Sequestration of host metabolism by an intracellular pathogen
Figures
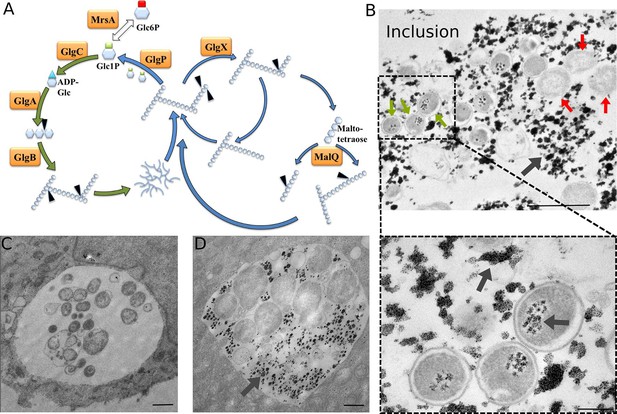
Glycogen accumulation in C. trachomatis inclusions.
(A) Glycogen metabolism in bacteria. In green: glycogen synthesis. In blue: glycogen degradation. Glc1P is the substrate of GlgC for ADP-Glc synthesis. GlgA (glycogen synthase) produces linear glycogen chains via α-1,4 glycosidic bonds, and the branching enzyme (GlgB) introduces branches through α-1,6 linkages. Glycogen depolymerization in Glc1P is the result of the activity of GlgP (glycogen phosphorylase), GlgX (debranching enzyme) and MalQ (4-α-glucanotransferase). The phosphoglucomutase MrsA converts Glc1P to Glc6P. The arrows point to the site within the polysaccharide that is subjected to enzymatic activity. Genes for all these enzymes are present in C. trachomatis. (B) HeLa cells were infected for 30 hr with C. trachomatis. Glycogen was visualized by TEM after PATAg stain. Grey arrows point at glycogen. Green arrows point at examples of EBs and red arrows at RBs. The picture on the bottom shows an enlargement of the boxed region containing three EBs. Scale bars: 1 µm (top), 200 nm (bottom). (C,D) Cells were Glc-deprived 48 hr prior to infection. 10 mg/ml Glc were added 24 hpi and cells were (C) fixed immediately or (D) 4 hr after Glc administration. Note that no glycogen is detectable in the bacteria while it is highly abundant in the inclusion lumen. Scale bar: 1 µm.
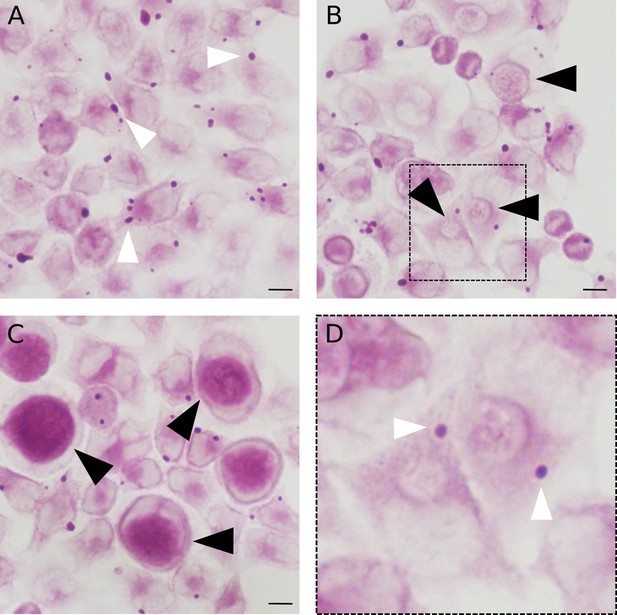
Relocation of glycogen stores to the inclusion during infection.
Cells were (A) non-infected, or infected with C. trachomatis for (B) 24 hr or (C) 48 hr, fixed in PFA and processed for PAS stain. (D) Enlargement of the boxed region in (B) Note that glycogen particles (white arrowheads) are still detected in cells infected for 24 hr but not later, while inclusion glycogen content strongly increases with infection time. Black arrowheads point to examples of inclusions. Scale bar: 10 µm.
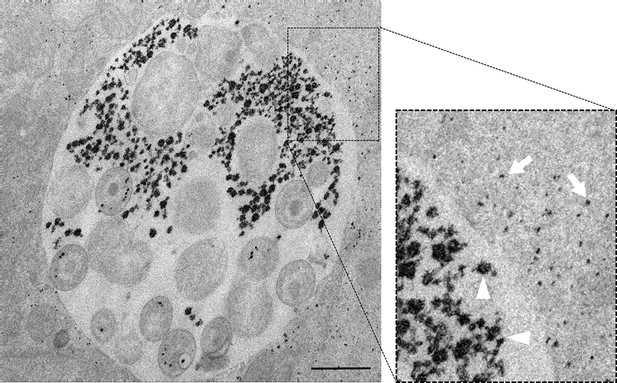
Luminal and cytoplasmic glycogen differs in size.
Cells were infected for 30 hr with C. trachomatis. The picture on the right shows an enlargement of the boxed region. Glycogen is visualized by TEM after PATAg stain. Glycogen deposits in the inclusion lumen (arrowheads) are on average of bigger size than in the host cell cytoplasm (arrows). Scale bar: 1 µm.
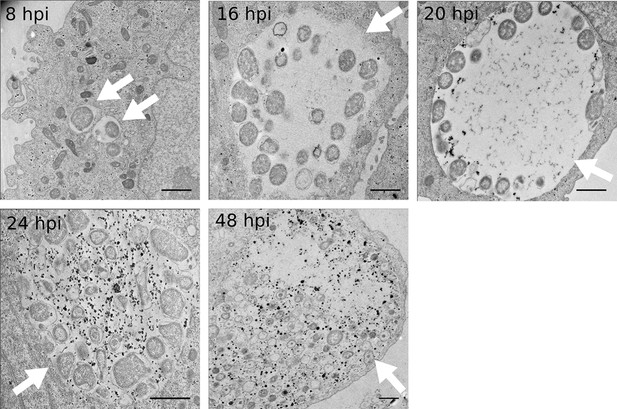
Kinetics of glycogen accumulation.
HeLa cells were infected with C. trachomatis for 8, 16, 20, 24 or 48 hr. Glycogen is visualized by TEM after PATAg stain. White arrows point to inclusions. Glycogen is first detected in the inclusion lumen at 20 hpi, and increases strongly after. Scale bar: 1 µm.
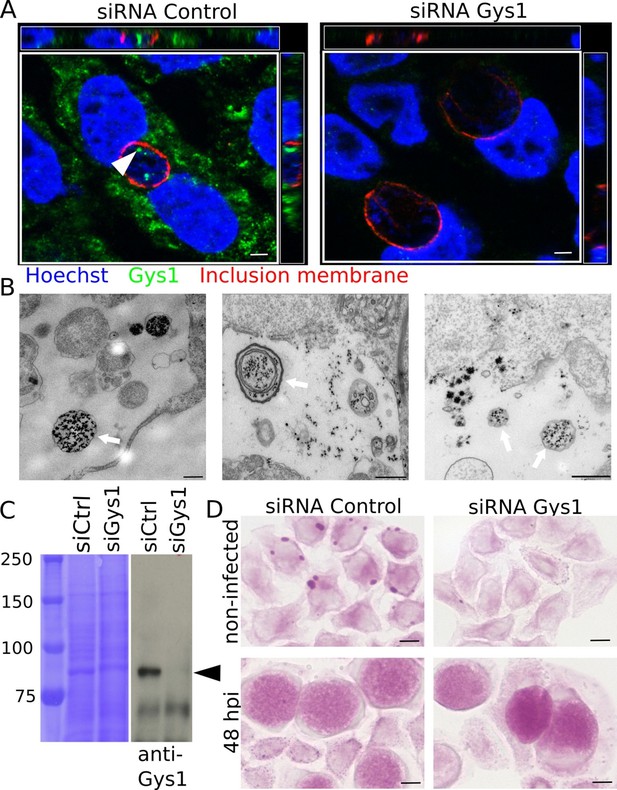
Bulk import of cytoplasmic glycogen contributes to the accumulation of glycogen in the inclusion.
(A) Gys1 is imported into the inclusion lumen. Cells were treated with siRNA control or against Gys1 prior to infection for 30 hr. DNA was stained in blue, Gys1 in green and the inclusion membrane in red (anti-CT813). The white arrowhead points to intraluminal Gys1, see also xz (top) and yz (right) projections. Scale bar: 10 µm. (B) Examples of TEM images of glycogen-filled vesicles (arrows) in inclusions 30 hpi. Glycogen is visualized through PATAg staining. Scale bar: 500 nm. (C,D) Cells were treated with either siRNA control or siRNA against Gys1 48 hr prior to infection. (C) Coomassie staining of whole cell lysates as loading control (left) and immunoblot with an anti-Gys1 antibody (right). The arrowhead points to Gys1 (MW = 85 kDa). (D) PAS on non-infected cells fixed 48 hr after siRNA treatment (top) and on siRNA treated cells infected for 48 hr (bottom). Scale bar: 10 µm.
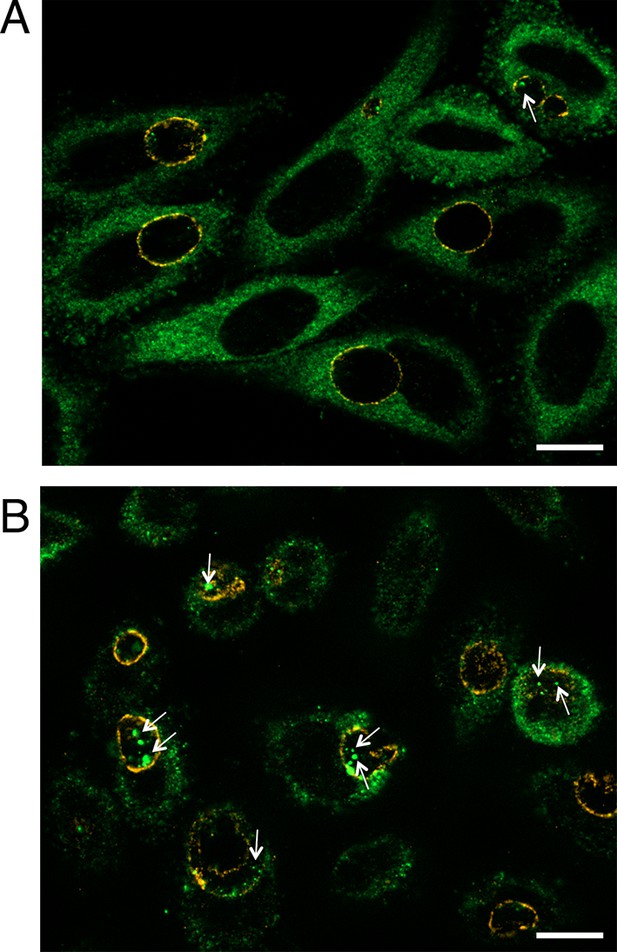
Luminal location of Gys1 strongly decreases in glucose-deprived cells.
HeLa cells were Glc-deprived (A) or not (B, same medium supplemented with 4.5 mg/ml Glc) for 48 hr, prior to infection with C. trachomatis. Gys1 was stained in green and the inclusion membrane in red (anti-CT813). The white arrowheads point to intraluminal Gys1. Scale bar: 5 µm.
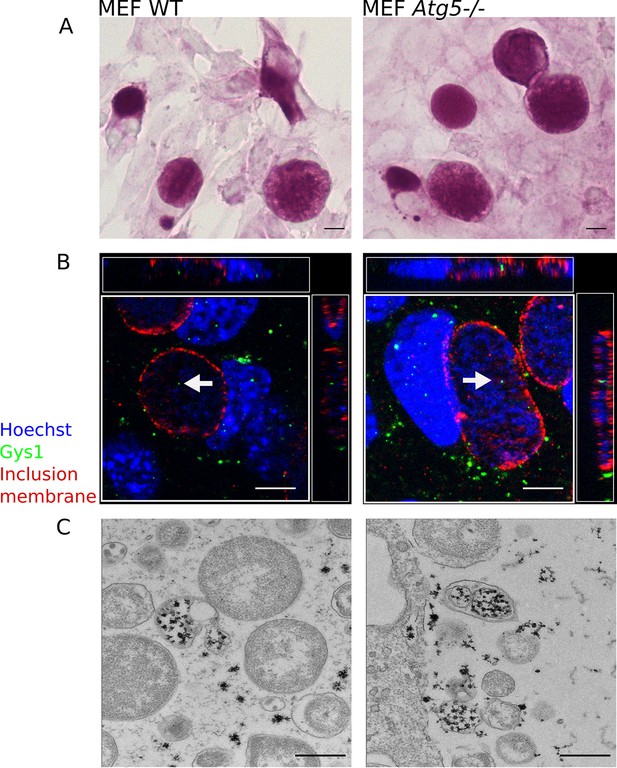
Host glycogen imported in bulk from the host is not of autophagic origin.
Wild-type (WT) or Atg5-/- mouse embryonic fibroblasts (MEFs) were infected for 30 hr with C. trachomatis. (A) PAS staining. Glycogen accumulation was identical in both cell lines. Scale bar: 10 µm. (B) DNA is stained in blue, Gys1 in green and the inclusion membrane in red (anti-CT813). While less abundant than in HeLa cells, Gys1 (white arrows) was detected inside inclusions of both cell lines. Scale bar: 10 µm. (C) Glycogen is visualized by TEM after PATAg stain. Glycogen-filled vesicles were observed in inclusions of both cell lines. Scale bar: 500 nm.
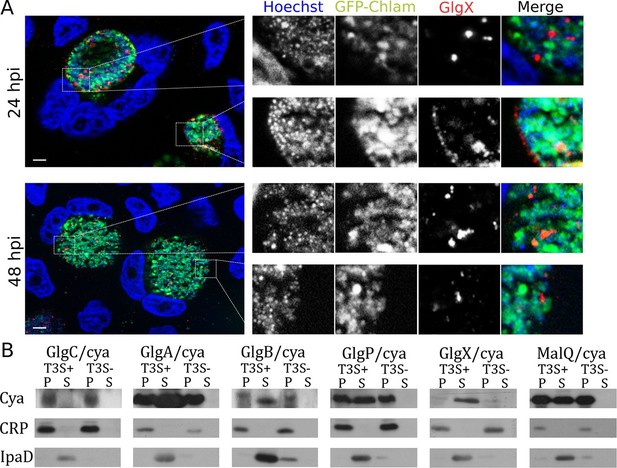
Bacterial glycogen metabolism enzymes are secreted in the inclusion lumen.
(A) HeLa cells were infected for 24 hr or 48 hr with GFP expressing L2 (GFP-Chlam, green) and stained with an anti-GlgX antibody (red) and Hoechst (blue). Insets to the right show enlargements of the boxed areas. Scale bar: 5 µm. (B) Heterologous test of secretion: the N-terminal 20 amino acids of the indicated proteins were fused to the reporter Cya, and constructs were transformed into the S. flexneri strains ipaB (T3S+) and mxiD (T3S-). Liquid cultures were fractionated into pellet (P) and supernatant (S). All chimeras except GlgC/cya were detected in the supernatant in T3S competent bacteria and not in T3S defective bacteria, indicative of a functional T3S signal. The endogenous T3S substrate of S. flexneri, IpaD, serves as a positive control, while the non-secreted cAMP receptor protein (CRP) controls for non-specific leakage into the supernatant.
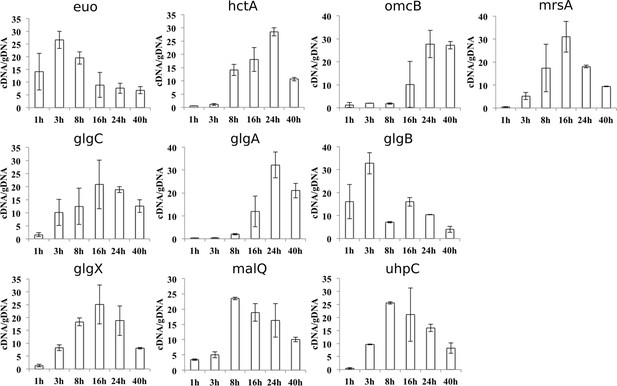
Expression profiles of genes related to glycogen metabolism.
qRT-PCR of selected genes related to glycogen metabolism was performed. Values were plotted against standard curves and cDNA was normalized with the overall chlamydial genomic DNA (gDNA) quantified in parallel samples (cDNA/gDNA). An early chlamydial gene, euo, and two late genes, hctA and omcB, served as controls. Values were obtained from three independent experiments, each performed in triplicates. Error bars depict the standard deviation (n=3).
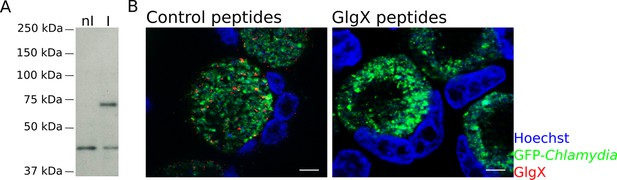
Specificity of the anti-GlgX antibody.
(A) Western Blot of HeLa cells non-infected (nI) or infected (I) with LGV for 24 hr. The anti-GlgX antibody detected a band of the expected molecular weight (73 kDa), only present in the infected sample. The lower band observed does not correspond to a bacterial antigen since it is also present in non-infected cells. (B) Cells were infected for 48 hr and processed for immunofluorescence as described in Figure 3, except that the anti-GlgX antibody was preincubated with either unrelated control peptides, or with the peptides the antibody was raised against. DNA is in blue, bacteria in green and GlgX in red. The GlgX staining disappears upon preincubation with the GlgX peptides, demonstrating its specificity. Scale bar: 10 µm.
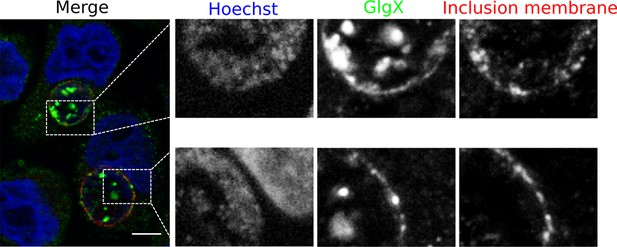
GlgX accumulates at the inclusion membrane.
HeLa cells were infected for 24 hr with LGV, fixed with 3% PFA and stained with the anti-GlgX antibody (green), an antibody against the inclusion protein CT813 (red) and Hoechst (blue). Insets to the right show enlargements of the boxed areas. Scale bar: 10 µm.
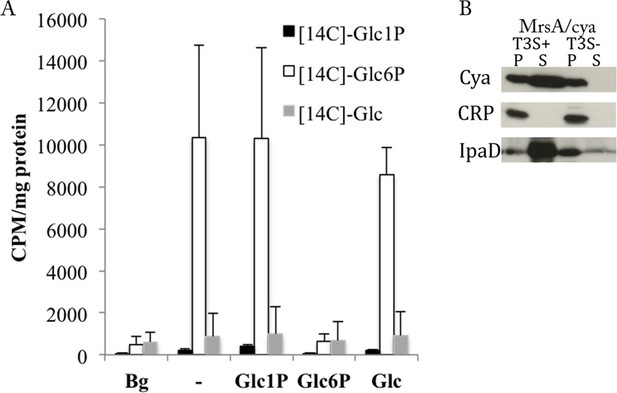
C. trachomatis takes up Glc6P and secretes phosphoglucomutase (MrsA).
(A) Purified EBs were incubated for 2 hr with [14C]-Glc, [14C]-Glc6P or [14C]-Glc1P in absence or presence of a 50-fold excess of non-radioactive Glc, Glc6P or Glc1P. Bacteria were subsequently washed and pelleted and radioactivity measured. Background (Bg) was measured after 1 min of incubation instead of 2 hr. CPM: counts per minute. Results show mean and S.D. (n=3). (B) Heterologous test of secretion was performed on MrsA as in Figure 3B.
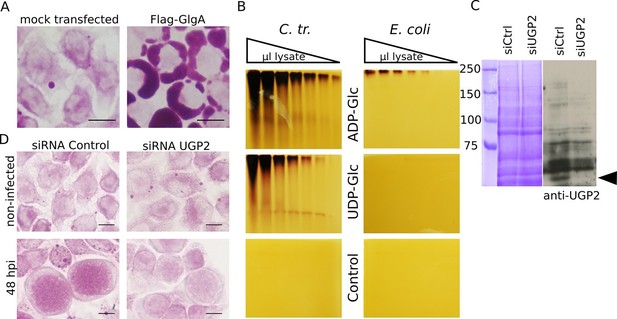
Host UDP-Glc is the substrate for intraluminal glycogen synthesis.
(A) PAS staining was performed 24 hr after transfection with chlamydial Flag-GlgA. (B) Zymogram analysis. Serial dilutions of lysates of E. coli lacking endogenous glgA and transformed with chlamydial glgA (C. tr.) or E. coli glgA (E. coli) were separated by native polyacrylamide electrophoresis and incubated in either 1.2 mM ADP-Glc, UDP-Glc or buffer only (control). Glycogen production was visualized by iodine staining. (C,D) Cells were treated with either siRNA control or siRNA against UGP2 48 hr prior to infection. (C) Coomassie staining of whole cell lysates as loading control (left) and immunoblot with an anti-UGP2 antibody (right). The arrowhead points to UGP2 (MW = 56 kDa). (D) PAS on siRNA treated cells at time of infection (top) and at 48 hpi (bottom). Knocking down UGP2 expression results in the decrease of luminal glycogen accumulation.
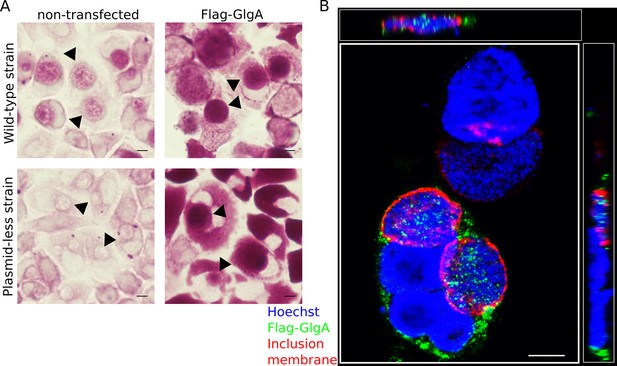
Flag-GlgA is imported into the inclusion lumen and enhances luminal glycogen accumulation.
(A) HeLa cells were transfected (top right) or not (top left) with Flag-GlgA before infection, and fixed 24 hpi. PAS staining revealed an increase of intraluminal glycogen (arrowheads) in Flag-GlgA expressing cells. C. trachomatis has a 8 kb plasmid, and its loss is associated with decreased GlgA expression and impaired glycogen accumulation (Carlson et al., 2008) (bottom left). Remarkably, transfection of Flag-GlgA restored luminal glycogen accumulation in cells infected with the plasmid-less strain LGV 25667R (bottom right). (B) Immunofluorescence on Flag-GlgA transfected cells infected with the wild-type LGV strain. DNA is stained in blue, Flag-GlgA in green and the inclusion membrane in red (anti-Cap1). Flag-GlgA is abundant in the host cytoplasm and the inclusion lumen (see also xz (top) and yz (right) projections). Scale bars: 10 µm.
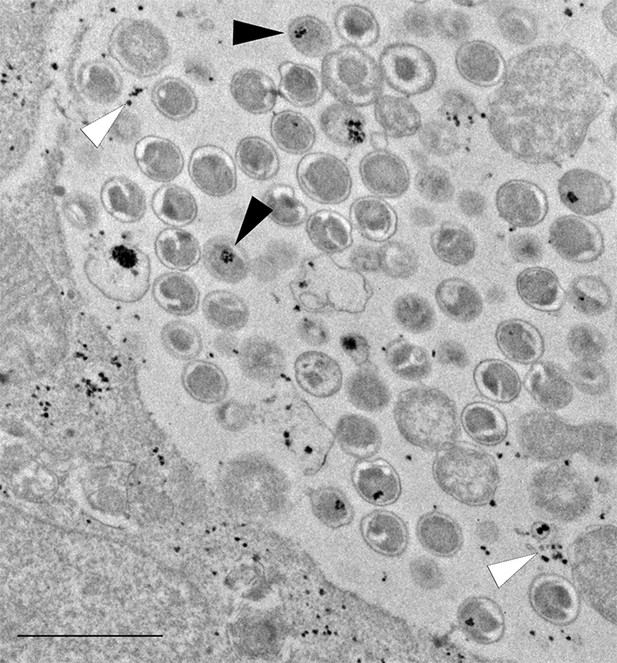
The plasmid-less strain accumulates glycogen in EBs and - to a lesser extent than the wild-type strain - in the inclusion lumen.
Cells were infected for 40 hr with the plasmid-less strain LGV 25667R. Glycogen is visualized by TEM after PATAg stain and is found in EBs (black arrow heads) and free in the inclusion lumen (white arrow heads). Scale bar: 1 µm.
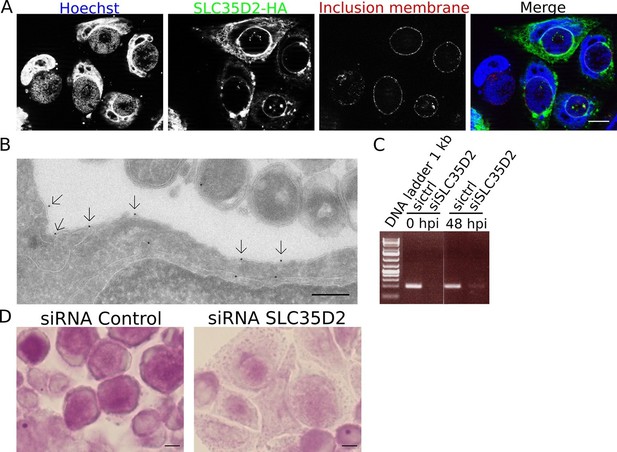
SLC35D2 imports UDP-Glc into the inclusion lumen.
(A, B) Prior to infection cells were transfected with SLC35D2-HA, and fixed 24 hpi. (A) Labelling of the HA tag (green), inclusion membrane marker Cap1 (red) and DNA (blue) show recruitment of SLC35D2 to the inclusion periphery. Scale bar: 10 µm. (B) Duplicate samples were processed for TEM. Staining with anti-HA antibodies, followed with gold-coupled secondary antibodies, showed that SLC35D2-HA is detected on the inclusion membrane (arrows). Scale bar: 250 nm. (C) Cells were treated with siRNA control (ctrl) or siRNA SLC35D2 48 hr and 4 hr prior to infection. Samples were taken before infection (0 hpi) and 48 hpi, and RT-PCR was performed with primers specific to SLC35D2. (D) PAS staining of siRNA ctrl or siRNA SLC35D2 treated cells 48 hpi. Scale bar: 10 µm.
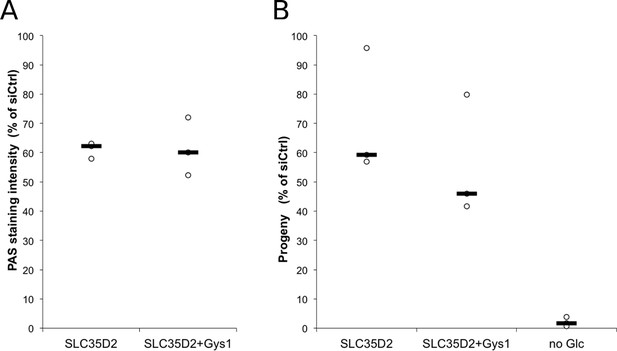
Depletion of Gys1 does not further decrease luminal glycogen content in cells depleted for SLC35D2.
Cells were treated with control siRNA, or siRNA against SLC35D2, or SLC35D2 and Gys1 48 hr and 4 hr prior to infection. (A) Cells were fixed 48 hr after infection and PAS staining was applied. Mean intensity of glycogen staining in inclusions is expressed as a percentage of the mean value measured in cells treated with control siRNA. The median of three independent experiments is shown. (B) Reinfection assays assessing the effect of the indicated knockdowns on infectious progeny formation 30 hpi. Results are expressed as the percentage of progeny measured in cells treated with control siRNA. The median of three independent experiments is shown.
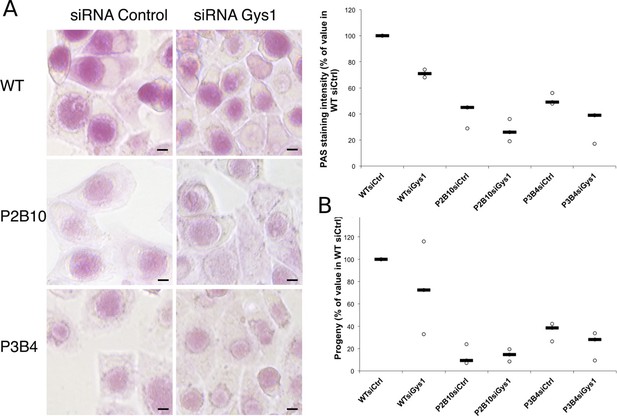
Mutations in GlgA result in defect in glycogen storage capacity and loss of infectivity.
Cells were treated with either siRNA control or siRNA against Gys1 48 hr and 4 hr prior to infection with glgA mutants P2B10, P3B4 or with the parental wild-type strain. (A) PAS staining was performed 48 hpi. Pictures show representative fields for each strain. Mean intensity of glycogen staining in inclusions was quantified and expressed as a percentage of the mean value measured in cells treated with control siRNA and infected with the parental strain. The median of three independent experiments is shown. Mutations in GlgA resulted in reduced glycogen stores, which were decreased further upon depletion of Gys1. Scale bar: 10 µm. (B) Progeny was determined 48 hpi in a reinfection assay. Results are expressed as the percentage of progeny measured in cells treated with control siRNA and infected with the parental strain. The median of three independent experiments is shown.

Alignment of GlgA sequences of different bacteria.
Alignments were obtained using Clustal W. An asterisk indicates positions which have a single, fully conserved residue. A colon points to conservation between groups of strongly similar properties and the period indicates conservation between groups of weakly similar properties. The amino acids mutated in the strains used in this report are highlighted in grey (H131Y for P2B10 and G386E for P3B4).
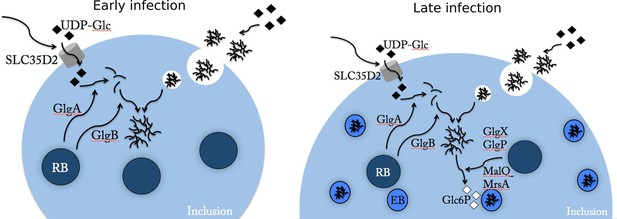
Glc flux in C. trachomatis infected cells.
Early during the infectious cycle (left) the inclusion contains mostly RBs. This developmental form does not accumulate glycogen and uses ATP rather than Glc6P (Omsland et al., 2012). SLC35D2, and possibly other transporters, are recruited to the inclusion membrane and UDP-Glc is translocated into the inclusion lumen. The activity of chlamydial glycogen metabolism enzymes, secreted by RBs into the inclusion lumen, leads to the onset of luminal glycogen synthesis between 16 and 20 hpi. In addition, host glycogen is imported into the inclusion lumen through invagination of the inclusion membrane. In culture cells de novo synthesis predominates over bulk glycogen import. Later on (right), RBs start converting into EBs, which rely on Glc6P as energy source. EBs obtain Glc6P via the degradation of luminal glycogen into Glc1P, subsequently converted to Glc6P by the phosphoglucomutase (MrsA) and imported by UhpC. During RB to EB conversion T3S is turned off, allowing for intrabacterial activity of the glycogen metabolism enzymes, and glycogen accumulation in the bacteria.
Additional files
-
Supplementary file 1
Primers used for cloning purposes.
- https://doi.org/10.7554/eLife.12552.023
-
Supplementary file 2
List of siRNAs.
- https://doi.org/10.7554/eLife.12552.024
-
Supplementary file 3
List of primers used in qRT-PCR and RT-PCR.
- https://doi.org/10.7554/eLife.12552.025





