Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue
Figures
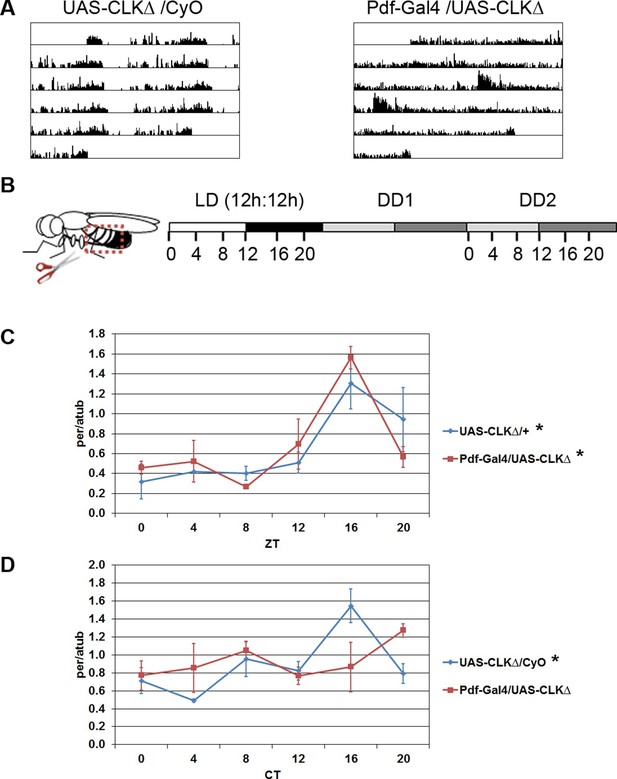
Oscillations of per in the fat body require an intact central clock in the absence of external cues.
(A) Representative double-plotted activity records of individual control UAS-CLKΔ/CyO (left) and Pdf-GAL4/UAS-CLKΔ (right) flies over the course of 5 days in constant darkness. (B) Schematic of experimental design. Male flies, aged 7–10 days, were entrained for several days in 12 hr light: 12 hr dark cycles (LD). Male flies were dissected to obtain abdominal fat bodies (dotted red box) either on the last day in LD or on the second day of constant darkness (DD2). Graphs depict mRNA levels, normalized to α−tubulin (atub), over the course of the day in the presence of light (LD; Zeitgeber Time, ZT) or in constant darkness (DD2; Circadian Time, CT). Ablating the central clock (Pdf-GAL4/UAS-CLKΔ) (red line) does not affect per rhythms in LD (C) but abolishes per rhythms in DD2 compared to controls (blue line) (D). Each experiment was repeated independently three times. The average value for each timepoint is plotted with error bars denoting the standard error of the mean (SEM). Significant rhythmicity was determined using JTK_cycle. Asterisk (*) adjacent to genotype label indicates JTK_cycle p value <0.05. See Table 3 for JTK cycle values.
-
Figure 1—source data 1
Data for behavioral analysis and for qPCR analysis of per in Pdf-GAL4/UAS-CLKΔ flies.
- https://doi.org/10.7554/eLife.13552.004
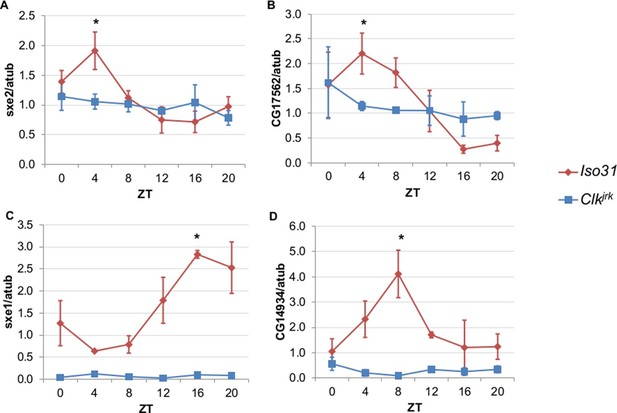
Rhythmic expression of genes that cycle independently of the fat body clock requires clocks in other tissues.
Daily oscillations of several fat body clock-independent genes were tested in male mutants lacking functional clocks in all tissues, Clkjrk mutants, in LD. Rhythmicity of sxe2 (A), CG17562 (B), sxe1 (C), and CG14934 (D) is abolished in Clkjrk mutants but is intact in Iso31 wild type controls. All genes were normalized to α−tubulin (atub) levels. Each experiment was repeated independently three times. The average value for each timepoint is plotted with error bars denoting SEM. JTK_cycle p value <0.05 is indicated by an asterisk (*) at the time of peak expression. See Table 3 for JTK_cycle p values. ZT- Zeitgeber Time.
-
Figure 2—source data 1
Data for qPCR analysis of fat body clock-independent genes in Clkjrk mutants.
- https://doi.org/10.7554/eLife.13552.008
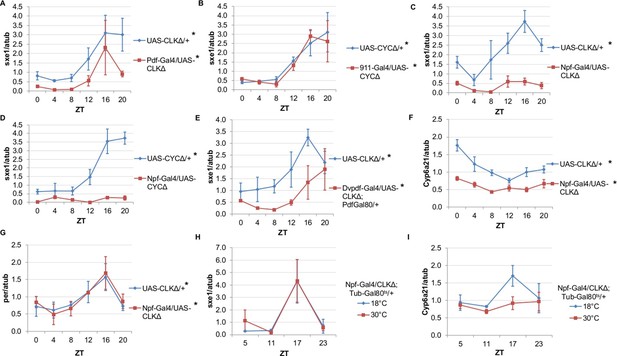
NPF-expressing clock neurons regulate rhythmic expression of fat body genes, sxe1 and Cyp6a21.
(A, B) Ablating the molecular clock by expressing CLKΔ or CYCΔ in either the LNvs (Pdf-GAL4) (A) or DN1s (911-GAL4) (B) does not eliminate rhythmic sxe1 expression in the fat body. (C, D) Expressing CLKΔ (C) or CYCΔ (D) using Npf-GAL4 abolishes rhythmic sxe1 expression in the fat body. (E) Expressing CLKΔ in a subset of LNds (Dvpdf-GAL4;Pdf-GAL80) also does not eliminate cycling but reduces sxe1 expression in the fat body. (F) Npf-GAL4>UAS-CLKΔ abolishes rhythmic Cyp6a21 expression in the fat body. (G) per expression is rhythmic in flies expressing UAS-CLKΔ under Npf-GAL4. (H, I) CLKΔ expression in NPF cells is restricted to adulthood using Tub-GAL80ts. (H) sxe1 expression is not affected with adult-specific clock ablation in NPF cells. (I) Rhythmic Cyp6a21 expression is affected in the fat body when Npf-GAL4>UAS-CLKΔ expression is induced in adult at 30°C. Each experiment was repeated independently at least twice. The average value for each timepoint is plotted with error bars denoting SEM. JTK_cycle p value <0.05 is indicated by an asterisk (*) next to the genotype label. See Table 3 for JTK_cycle p values. ZT- Zeitgeber Time.
-
Figure 3—source data 1
Data for qPCR analysis of fat body clock-independent genes and clock genes in flies with ablated clock neurons.
- https://doi.org/10.7554/eLife.13552.010
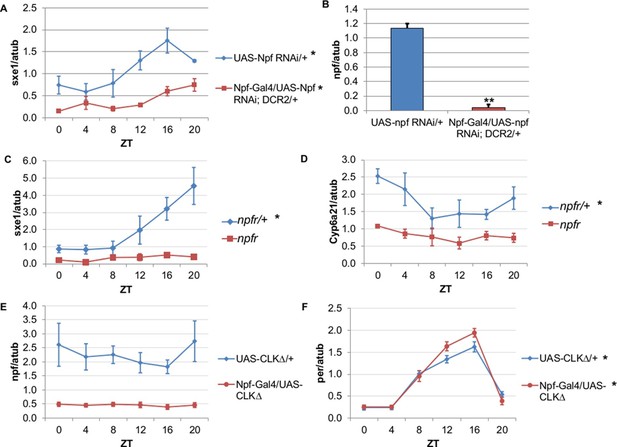
NPF is a critical circadian signal for sxe1 and Cyp6a21 rhythms in the fat body.
(A) Knockdown of npf in all NPF-positive cells does not eliminate rhythmicity but reduces expression of sxe1 in the fat body at all times. (B) Analysis of npf knockdown efficiency in heads of Npf-GAL4/UAS-npf RNAi; DCR2 (UAS-Dicer2) flies showed a significant reduction in npf levels by Student’s t-test (**p<0.001). (C, D) sxe1 and Cyp6a21 expression in the fat body are reduced and do not cycle in homozygous npfr mutants compared to heterozygous controls. (E) npf levels in the heads of Npf-GAL4/UAS-CLKΔ are reduced compared to controls (UAS-CLKΔ/+). (F) Total per levels are not altered in the heads of Npf-GAL4/UAS-CLKΔ compared to controls. Each experiment was repeated independently three times except for (B) which n=6 for each genotype. The average value ± SEM for each timepoint is plotted. JTK_cycle p value <0.05 is indicated by an asterisk (*) next to the genotype label. See Table 3 for JTK_cycle p values. ZT- Zeitgeber Time.
-
Figure 4—source data 1
Data for qPCR analysis of fat body clock-independent genes and clock genes in flies with perturbed NPF signaling.
- https://doi.org/10.7554/eLife.13552.012
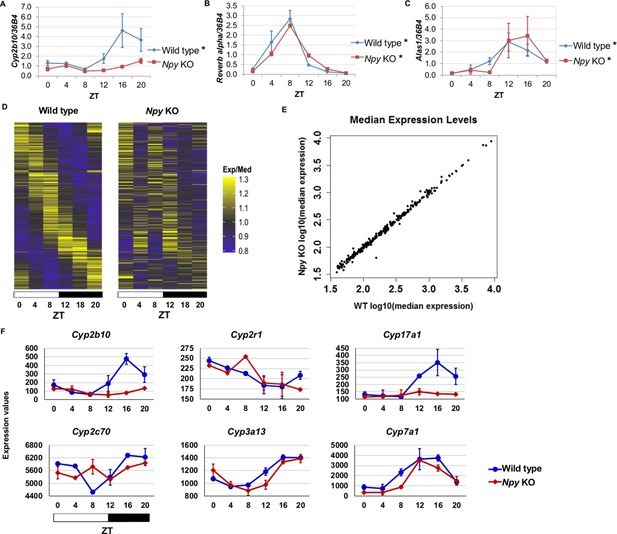
Npy regulates circadian expression of cytochrome P450 genes in the murine liver.
(A–C) Quantitative PCR analysis in murine livers. Daily oscillations of Cyp2b10 expression (A) are abolished in Npy KOs compared to their background controls (wild type), while oscillations of the circadian gene, Reverb alpha (B), are unaffected. (C) Levels of another liver clock-independent gene, Alas1, are similar in wild type and Npy KO, suggesting Npy does not regulate its rhythmicity. For qPCR data, n=3–4 mice for each genotype and time point. Transcript levels were normalized to the housekeeping gene 36B4. (D–F) Microarray analysis was used to detect transcript expression in livers of Npy KO and their background controls collected over the course of 24 hr in LD. (D) The heatmap includes transcripts that oscillate in wild type but not in Npy KO liver. Data represent the average transcript abundance from n=2 samples for each genotype and timepoint. Here, the MetaCycle p-value cutoff of p<0.01 was used to identify cyclic transcripts; p>0.8 was considered not cyclic. (E) The median expression values of the wild type-only cyclic transcripts are not different between Npy KO and wild type. (F) Daily expression values of cytochrome P450 genes from microarrays. Cytochrome P450 genes Cyp2b10, Cyp2r1, Cyp17a1, and Cyp2c70 are cylic in wild type liver but are not cyclic in Npy KO liver. Cyp3a13 and Cyp7a1 cycle robustly in both wild type and Npy KO. Graphs show average ± SEM. ZT- Zeitgeber Time.
-
Figure 5—source data 1
Data for qPCR and microarray analysis in wild type and Npy knockout mouse liver.
- https://doi.org/10.7554/eLife.13552.014
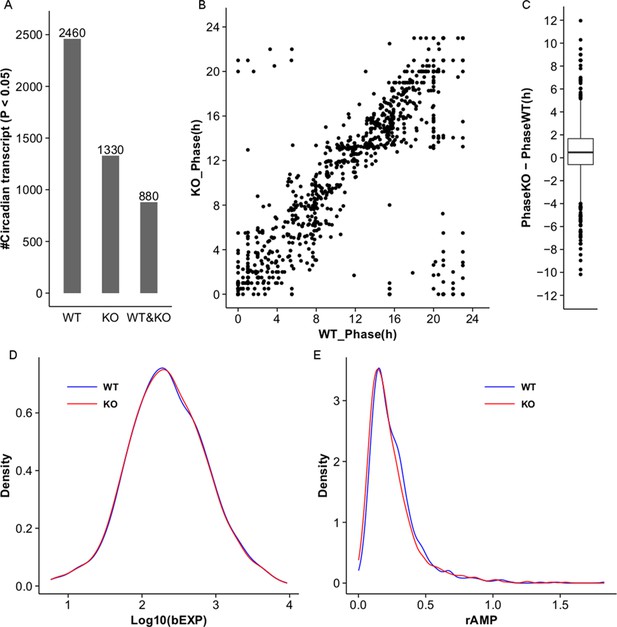
MetaCycle analysis of cycling liver transcripts in wild type and NPY KO.
(A) Microarray analysis detects 2460 and 1330 cycling transcripts (MetaCycle p<0.05) in WT and Npy KO liver datasets respectively. 880 cycling transcripts are common between the two datasets. (B,C) Scatter plot and box plot graph the phase in Npy KO relative to WT for the 880 genes with cycling expression patterns in both datasets. In general, the phase in Npy KO is delayed compared to that in WT. (D) Density plot graphs the distribution of baseline expression levels, bEXP (see Materials and methods), for the 880 cycling transcripts in WT and KO datasets. X-axis graphs the log base 10 of bEXP value. (E) Density plot graphs the distribution of relative amplitudes, rAMP (see Materials and methods), for the 880 cycling transcripts in WT and KO datasets.
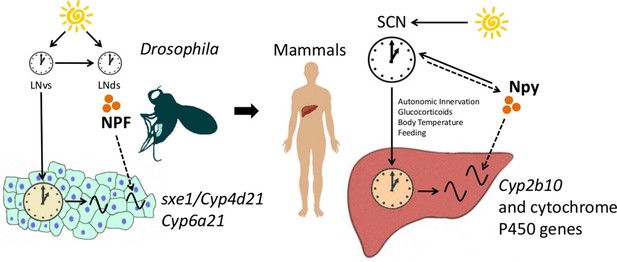
NPF/Npy regulate rhythmically expressed P450 enzymes in the periphery of flies and mammals.
A model of brain clock regulation of peripheral cycling. Brain clocks regulate clocks in peripheral tissues. In Drosophila, clocks in PDF-positive neurons (LNvs) regulate the clock in the fat body. Similarly, in mammals, clocks in the suprachiasmatic nuclei (SCN) have been shown to regulate peripheral clocks such as the liver clock via autonomic innervation, glucocorticoids, body temperature, and feeding. In both the fat body and liver, not all circadian transcripts depend on the local-tissue clock. Clocks in NPF-positive LNds and NPF itself regulate circadian expression of cytochrome P450 enzymes in the fly fat body. The LNvs can influence other brain clocks (such as the LNds), but are not required for rhythms of fat body transcripts in LD as LNds may entrain directly to light. In mammals, Npy was previously known to be a non-photic signal involved in entraining the SCN. However, the SCN could also influence Npy production or release, which in turn drives rhythmic expression of cytochrome P450 enzymes in the liver.
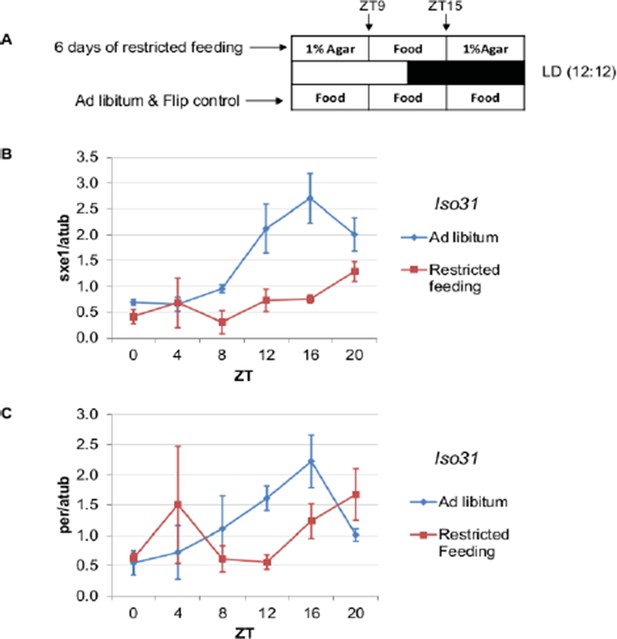
Restricted feeding between ZT9 and ZT15 alters the rhythmic expression of sxe1 and clock gene per in the Drosophila fat body.
https://doi.org/10.7554/eLife.13552.021Tables
Free-running rest:activity rhythms. Clock ablation in Pdf neurons (Pdf-GAL4/UAS-CLKΔ) or in LNd neurons (Dvpdf-GAL4/UAS-CLKΔ; pdf-GAL80/+) disrupts free-running behavioral rhythms in flies. Flies with clock ablation in Npf neurons (Npf-GAL4/UAS-CLKΔ) and npfr mutants have normal free-running rhythms.
| Genotype | n | % Rhythmic | Period (hr) | FFT |
|---|---|---|---|---|
| Pdf-GAL4/UAS-CLKΔ | 39 | 36 | 23.51 | 0.04 |
| UAS- CLKΔ/CyO | 48 | 90 | 23.71 | 0.06 |
| Npf-GAL4/UAS-CLKΔ | 62 | 98 | 24.01 | 0.06 |
| UAS-CLKΔ/+ | 58 | 100 | 23.70 | 0.05 |
| npfr | 39 | 95 | 23.66 | 0.05 |
| npf/+ | 46 | 100 | 23.44 | 0.11 |
| Dvpdf-GAL4/UAS-CLKΔ; pdf-GAL80/+ | 63 | 68 | 25.51 | 0.06 |
| UAS-CLKΔ/+ | 63 | 100 | 23.91 | 0.08 |
-
Table 1—source data 1
Data for circadian analysis of fly behavior in constant darkness.
- https://doi.org/10.7554/eLife.13552.006
MetaCycle statistics for cycling of liver clock-independent genes in wild type and Npy knockout mice. Our Npy KO data compared to the previously reported set of liver transcripts whose expression oscillates independently of the liver clock (Kornmann et al., 2007b). Ten liver clock-independent transcripts require Npy for robust rhythmic expression.
| Liver clock-independent genes with disrupted cycling of expression in Npy KO | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Affymetrix transcript ID | Gene | WT MetaCycle P-value | WT Phase | WT Median Exp | WT Relative Amp | KO MetaCycle P-value | KO Phase | KO Median Exp | KO Relative Amp |
| 17503756 | Rbl2 | 0.0003 | 8.94 | 367.0 | 0.217 | 0.0753 | 9.20 | 338.7 | 0.132 |
| 17287733 | Ddx46 | 0.0007 | 7.77 | 264.2 | 0.229 | 0.1756 | 11.46 | 262.8 | 0.158 |
| 17235227 | Cirbp | 0.0037 | 6.28 | 58.1 | 0.188 | 0.0924 | 7.83 | 54.3 | 0.056 |
| 17311807 | Sqle | 0.0039 | 21.00 | 193.4 | 0.790 | 0.7685 | 9.43 | 229.9 | 0.310 |
| 17365314 | Ldb1 | 0.0067 | 10.38 | 234.1 | 0.164 | 0.1503 | 12.05 | 216.2 | 0.171 |
| 17475360 | Cyp2b10 | 0.0102 | 17.75 | 155.9 | 0.869 | 0.0782 | 0.00 | 85.7 | 0.486 |
| 17331429 | Actg1 | 0.0153 | 18.56 | 155.7 | 0.470 | 0.3570 | 1.52 | 124.5 | 0.512 |
| 17290173 | Hmgcs1 | 0.0230 | 0.50 | 751.5 | 0.687 | 0.1625 | 4.83 | 695.2 | 0.083 |
| 17239493 | Heca | 0.0265 | 9.30 | 255.6 | 0.132 | 0.1921 | 10.93 | 258.6 | 0.086 |
| 17232235 | Ctgf | 0.0331 | 13.18 | 37.9 | 0.364 | 0.9984 | 11.52 | 42.8 | 0.152 |
| Liver clock-independent gene with altered phase of expression in Npy KO | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Affymetrix transcript ID | Gene | WT MetaCycle P-value | WT Phase | WT Median Exp | WT Relative Amp | KO MetaCycle P-value | KO Phase | KO Median Exp | KO Relative Amp |
| 17268729 | Fbxl20 | 0.0001 | 7.66 | 72.8 | 0.338 | 0.0287 | 3.35 | 74.6 | 0.024 |
-
Exp, expression level; Amp, amplitude.
-
Table 2—source data 1
Microarray data for MetaCycle analysis.
- https://doi.org/10.7554/eLife.13552.017
JTK_Cycle Statistics and Two-factor ANOVA Results. All qPCR data were tested for circadian rhythmicity with JTK_cycle test and two-way ANOVA for repeated measures and a Tukey’s post hoc test. P-values from these tests are summarized.
| Figure | Genotype | Gene | Tissue | JTK_cycle P-value | Time P-value | Genotype P-value | Time X Genotype P-value |
|---|---|---|---|---|---|---|---|
| 1C | [ZT] UAS-CLK∆/+ | per | Fat Body (FB) | 0.0194 | < 0.0001 | 0.7826 | 0.4679 |
| 1C | [ZT] Pdf-GAL4/UAS-CLK∆ | per | FB | 0.0094 | |||
| 1D | [CT] UAS-CLK∆/+ | per | FB | 0.0041 | 0.0313 | 0.6650 | 0.0281 |
| 1D | [CT] Pdf-GAL4/UAS-CLK∆ | per | FB | 1 | |||
| 2A | Iso31 | sxe2 | FB | 0.0014 | 0.0131 | 0.1852 | 0.0843 |
| 2A | Clkjrk | sxe2 | FB | 1 | |||
| 2B | Iso31 | CG17562 | FB | 0.0014 | 0.0223 | 0.6409 | 0.1746 |
| 2B | Clkjrk | CG17562 | FB | 0.6945 | |||
| 2C | Iso31 | sxe1 | FB | 0.0020 | 0.0019 | <0.0001 | 0.0020 |
| 2C | Clkjrk | sxe1 | FB | 1 | |||
| 2D | Iso31 | CG14934 | FB | 0.0820 | 0.1146 | <0.0001 | 0.0266 |
| 2D | Clkjrk | CG14934 | FB | 0.5429 | |||
| 3A | UAS-CLK∆/+ | sxe1 | FB | 0.0007 | 0.0003 | 0.0049 | 0.7061 |
| 3A | Pdf-GAL4/UAS-CLK∆ | sxe1 | FB | 9.00E-07 | |||
| 3B | UAS-CYC∆/+ | sxe1 | FB | 0.0363 | 0.0006 | 0.7995 | 0.9592 |
| 3B | 911-GAL4/UAS-CYC∆ | sxe1 | FB | 0.0044 | |||
| 3C | UAS-CLK∆/+ | sxe1 | FB | 0.0014 | 0.0060 | <0.0001 | 0.0749 |
| 3C | Npf-GAL4/UAS-CLK∆ | sxe1 | FB | 0.1969 | |||
| 3D | UAS-CYC∆/+ | sxe1 | FB | 0.0029 | <0.0001 | <0.0001 | <0.0001 |
| 3D | Npf-GAL4/UAS-CYC∆ | sxe1 | FB | 1 | |||
| 3E | UAS-CLK∆/+ | sxe1 | FB | 0.0196 | 0.0038 | 0.0017 | 0.5326 |
| 3E | Dvpdf-GAL4/UAS-CLK∆;Pdf-GAL80/+ | sxe1 | FB | 0.0001 | |||
| 3F | UAS-CLK∆/+ | Cyp6a21 | FB | 0.0009 | <0.0001 | <0.0001 | 0.0574 |
| 3F | Npf-GAL4/UAS-CLK∆ | Cyp6a21 | FB | 0.0259 | |||
| 3G | UAS-CLK∆/+ | per | FB | 0.0568 | 0.0057 | 0.8767 | 0.9902 |
| 3G | Npf-GAL4/UAS-CLK∆ | per | FB | 0.0441 | |||
| 4A | UAS-Npf RNAi/+ | sxe1 | FB | 0.0020 | 0.0005 | <0.0001 | 0.1421 |
| 4A | Npf-GAL4/UAS-Npf RNAi | sxe1 | FB | 0.0441 | |||
| 4B | Not analyzed with JTK | npf | Head | - | |||
| 4C | npfr/+ | sxe1 | FB | 0.0128 | 0.0008 | <0.0001 | 0.0038 |
| 4C | npfr | sxe1 | FB | 0.1969 | |||
| 4D | npfr/+ | Cyp6a21 | FB | 0.0916 | 0.0319 | <0.0001 | 0.4182 |
| 4D | npfr | Cyp6a21 | FB | 0.1150 | |||
| 4E | UAS-CLK∆/+ | npf | Head | 1 | 0.7588 | <0.0001 | 0.846 |
| 4E | Npf-GAL4/UAS-CLK∆ | npf | Head | 1 | |||
| 4F | UAS-CLK∆/+ | per | Head | 0.0001 | <0.0001 | 0.1410 | 0.0420 |
| 4F | Npf-GAL4/UAS-CLK∆ | per | Head | 1.19E-05 | |||
| 5A | Wild type | Cyp2b10 | Liver | 0.0010 | 0.0209 | 0.0034 | 0.1640 |
| 5A | Npy Knockout (KO) | Cyp2b10 | Liver | 0.0510 | |||
| 5B | Wild type | Reverbα | Liver | 7.46E-12 | <0.0001 | 0.4979 | 0.0661 |
| 5B | Npy KO | Reverbα | Liver | 7.17E-09 | |||
| 5C | Wild type | Alas1 | Liver | 9.81E-06 | 0.0007 | 0.8476 | 0.8202 |
| 5C | Npy KO | Alas1 | Liver | 0.0018 |
Additional files
-
Supplementary file 1
MetaCycle statistics of microarray data generated in this study.
MetaCycle p-value, phase, baseline expression (baselineEXP), and relative amplitude (relativeAMP) are calculated for each gene in wild type and NPY KO animals.
- https://doi.org/10.7554/eLife.13552.020





