The postsynaptic t-SNARE Syntaxin 4 controls traffic of Neuroligin 1 and Synaptotagmin 4 to regulate retrograde signaling
Figures
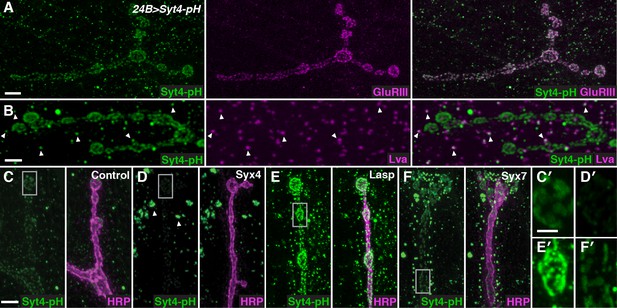
A candidate RNAi screen for regulators of postsynaptic exocytosis.
(A,B) Representative images of Syt4-pH expressed with the postsynaptic muscle driver 24B-GAL4. Syt4-pH (green) accumulates in postsynaptic membrane that also contains domains of GluRIII (magenta) (A). Syt4-pH also decorates numerous cytoplasmic puncta, many of which overlap with the Golgi marker Lva (magenta), arrowheads (B). (C–F) Examples of candidate RNAis affecting Syt4-pH localization: control (C); Syx4-RNAi reduces Syt4-pH at the membrane and causes a redistribution to prominent cytoplasmic puncta, arrowheads (D); Lasp-RNAi increases Syt4-pH at the membrane (E); and Syx7-RNAi causes a redistribution of Syt4-pH puncta around the NMJ without affecting the intensity at the membrane (F). (C′–F′) Close-ups of C–F. Scale bars = 7 μm (A), 5 μm (B–F), 2 μm (C′–F′).
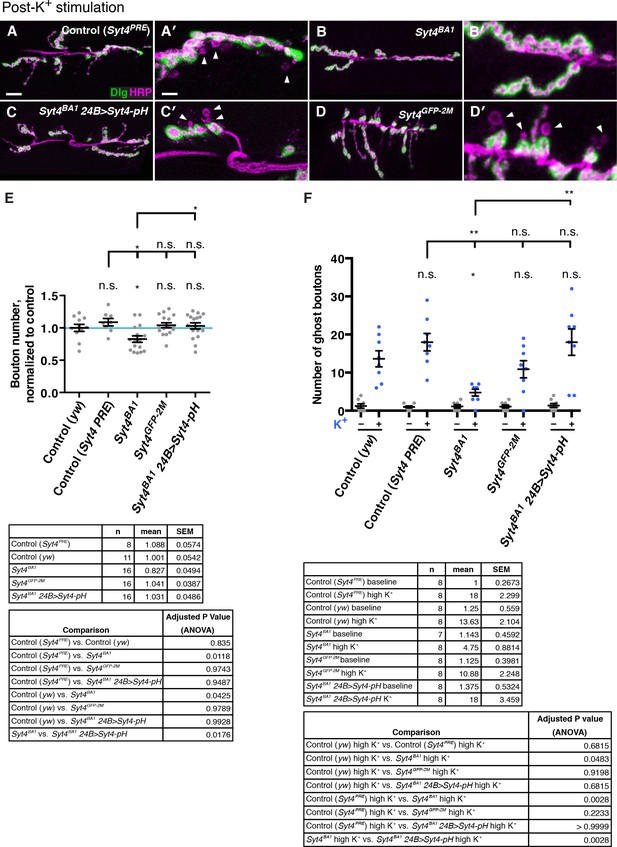
Both Syt4-GFP CRISPR knock-in and overexpression of Syt4-pH can replace endogenous Syt4 with respect to synaptic architecture and plasticity.
(A–D) Representative images of NMJs stained with antibodies to HRP (magenta) and the postsynaptic marker Dlg (green) to highlight synaptic boutons. Acute budding of new varicosities (“ghost boutons”) was stimulated with spaced incubations in high K+. Ghost boutons are identified as round HRP+ structures lacking Dlg signal (arrowheads). Images are shown from the control genotype Syt4PRE (A,A′), a precise excision line that serves as a genetic background control for the Syt4BA1 allele (B,B′). Also shown are images from animals expressing Syt4-pH postsynaptically in the Syt4 null background (Syt4BA1 24B>Syt4-pH; C,C′), and the CRISPR GFP knock-in line Syt4GFP-2M (D,D′). (E) Quantification of bouton number normalized to yw, a genetic background control for Syt4GFP-2M. Blue line indicates the yw control mean. Data are presented as mean ± SEM. (F) Quantification of ghost bouton number per NMJ from animals without (−) or with (+) high K+ stimulation. Data are presented as mean ± SEM. Syt4BA1 24B>Syt4-pH animals have a normal number of boutons and exhibit normal budding of ghost boutons compared to the Syt4PRE control, and are significantly rescued compared to Syt4BA1. Syt4GFP-2M animals have a normal number of boutons and exhibit normal budding of ghost boutons compared to the yw control line. Scale bars = 20 μm (A–D), 6.7 μm (A′–D′). Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in (E) and (F).
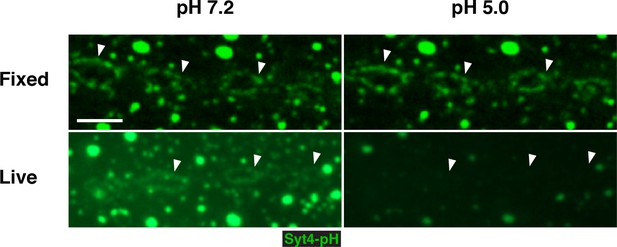
pHluorin is quenched in live but not fixed preparations.
Animals expressing Syt4-pH in the postsynaptic cell (24B>Syt4-pH) were dissected and imaged live or following fixation in paraformaldehyde. The same animal was imaged first following incubation in pH 7.2 HL3.1 buffer and second following incubation in pH 5.0 HL3.1 buffer. Arrows indicate plasma membrane accumulations of Syt4-pH. Scale bars = 2.5 μm.
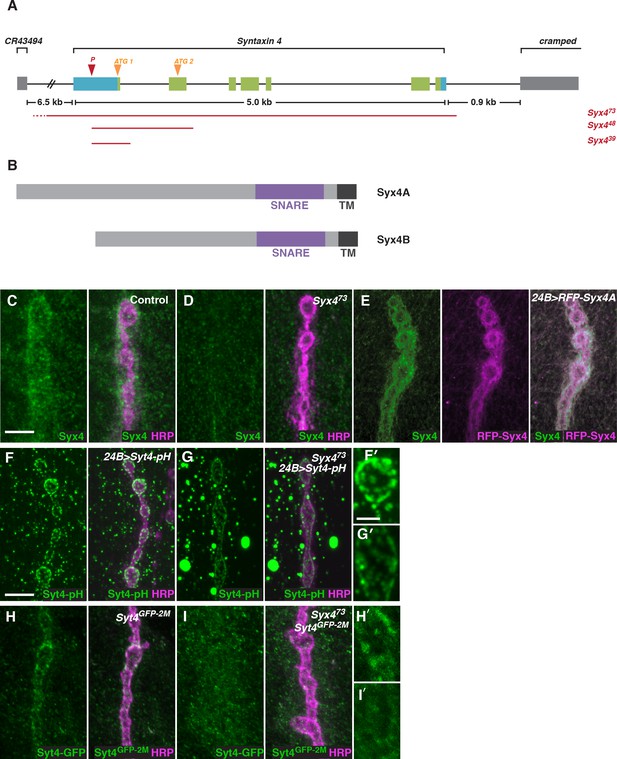
Syntaxin 4 is a postsynaptic plasma membrane SNARE.
(A) Syx4 genomic region. Coding exons are indicated in green while non-coding exons are in blue. Two predicted start sites (ATG) are indicated in orange. The location of the P-element used for mutagenesis (P) is indicated in red. Three alleles of Syx4 were isolated. Deleted regions are indicated in red. Solid lines indicate regions known to be deleted from PCR analysis and sequencing, while dotted lines indicate regions within which breakpoints have been mapped. (B) Syx4 encodes a protein with an N-terminal domain, a SNARE domain and a C-terminal transmembrane domain. There are two predicted isoforms that differ in the size of the N-terminal domain. (C,D) Representative images of NMJs stained for Syx4 (green) and the neuronal membrane marker HRP (magenta). Syx4 staining at the synapse in precise excision control animals (C) is absent in Syx473 mutant animals (D). (E) Representative image from an animal stained for Syx4 (green) and expressing RFP-Syx4 (magenta) with 24B-GAL4. (F,G) Representative images from animals expressing Syt4-pH with 24B-GAL4 in a control (F) or Syx473 (G) background. Syt4-pH is reduced at the postsynaptic membrane and redistributed to large cytoplasmic accumulations in Syx473 mutants. (F′,G′) Close-ups of F and G. (H,I) Representative images from Syt4GFP-2M knock-in animals in a control (H) or Syx473 (I) background. Synaptic localization of Syt4GFP-2M is reduced in Syx473 mutants. (H′,I′) Close-ups of H and I. Scale bars = 5 μm (C–I), 2 μm (F′,G′,H′,I′).
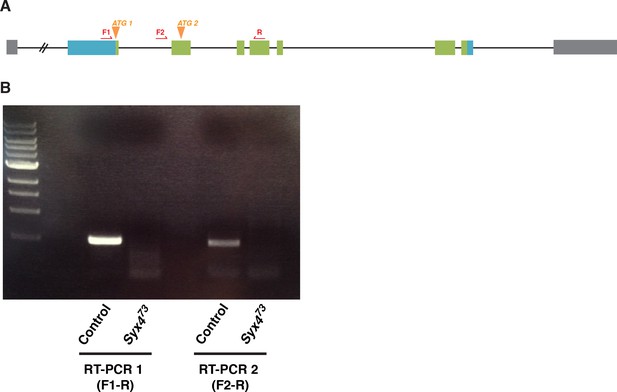
RT-PCR analysis of Syntaxin 4.
Primers (red arrows) were designed to distinguish Syx4A and Syx4B transcripts by RT-PCR. F1 and R amplify a product from Syx4A transcript and F2 and R amplify a product from Syx4B transcript. Both transcripts are detected in control animals and both are absent from Syx473 nulls.
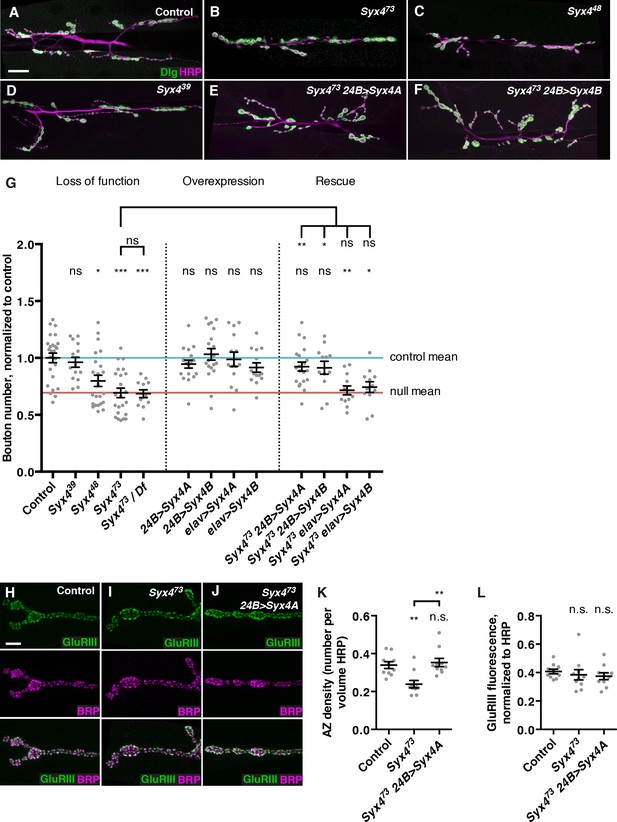
Syntaxin 4 regulates synaptic growth at the NMJ.
(A–F) Representative images of NMJs stained with antibodies to the postsynaptic marker Dlg (green) and the neuronal membrane marker HRP (magenta) to highlight the number of synaptic boutons; images are shown from precise excision control (A), Syx473 (B), Syx448 (C), Syx439 (D), Syx473 24B>Syx4A (E), and Syx473 24B>Syx4B (F) animals. (G) Quantification of bouton number, normalized to controls. Blue line indicates the control mean. Red line indicates Syx473 null mean. Data are presented as mean ± SEM. (H–J), Representative images of NMJs stained with antibodies to GluRIII (green) and the AZ marker Brp (magenta); images are shown from precise excision control (H), Syx473 (I), and Syx473 24B>Syx4A (J) animals. (K), Quantification of AZ density, calculated as the number of AZs per volume HRP. Data are presented as mean ± SEM. (L) Quantification of GluRIII fluorescence per HRP fluorescence. Data are presented as mean ± SEM. Scale bars = 20 μm (A–F), 5 μm (H–J). Statistical comparisons are fully described in Figure 3—source data 1, and are indicated here as ***p<0.001, **p<0.01, *p<0.05, ns = not significant; comparisons are with control unless indicated.
-
Figure 3—source data 1
Statistical data for Figure 3.
Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in Figure 3G,K, and L.
- https://doi.org/10.7554/eLife.13881.010
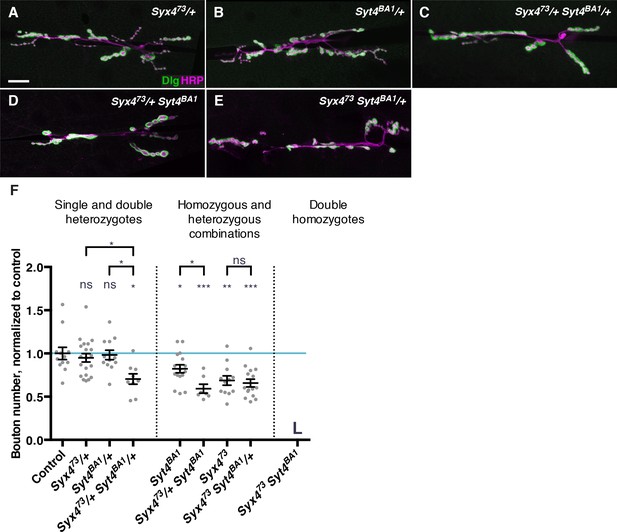
Genetic interactions between Syntaxin 4 and Synaptotagmin 4.
(A–E) Representative images of NMJs stained with antibodies to the postsynaptic marker Dlg (green) and the neuronal membrane marker HRP (magenta) to highlight the number of synaptic boutons; images are shown from Syx473/+ (A), Syt4BA1/+ (B), Syx473/+ Syt4BA1/+ (C), Syx473/+ Syt4BA1 (D), and Syx473 Syt4BA1/+ (E) animals. (F) Quantification of bouton number, normalized to controls. Blue line indicates the control mean. Data are presented as mean ± SEM. L = lethal. Scale bars = 20 μm (A–E). Statistical comparisons are fully described in Figure 4—source data 1, and are indicated here as ***p<0.001, **p<0.01, *p<0.05, ns = not significant; comparisons are with control unless indicated.
-
Figure 4—source data 1
Statistical data for Figure 4.
Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in Figure 4F.
- https://doi.org/10.7554/eLife.13881.012
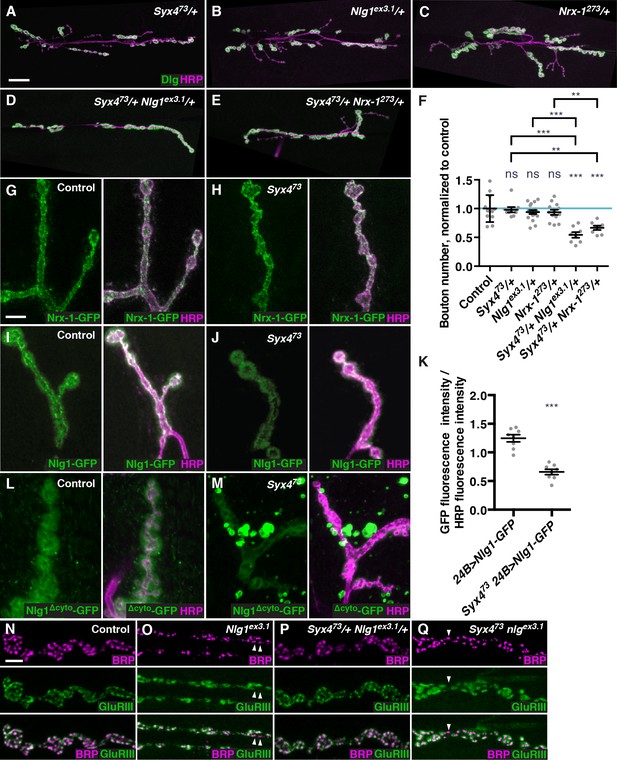
Syntaxin 4 interacts with Neuroligin 1 and regulates its membrane localization.
(A–E), Representative images of NMJs stained with antibodies to the postsynaptic density marker Dlg (green) and the neuronal membrane marker HRP (magenta) to highlight the number of synaptic boutons; images are shown from Syx473/+ (A), Nlg1ex3.1/+ (B), Nrx-1273/+ (C), Syx473/+ Nlg1ex3.1/+ (D), and Syx473/+ Nrx-1273/+ (E) animals. (F) Quantification of bouton number, normalized to controls. Blue line indicates the control mean. Data are presented as mean ± SEM. (G–H), Representative images of NMJs stained with antibodies against HRP (magenta) and expressing Nrx-1-GFP in a control (G) or Syx473 mutant (H) background. (I–J) Representative images of NMJs stained with antibodies against HRP (magenta) and expressing Nlg1-GFP in a control (I) or Syx473 mutant (J) background. (K) Quantification of GFP fluorescence per HRP fluorescence from animals expressing Nlg1-GFP in a control or Syx473 mutant background. Data are presented as mean ± SEM. (L–M) Representative images of NMJs stained with antibodies against HRP (magenta) and expressing Nlg1Δcyto-GFP in a control (L) or Syx473 mutant (M) background. (N–Q) Representative images of NMJs stained with antibodies against Brp (magenta) and GluRIII (green), from precise excision control (N), Nlg1ex3.1 (O), Syx473/+ Nlg1ex3.1/+ (P), and Syx473 Nlg1ex3.1 (Q) animals. Arrowheads indicate AZs lacking an apposed GluR field. Scale bars = 20 μm (A–E), 5 μm (I,J,L–Q). Statistical comparisons are fully described in Figure 5—source data 1, and are indicated here as ***p<0.001, **p<0.01, *p<0.05, ns = not significant; comparisons are with control unless indicated.
-
Figure 5—source data 1
Statistical data for Figure 5.
Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in Figure 5F and K.
- https://doi.org/10.7554/eLife.13881.014
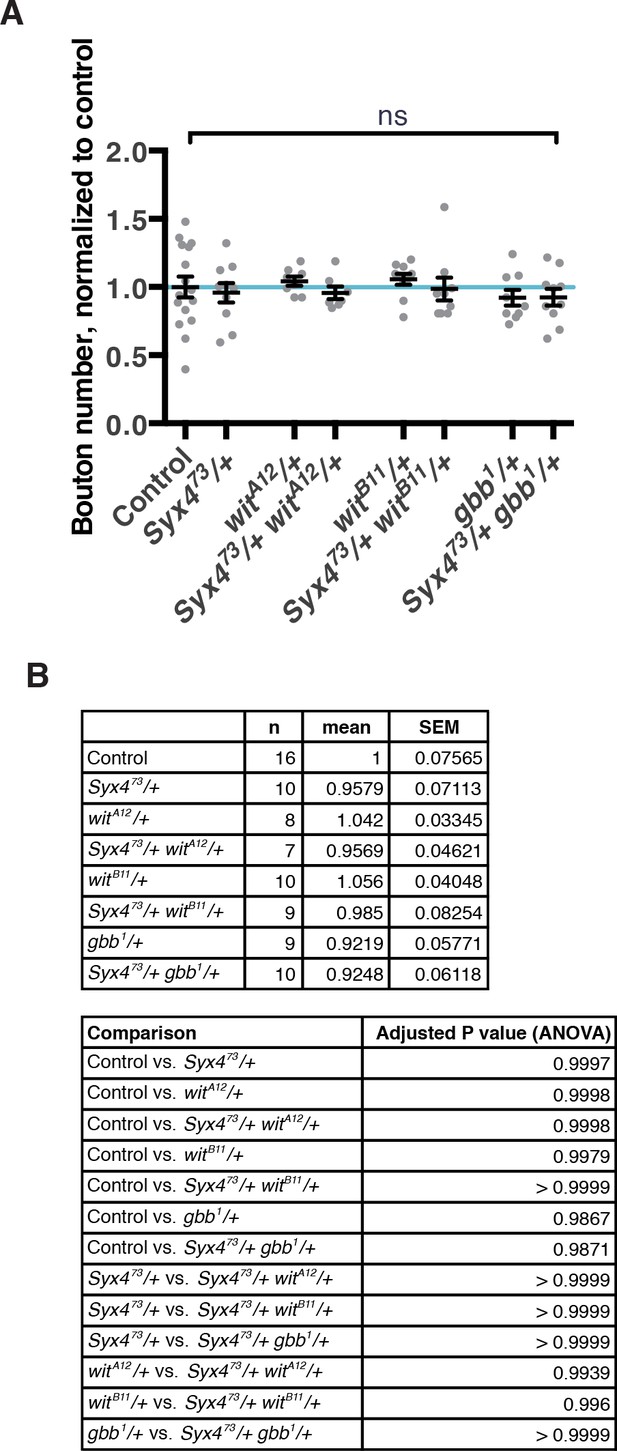
Genetic interaction experiments between Syx4 and BMP pathway components.
(A) No genetic interactions are detected between Syx473 and components of the BMP pathway: witA12, witB11, or gbb1. Single and double heterozygous combinations are shown. Data are presented as mean ± SEM, ns = not significant. (B) Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in (A).
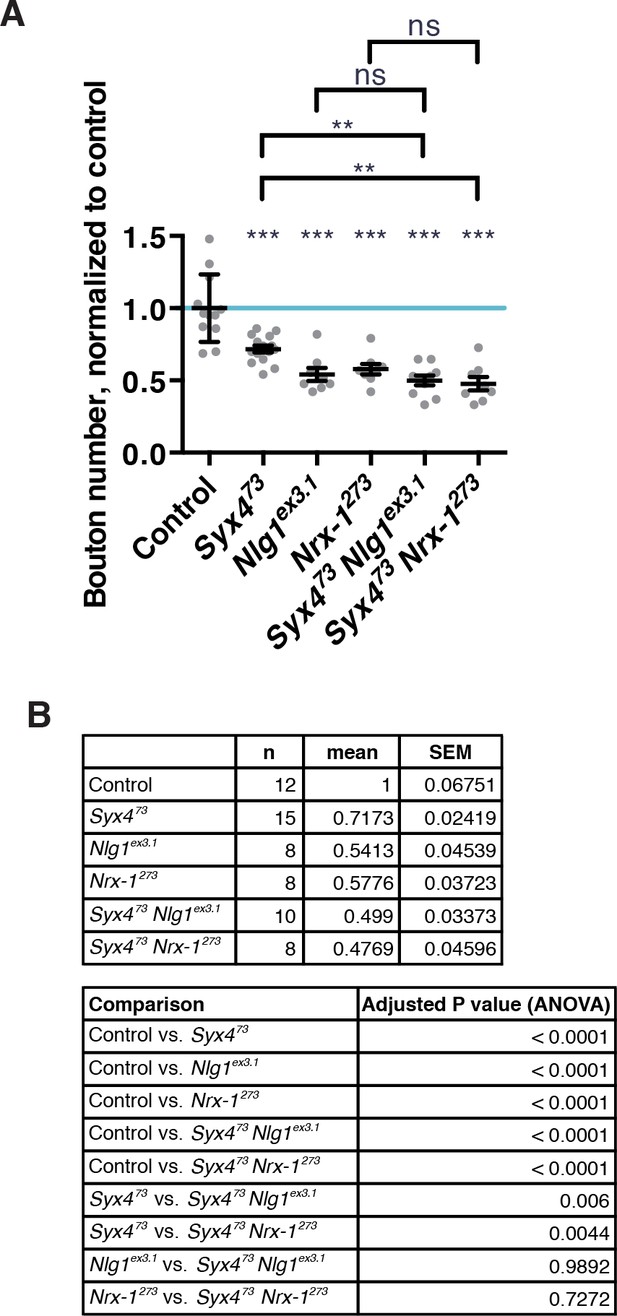
Genetic interaction experiments between single and double null mutants of Syx4, Nlg1, and Nrx-1.
(A) Double mutants combinations between Syx473, Nlg1ex3.1, and Nrx-1273 have severe synaptic growth defects. Data are presented as mean ± SEM. Statistical comparisons are indicated here as ***p<0.001, **p<0.01, *p<0.05, ns = not significant; comparisons are with control unless indicated. (B) Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in (A).
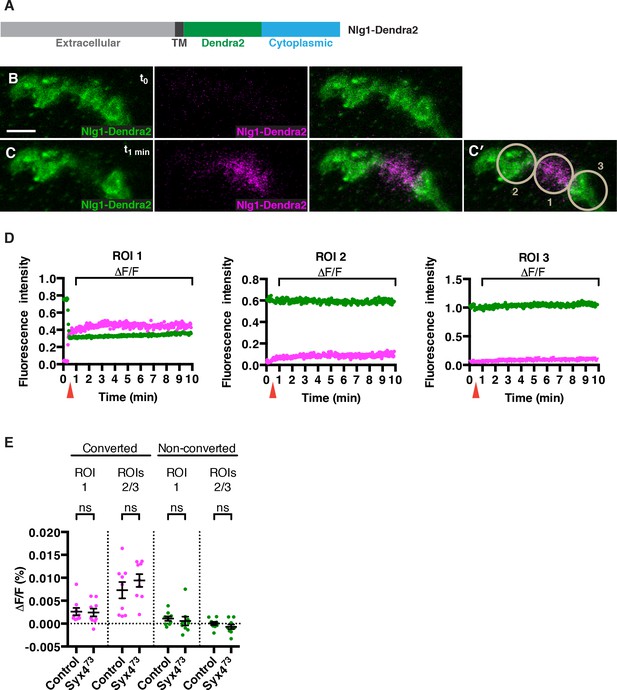
No change in mobility of Neuroligin 1 is observed in Syntaxin 4 mutants.
(A) Nlg1-Dendra2 construct. The Dendra2 tag was placed between the transmembrane domain and the cytoplasmic tail. (B–C) Representative image from a single animal expressing Nlg1-Dendra2 in the postsynaptic cell. One bouton (ROI1) was targeted with a 405 nm laser for photoconversion of the Dendra2 tag after 1 min. Non-photoconverted Nlg1-Dendra2 is shown in green and photoconverted Nlg1-Dendra2 is shown in magenta before (B) and immediately after (C) photoconversion. (C′) Regions of interest: ROI1, photoconverted region; ROIs 2 and 3, adjacent regions. (D) Fluorescent intensity over time for photoconverted and non-photoconverted molecules in all three ROIs. Red arrows indicate time of photoconversion. (E) Quantification of ΔF/F of both photoconverted and non-photoconverted molecules, in all three ROIs, in both the control and Syx473 mutant backgrounds. Data are presented as mean ± SEM. Scale bars = 5 μm. Statistical comparisons are fully described in Figure 6—source data 1; no significant differences found (ns = not significant).
-
Figure 6—source data 1
Statistical data for Figure 6.
Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in Figure 6E.
- https://doi.org/10.7554/eLife.13881.018
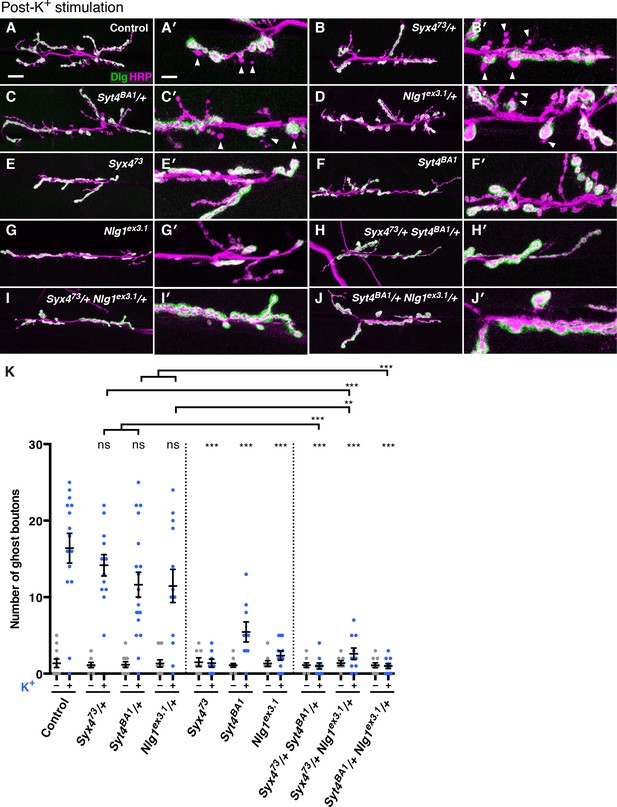
Syntaxin 4, Synaptotagmin 4, and Neuroligin 1 regulate acute structural plasticity at the NMJ.
(A–J) Representative images of NMJs stained with antibodies to HRP (magenta) and the postsynaptic marker Dlg (green) to highlight synaptic boutons. Ghost bouton budding was stimulated with spaced incubations in high K+. Ghost boutons are identified as round HRP+ structures lacking Dlg signal (arrowheads); images are shown from precise excision control (A), Syx473/+ (B), Syt4BA1/+ (C), Nlg1ex3.1/+ (D), Syx473 (E), Syt4BA1 (F), Nlg1ex3.1 (G), Syx473/+ Syt4BA1/+ (H), Syx473/+ Nlg1ex3.1/+ (I), and Syt4BA1/+ Nlg1ex3.1/+ (J) animals. (K) Quantification of ghost bouton number per NMJ from animals without (−) or with (+) high K+ stimulation. Data are presented as mean ± SEM. Scale bars = 20 μm (A–J), 6.7 μm (A′–J′). Statistical comparisons are fully described in Figure 7—source data 1, and are indicated here as ***p<0.001, **p<0.01, *p<0.05, ns = not significant; comparisons are with control unless indicated.
-
Figure 7—source data 1
Statistical data for Figure 5.
Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in Figure 7K.
- https://doi.org/10.7554/eLife.13881.022
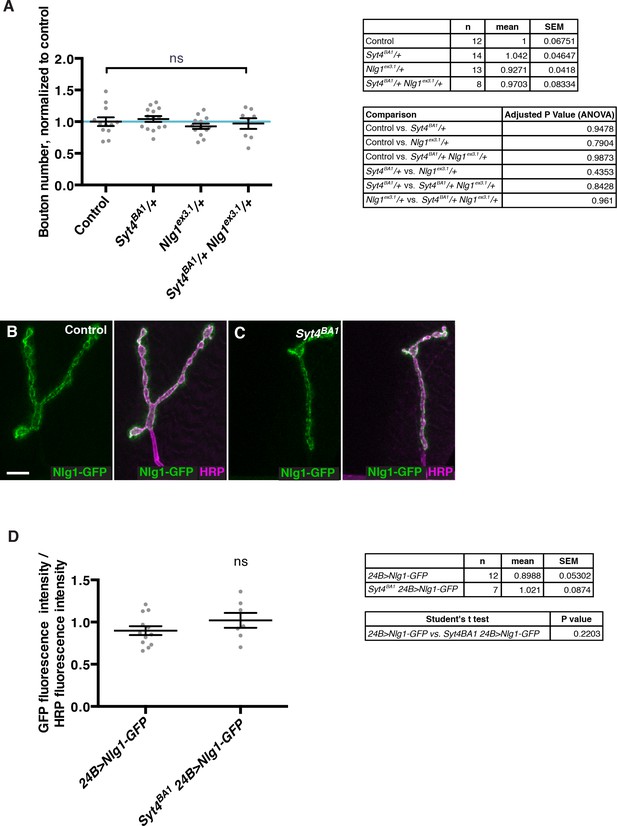
Interaction experiments between Syt4 and Nlg1.
(A) No genetic interactions are detected between Syt4 and Nlg1 with respect to bouton number. Data are presented as mean ± SEM, ns = not significant, ANOVA. Control refers to a precise excision control line for the Syt4BA1 allele. (B–C) Representative NMJs expressing Nlg1-GFP in control (B) or Syt4BA1 (C) backgrounds. (D) Quantification of GFP fluorescence intensity per HRP fluorescence intensity, in animals expressing Nlg1-GFP in control or Syt4BA1 backgrounds. Data are presented as mean ± SEM, ns = not significant, Student’s t test. Scale bars = 5 μm. Sample size (n), mean, SEM, and pairwise statistical comparisons are presented for the data in (A) and (D).
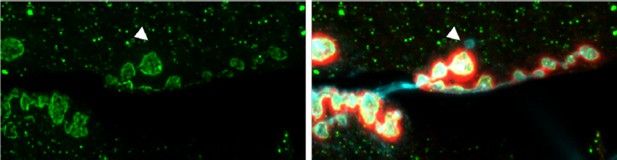
NMJ from an animal expressing Syt4-pH postsynaptically (green).
The postsynaptic scaffold is labeled with Dlg (red). The neuronal membrane is labeled with HRP (blue). A single ghost bouton is indicated with an arrow. The ghost bouton is devoid of Syt4-pH signal, and there is no obvious change in Syt4-pH at or near the bouton from which the ghost bouton emerged.
Videos
Photoconversion of Nlg1-Dendra2 in control animals.
Visualization of a synaptic arbor expressing postsynaptic Nlg1-Dendra2 at muscle 4 in a dissected third instar larva. One bouton is photoconverted after 20 sec, with about 50% of the green molecules converted to red (shown here as magenta). Over the next 9 min of imaging, very little movement of photoconverted molecules is observed. Scale bar = 2.5 μm.
Photoconversion of Nlg1-Dendra2 in Syx473 animals.
Visualization of a synaptic arbor expressing postsynaptic Nlg1-Dendra2 at muscle 4 in a dissected third instar larva mutant for Syx4. One bouton is photoconverted after 20 sec, with about 50% of the green molecules converted to red (shown here as magenta). Over the next 9 min of imaging, very little movement of photoconverted molecules is observed. Scale bar = 2.5 μm.
Tables
RNAis that alter the localization of Syt4-pH Candidate gene products are listed, along with the predicted gene function, the specific effect on Syt4-pH, and the RNAi constructs tested. RNAi lines were obtained from the Transgenic RNAi Project (TRiP) at Harvard Medical School (Perkins et al., 2015)a or the Vienna Drosophila RNAi Center (Dietzl et al., 2007)b.
| Gene product | CG | Function | Syt4-pH distribution | RNAis |
|---|---|---|---|---|
| Syntaxin 4 | CG2715 | t-SNARE | Reduced intensity at NMJ, large clusters in cytoplasm | JF01714a V32413b |
| Syntaxin 6 | CG7736 | t-SNARE | Reduced intensity at NMJ, large clusters in cytoplasm | V1579b V1501b |
| Syntaxin 18 (Gtaxin) | CG13626 | t-SNARE | Reduced intensity at NMJ, large clusters in cytoplasm | JF02263a |
| MyoV | CG2146 | Dilute class unconventional myosin | Reduced intensity at NMJ, large clusters in cytoplasm | JF03035a V16902b |
| Actin-related protein 2/3 complex, subunit 3A | CG4560 | Arp2/3 complex-mediated actin nucleation | Reduced intensity and size at NMJ, smaller cytoplasmic puncta | JF02370a |
| Gdi | CG4422 | Rab GDP-dissociation inhibitor | Reduced intensity at NMJ, large clusters in cytoplasm | JF02617a V26537b |
| Rabex-5 | CG9139 | Rab5 guanyl-nucleotide exchange factor activity | Reduced intensity at NMJ, large clusters in cytoplasm | JF02521a |
| Lasp | CG3849 | Actin binding | Increased intensity at NMJ | JF02075a |
| Neuroglian | CG1634 | Cell adhesion; axon guidance; synapse organization | Increased intensity at NMJ | JF03151a V27201b |
| Contactin | CG1084 | Cell adhesion | Increased intensity at NMJ | HM05134a HMS00186a |
| Syntaxin 7 | CG5081 | t-SNARE, early endosomal regulation | Intensity at NMJ normal, many small bright puncta cluster adjacent to NMJ | JF02436a V5413b |
| Dynamin associated protein 160 | CG1099 | Synaptic vesicle endocytosis; cell polarity | Intensity at NMJ normal, many small bright puncta cluster adjacent to NMJ | JF01918a V16158b |
| Adaptor Protein complex 2, σ subunit | CG6056 | Endocytosis | Intensity at NMJ normal, many small bright puncta cluster adjacent to NMJ | JF02631a |
| Adaptor Protein complex 2, α subunit | CG4260 | Endocytosis | Intensity at NMJ normal, many small bright puncta cluster adjacent to NMJ | HMS00653a |
| β-spectrin | CG5870 | Cytoskeleton; synapse organization | Irregular size and spacing at NMJ | HMS01746a V42053b |
Additional files
-
Supplementary file 1
Candidate list for RNAi screen Complete list of transgenic lines tested.
Each carries a UAS-RNAi to knock down the gene product (indicated with CG and gene symbol). RNAi stock ID refers to the unique ID from either the TRiP center at Harvard Medical School or the Vienna Drosophila Resource Center.
- https://doi.org/10.7554/eLife.13881.024





