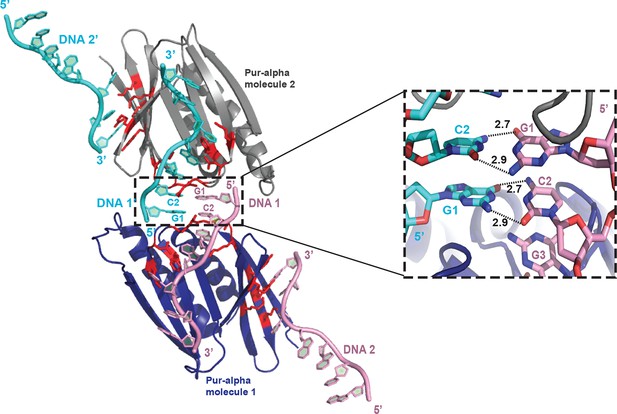
Within the crystal structure the protein-bound DNA anneals with another symmetry-related DNA molecule.
Cartoon representation shows Pur-alpha repeat I-II (blue) bound to two DNA molecules (pink) and the symmetry-related Pur-alpha-DNA complex (grey and cyan, respectively). The 5’ ends of the symmetry-related DNA molecules (DNA 1 and 1’) are base pairing (see close-up on the right side).
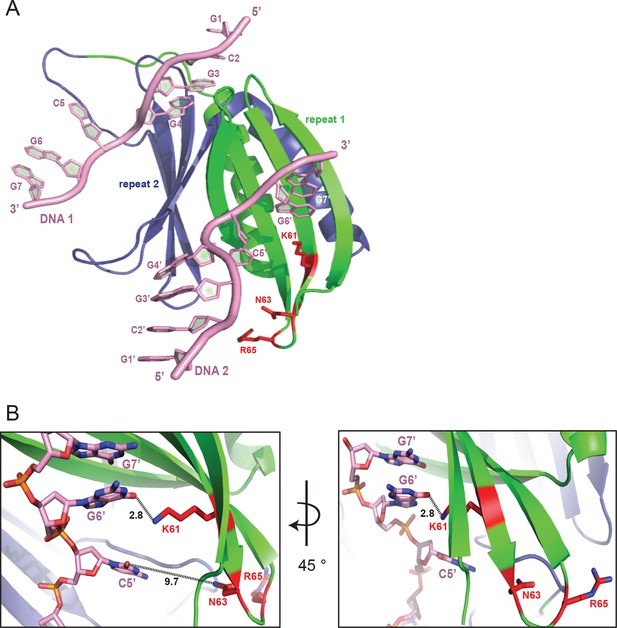
The DNA-binding site consisting of K138, N140, and R142 (KNR II) on PUR repeat II has its equivalent at the positions K61, N63, and R65 on PUR repeat I (KNR I).
(A) Cartoon representation of Pur-alpha repeat I (green) and II (blue) bound to two molecules of DNA (pink). Side chains of KNR I are indicated as red sticks. Only K61 interacts directly with the G6’ base of DNA molecule 2. (B) Zoom-in of (A) with indicated distance measurements. A close-up of KNR II binding to DNA 1 is shown in Figure 2C.

Amino acid sequence alignment of Pur-alpha.
(A) Sequence alignment from different species. D.m., Drosophila melanogaster; H.s., Homo sapiens; D.r., Danio rerio; C.e., Caenorhabditis elegans. (B) Sequence alignment between the three PUR repeats of Drosophila Pur-alpha. Color-coding from blue to red reflects the range of sequence conservation from 0 to 100%. Asterisk indicates positions, which have a single, fully conserved residue. Colon indicates conservation between groups of strongly similar properties. Period indicates conservation between groups of weakly similar properties. Secondary structure assignment is based on the crystal structure of Pur-alpha repeat I-II and Pur-alpha repeat III. Red boxes indicate DNA interaction sites seen in the crystal structure. Blue boxes indicate additional residues used in mutational analyses.
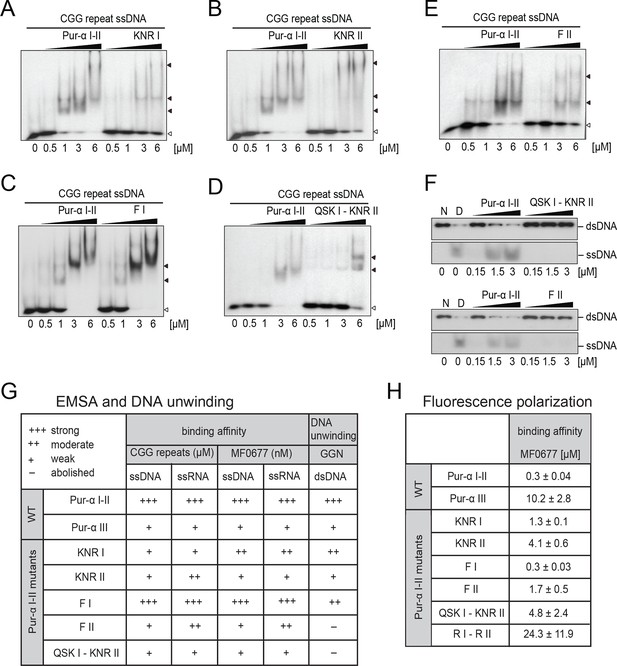
Mutations in Pur-alpha repeat I-II decrease nucleic-acid binding and dsDNA unwinding.
(A–E) Radioactive EMSA with wild-type and mutant Pur-alpha repeat I-II. All mutants show a decrease in binding affinity, except for the F I mutant in C. Open arrowheads indicate free and filled arrowheads indicate protein-bound DNA/RNA oligonucleotides. (F) Unwinding assays with wild-type and mutant Pur-alpha repeat I-II. Protein was titrated to a dsDNA substrate containing a GGN motif. Pur-alpha repeat I-II is able to separate the DNA strands, whereas mutations in both repeats (QSK I – KNR II) (top) and the F II mutation (bottom) abolish the unwinding activity. (G) Summary of the results of all EMSA and unwinding experiments of Pur-alpha derivatives and mutants. Original data are shown in Figure 3A–F and Figure 3—figure supplement 1. (H) Summary of the results of fluorescence-polarization experiments of Pur-alpha derivatives and mutants with MF0677 ssDNA. Original data are shown in Figure 3—figure supplement 2.
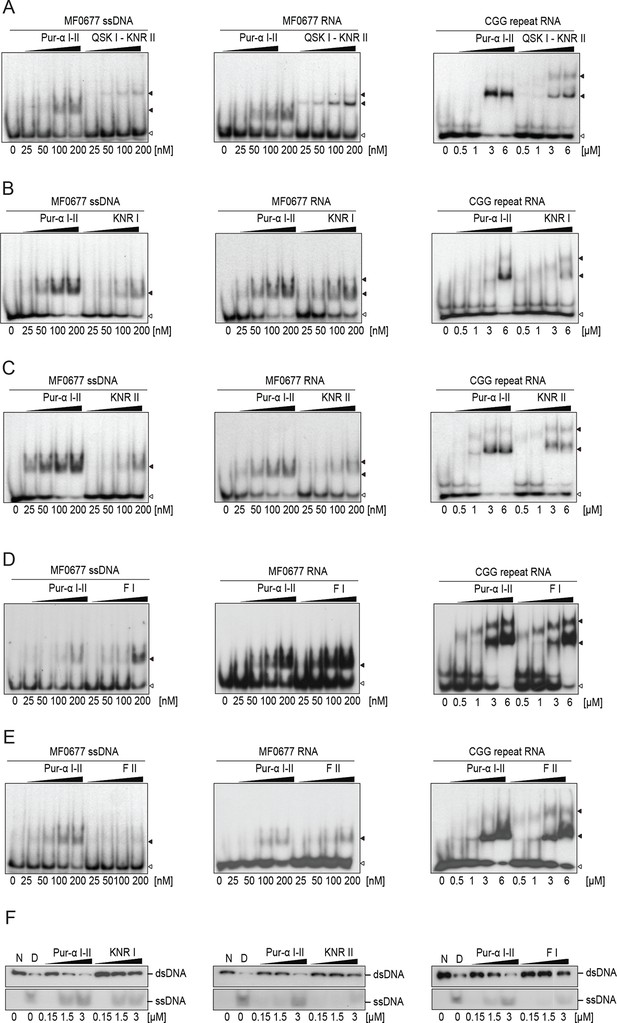
Drosophila Pur-alpha repeat I-II mutants show decreased binding affinity to DNA and RNA and decreased dsDNA-unwinding activity.
(A–E) Radioactive EMSA with wild-type or mutant Pur-alpha repeat I-II and with MF0677 ssDNA (left) or RNA (middle) and CGG-repeat RNA (right). EMSA with CGG-repeat DNA is shown in Figure 3A–E. All mutants show decreased nucleic acid binding, except for the F68 mutant (F I) (D), the counterpart to F145 on repeat II (F II). Open arrowheads indicate free and filled arrowheads indicate protein-bound DNA/RNA oligonucleotides. (F) Mutations in repeat I (KNR I) (left) or in the identical motif in repeat II (KNR II) (middle) decreased the unwinding activity. (Right) Decreased unwinding also occurs upon mutation of F I.
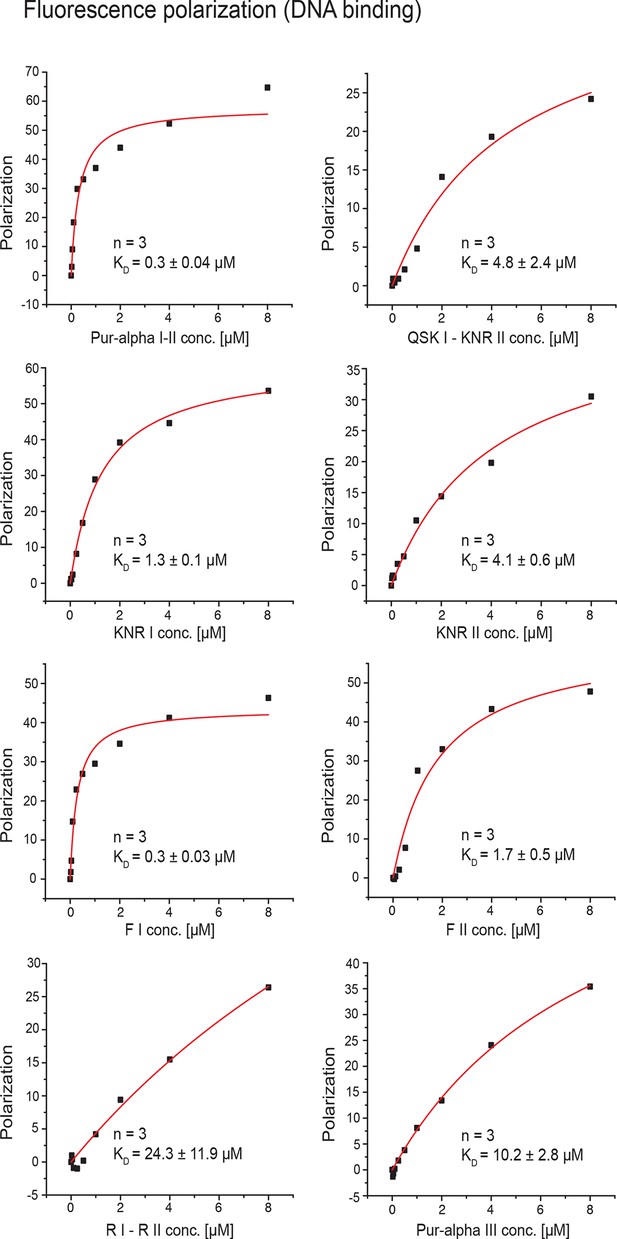
Fluorescence-polarization measurements with wild type or various mutants of Pur-alpha I-II and MF0677 ssDNA.
All experiments were performed as triplicates. KD values and standard deviations are given as insets in the plots. Curve fitting was performed as one-site binding.
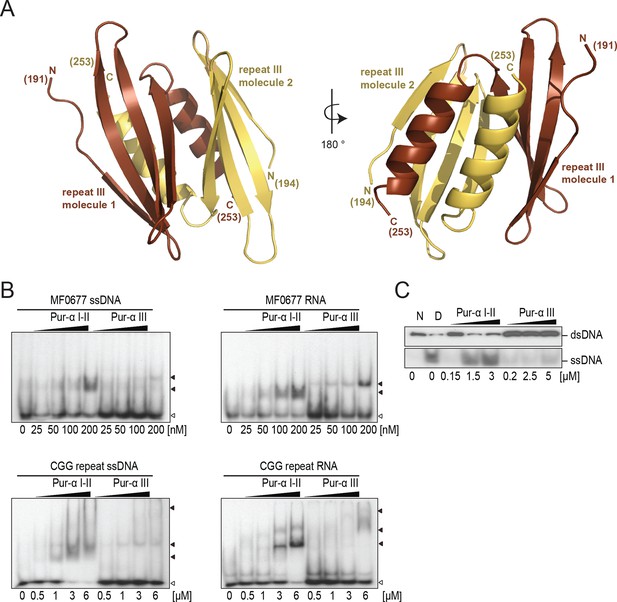
Crystal structure of PUR repeat III and assessment of its weak nucleic-acid-binding and unwinding activity.
(A) Crystal structure of Drosophila Pur-alpha repeat III. Two molecules (one depicted in brown, the other in yellow) of repeat III form a dimer with intertwined α-helices, very similar to Pur-alpha repeat I-II. (B) Radioactive EMSA with PUR repeat III and the MF0677 DNA/RNA (top) and the CGG DNA/RNA oligonucleotides (bottom). Repeat III shows only weak binding affinity to each of both sequences, regardless of whether they consist of DNA or RNA. Open arrowheads indicate free and filled arrowheads indicate protein-bound DNA/RNA oligonucleotides. (C) Pur-alpha repeat III shows only weak dsDNA-unwinding activity compared to PUR repeat I-II.
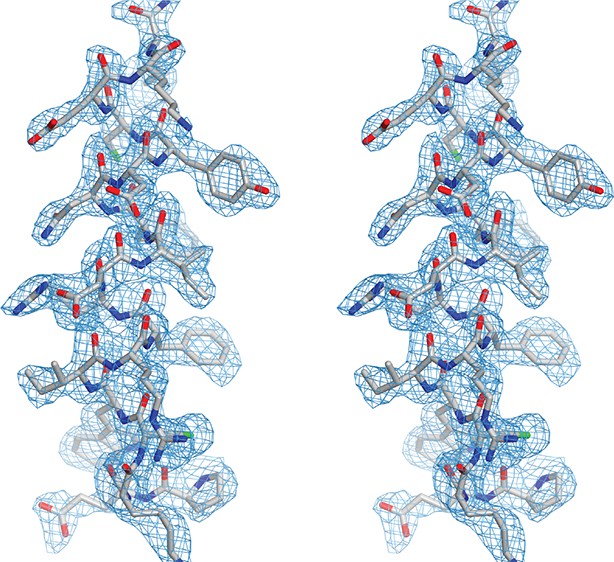
Analysis of the structural model of Drosophila Pur-alpha repeat III.
Stereo view of the helical region of chain B (grey), from lysine 254 to proline 235 with (2Fo-Fc) electron-density map (blue).
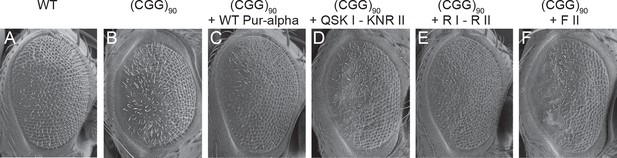
Mutations in Pur-alpha’s nucleic-acid-binding domain abolish rescue of CGG RNA-mediated neurodegeneration.
(A–F) Scanning electron microscope pictures of the eyes of adult flies. (A) Wild-type fly, (B) flies expressing (CGG)90-EGFP/+ alone, together with wild-type Pur-alpha (C), with the QSK I – KNR II mutant (D), with the previously published R I – R II mutant (E) (Graebsch et al., 2009) and with the F II mutant (F). Only wild-type Pur-alpha and the R I – R II mutant can rescue the neurodegeneration induced by rCGG repeats.
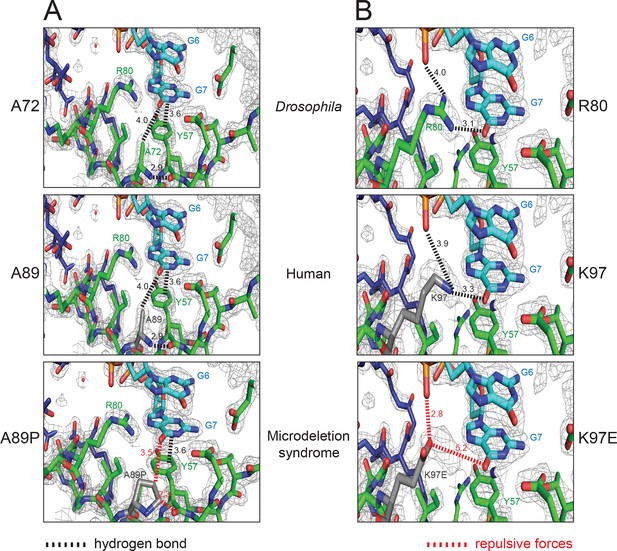
Pur-alpha mutations found in the 5q31.3 microdeletion syndrome can be modeled into the crystal structure of Drosophila Pur-alpha repeat I-II (green) in complex with DNA (cyan).
(A) Residue A72 of the Drosophila protein (top) corresponds to the residue A89 (grey) of the human protein modeled into the Drosophila crystal structure (middle). In both species, the alanines form backbone hydrogen bonds. In the microdeletion syndrome A89 is mutated to proline, which disrupts backbone interactions (bottom). (B) Residue R80 of the Drosophila protein (top) corresponds to the residue K97 (grey) of the human protein, which was modeled into the crystal structure (middle). Both R80 and K97 are positively charged residues. In Drosophila R80 interacts with the guanine G7. The same interaction is likely to be mediated by K97. In the microdeletion syndrome, K97 is mutated to a glutamate, which probably impairs nucleic-acid binding due to its negative charges (bottom). A list of all published mutations in human Pur-alpha leading to the 5q31.3 microdeletion syndrome is shown in Figure 6—source data 1. In this table, their predicted effects on the structure and function of Pur-alpha are also indicated.
-
Figure 6—source data 1
Mutations in the gene encoding for human Pur-alpha that result in the 5q31.3 microdeletion syndrome.
Amino acids in brackets indicate the corresponding positions in Drosophila Pur-alpha, based on the protein-sequence alignment shown in Figure 2—figure supplement 4. “p.” indicates a point mutation, “*” a stop codon, “fs” a frame shift followed by the number of residues produced until the next stop codon, “del” a deletion, and “delins” a combination of deletion and insertion.
- https://doi.org/10.7554/eLife.11297.020
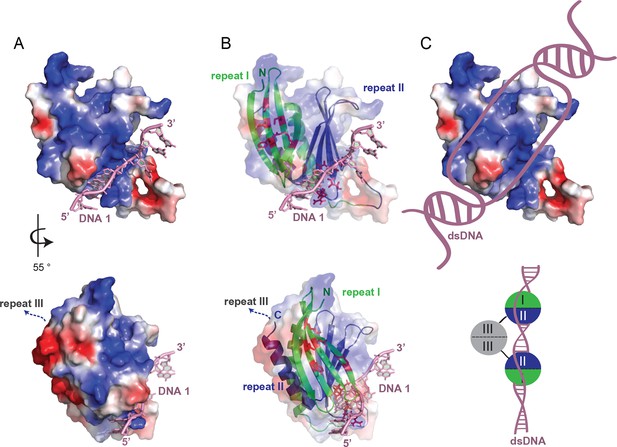
Model for unwinding of dsDNA by full-length Pur-alpha.
(A, top) Electrostatic surface model of Pur-alpha repeat I-II in complex with one ssDNA molecule (pink). Red and blue colorations of the surface indicate negative and positive electrostatic potentials, respectively. (B, top) Cartoon shows in addition the structure model of PUR repeat I (green) and II (blue). DNA interaction sites, seen in the crystal structure, are shown as red sticks and correspond to the residues highlighted in Figure 2A. (C, top) Model showing the most likely overall trajectory of dsDNA (pink) when bound to Pur-alpha repeat I-II. The double-strand is locally unwound and the two separated strands bind to the two opposing binding sites on the protein. (A, B, bottom) Representation as in (A, B, top), additionally showing the C-terminus connecting to PUR repeat III. PUR repeat III likely arranges at the opposing site of the nucleic-acid-binding region. (C, bottom) Schematic drawing of an intermolecular Pur-alpha dimer bound to dsDNA (pink). PUR repeat III (grey) mediates dimerization, potentially orienting both nucleic-acid-binding domains (repeat I, green and II, blue) to the dsDNA. There both PUR domains could unwind larger regions of the DNA.
Videos
Movie of the crystal structure of Drosophila Pur-alpha repeat I-II in complex with the GCGGCGG ssDNA.
https://doi.org/10.7554/eLife.11297.012Tables
Data collection/processing and refinement statistics (molecular replacement) for the two crystal structures of Drosophila Pur-alpha repeat I-II/DNA co-complex and Pur-alpha repeat III alone.
| Pur-alpha repeat I-II/DNA | Pur-alpha repeat III | |
| Data collection/processing | ||
| PDB ID Beamline Wavelength (Å) Detector Distance (mm) Number of images Oscillation range (°) Space group | 5FGP ESRF ID23-2 0.8726 265.433 144 2.5 P21212 | 5FGO ESRF ID14-1 0.9334 261.345 180 1.0 P 1 21 1 |
| Cell dimensions | ||
| a, b, c (Å) | 81.9, 40.2, 48.8 | 61.5, 55.5, 67.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 95.7, 90.0 |
| Resolution (Å) | 50.0-2.0 (2.05-2.0) | 50-2.6 (2.67-2.6) |
| Rsym or Rmerge | 12.5 (79.3) | 11.1 (68.0) |
| I / σI | 18.85 (2.61) | 10.4 (1.97) |
| Completeness (%) | 99.4 (94.3) | 96.5 (98.4) |
| Redundancy | 13.1 (7.6) | 1.9 (1.9) |
| Refinement | ||
| Resolution (Å) | 41.9-2.0 | 47.7-2.6 |
| No. reflections | 11,349 | 14,001 |
| Rwork / Rfree | 16.3 / 21.5 | 20.6 / 28.9 |
| No. atoms | ||
| Protein | 1207 | 3144 |
| Ligand/ion | 145 | - |
| Water | 126 | 112 |
| B-factors | ||
| Protein | 24.8 | 12.5 |
| Ligand | 30.4 | - |
| Water | 35.2 | 9.55 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | 0.01 |
| Bond angles (°) | 1.25 | 1.36 |
| Ramanchandran plot Allowed (%) Additionally allowed (%) Disallowed (%) | 96.0 3.3 0.7 | 93.0 6.5 0.5 |
-
Values in parentheses are for highest-resolution shell.





