DAPK interacts with Patronin and the microtubule cytoskeleton in epidermal development and wound repair
Figures
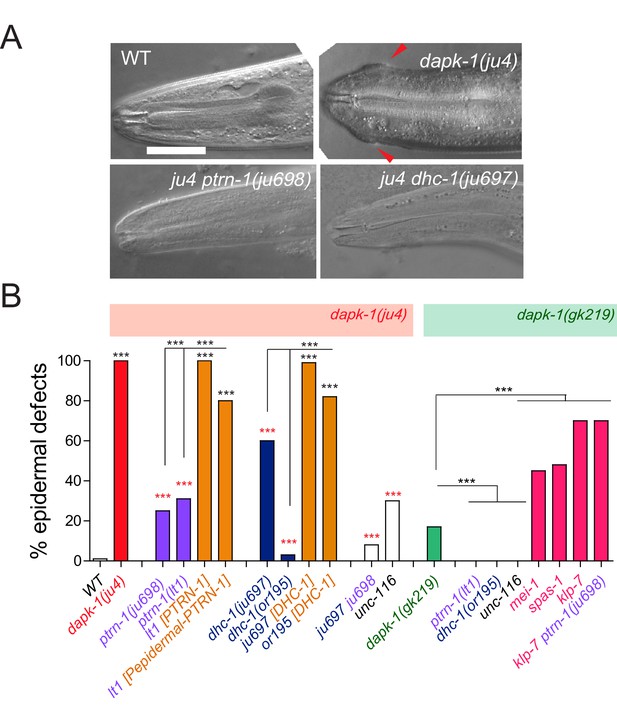
dapk-1 epidermal phenotypes are modified by mutations in cytoskeletal regulating genes.
(A) Anterior epidermal morphology of wild type adult and aberrant morphology (Mor phenotype, red arrowhead) of dapk-1(ju4) animals; suppression by ptrn-1(ju698) and dhc-1(ju697). DIC images; scale bar, 10 μm. Anterior is to the left and dorsal up. (B) Loss of function in ptrn-1 or dhc-1 suppresses the anterior epidermal morphology defects of dapk-1(ju4) and dapk-1(gk219). Mor penetrance at 20°C; N>100 per genotype. Fisher’s exact test; ***p<0.001. Black stars in B are comparisons to WT; red stars are comparisons to dapk-1(ju4)
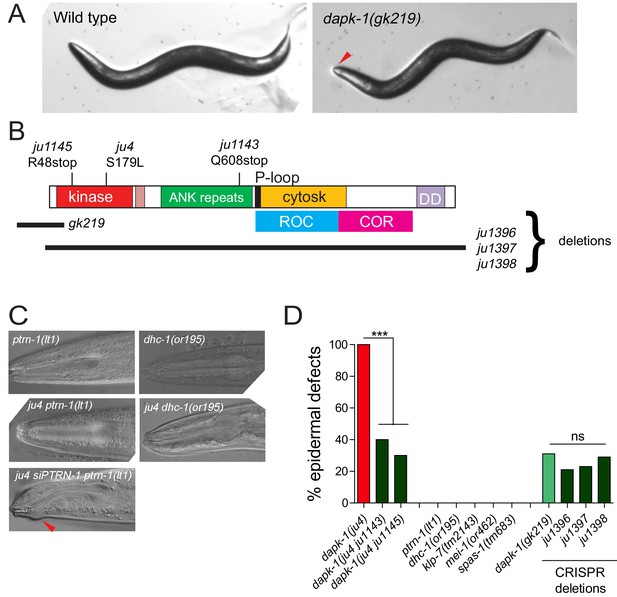
Partial suppression of dapk-1 phenotypes by loss of function in cytoskeletal regulators
(A) Dissection microscope images of WT and Mor animals. (B) Schematic of DAPK-1 protein showing domains and locations of previous and newly identified alleles. (C) Suppression of dapk-1(ju4) Mor phenotypes by additional alleles of ptrn-1 and dhc-1, and rescue by PTRN-1(+) single copy insertion. ptrn-1 and dhc-1 single mutants display normal head morphology. (D) Partial suppression of dapk-1(ju4) by intragenic stop codon mutations. Mutants that modify dapk-1(ju4) do not induce epidermal defects in a dapk-1(+) background. The penetrance of dapk-1(gk219) resembles that of mutations in which the entire dapk-1 locus is deleted. N>100. Fisher’s exact test; ***p<0.001.
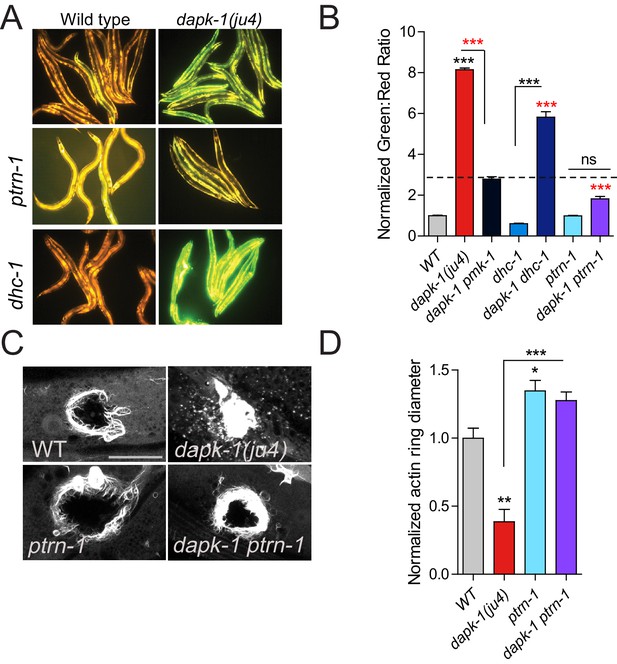
ptrn-1 suppresses dapk-1(ju4) innate immune and wound repair phenotypes.
(A) dapk-1(ju4) animals display an elevated expression of the antimicrobial peptide nlp-29 (Pnlp-29-GFP, frIs7); this is suppressed by ptrn-1(0), whereas dhc-1(or195) and other mutations do not suppress (see also Figure 2—figure supplement 1A). Day 1 adults. (B) Quantitation of fluorescence intensity ratio (Green:Red, normalized to WT = 1) of animals in A, using the COPAS Biosort. N>100. Fisher’s exact test; ***p<0.001. pmk-1 is a MAPK required for activation of nlp-29 transcription; pmk-1 mutants serve as a negative control. (C) ptrn-1(0) suppresses the accelerated actin ring closure in dapk-1(ju4) mutants after needle wounding. The wound-triggered actin ring is visualized using Pcol-19-GFP::moesin (juIs352); scale, 10 µm. (D) Quantitation of actin ring diameter, mean ± SEM. One-way ANOVA and Dunn’s post-test; *p<0.05; **p<0.01; ***p<0.001.
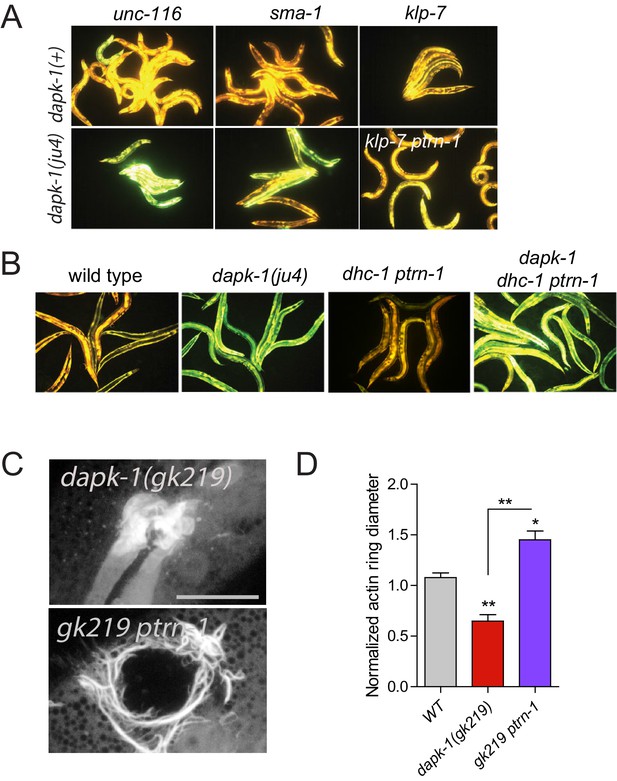
Most dapk-1 modifiers do not affect the dapk-1(ju4) constitutively active innate immune response.
(A) Loss of function in unc-116, sma-1, or klp-7 does not drastically modify the elevated expression of the Pnlp-29-GFP transcriptional reporter in dapk-1(ju4) mutants. (B) dhc-1(or195) is epistatic to ptrn-1 for the innate immune response phenotype in dapk-1(+) background. (C) ptrn-1(0) suppresses the accelerated actin ring closure in dapk-1(gk219) mutants after needle wounding (Pcol-19-GFP::moesin, juIs352); scale, 10 µm. (D) Quantitation of C, mean ± SEM. Kruskal-Wallis and Dunn’s post test; *p<0.05; **p<0.01; ***p<0.001.
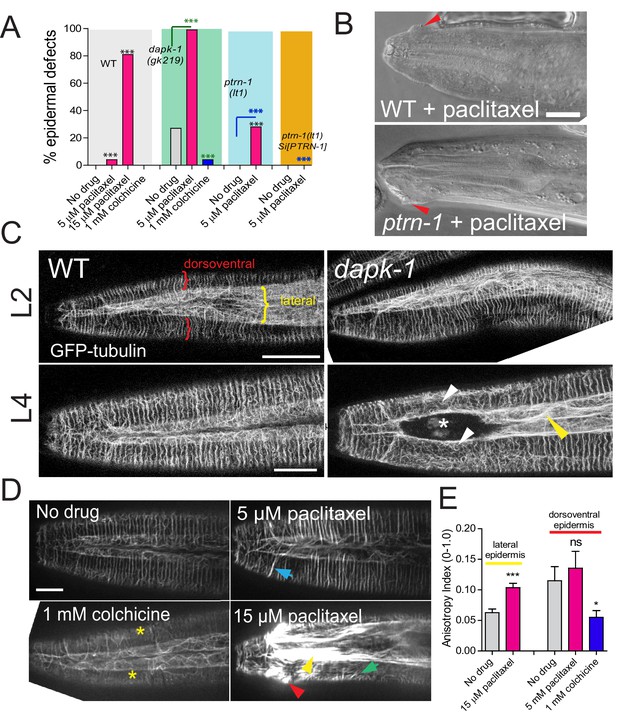
dapk-1 epidermal morphology defects are mimicked or enhanced by MT stabilization and suppressed by MT depolymerization.
(A) MT stabilization by paclitaxel treatment induces dapk-1-like morphological defects in the wild type and in ptrn-1(0) mutants, and enhances the morphological defects of dapk-1(gk219). Colchicine treatment suppresses dapk-1(gk219) morphological defects. Statistics: Fisher’s exact test; ***p<0.001; n > 100 animals per condition. (B) DIC images of epidermal defects in representative wild type and ptrn-1(0) animals after paclitaxel treatment. (C) Confocal images of epidermal MT architecture in the heads of WT and dapk-1(ju4) L2 and L4 larvae (Pdpy-7-GFP::TBB-2, ltSi570). Yellow bracket indicates lateral epidermal ridge, red brackets mark dorsoventral epidermis overlying muscles. dapk-1(ju4) L4 animals display bundling of MTs in the lateral epidermis (yellow arrowhead) and disorganized circumferential MTs (white arrowhead). (D) Effects of MT drugs on epidermal MT organization (ltSi570) in heads of L4 animals. Treatment with colchicine causes loss of circumferential MT bundles (yellow asterisks). Treatment with low concentrations of paclitaxel (5 µM) causes circumferential bundles to be brighter and straighter than in wild type (blue arrow); higher concentrations of paclitaxel (15 µM) induce overt MT bundling (yellow arrow), crossing of circumferential MTs (green arrow), and dapk-1-like morphological defects (red arrowhead). All scale bars, 10 µm. (E) Treatment with 15 µM paclitaxel significantly increases the anisotropy of MT bundles in the lateral epidermis. Statistics, Kruskal-Wallis and Dunn’s post test; *p<0.05, ***p<0.001; N>8 animals.
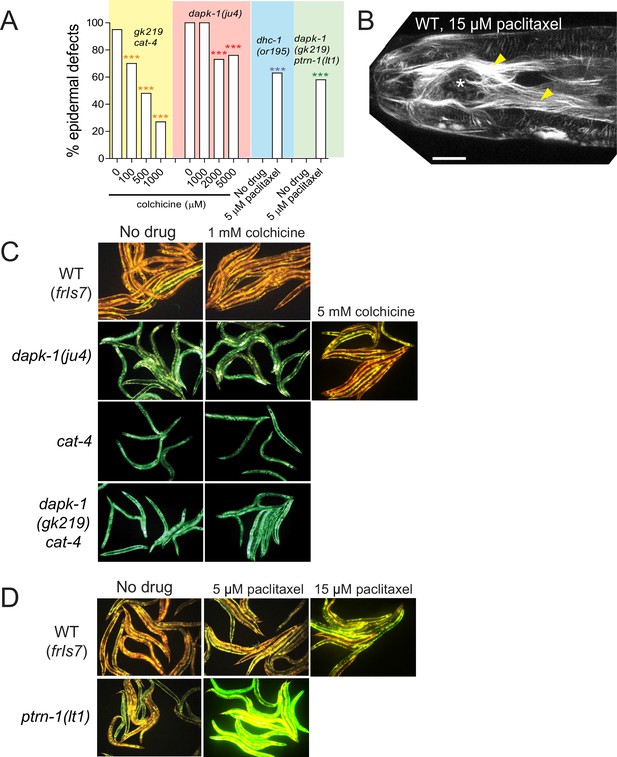
Effects of MT-altering drugs on epidermal morphology and innate immune responses in mutant backgrounds.
(A) MT modulation of epidermal morphology in dapk-1 mutants. Colchicine suppresses the epidermal morphology defects of dapk-1(gk219) cat-4 in a dose-dependent manner. dhc-1(or195) and dapk-1(gk219) ptrn-1(0) backgrounds are hypersensitive to effects of paclitaxel. N>100. Fisher’s exact test; ***p<0.001. (B) Adult animal with Mor defects (white asterisk) and bundled MTs (yellow arrowheads) after paclitaxel treatment (Pcol-19-GFP::TBB-2, juSi239). Scale, 10 µm. (C) Treatment with colchicine does not induce nlp-29 expression, while only higher concentrations of colchicine can suppress the constitutively active nlp-29 expression of dapk-1(ju4) mutants; marker: Pnlp-29-GFP, frIs7. (D) Treatment with paclitaxel induces nlp-29 expression in ptrn-1(0) mutants, and to a lesser extent, in WT animals.
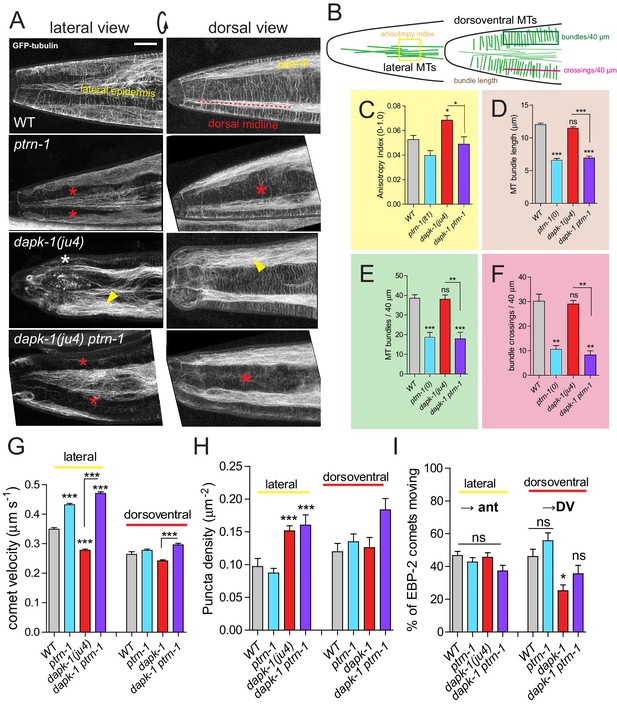
dapk-1 defects in epidermal MT architecture are suppressed by ptrn-1.
(A) ptrn-1(0) and dapk-1(ju4) display distinct effects on epidermal MT organization; MTs visualized in young adults (Pcol-19-GFP::TBB-2, juSi239). Left column, lateral views of the head; right column, dorsal view. ptrn-1(0) mutant adults display fewer circumferential MT bundles in the dorsoventral epidermis (red asterisks). dapk-1(ju4) mutants display increased bundling in the lateral epidermis (yellow arrowhead), quantified by anisotropy index in panel C. dapk-1(ju4) ptrn-1(0) double mutants display normal MT bundling in the lateral epidermis and reduced dorsoventral MTs (red asterisks). Mor phenotype, white asterisk. Scale, 10 µm. (B) Cartoon of MT organization in the C. elegans lateral and dorsoventral epidermis; colored boxes indicate ROIs used for quantitation of MT parameters in panels C–F. (C) dapk-1(ju4) animals display elevated MT bundle anisotropy in the lateral epidermis, which is suppressed by ptrn-1(0). (D–F) MT bundle length, density and crossing frequency in the dorsoventral epidermis is reduced in ptrn-1(0) and in dapk-1(ju4) ptrn-1(0) double mutants. N>8 animals per genotype. Bars show mean ± SEM. Kruskal-Wallis and Dunn’s post test; *p<0.05; **p<0.01; ***p<0.001. (G–I) Quantitation of epidermal EBP-2::GFP comet dynamics in the lateral and dorsoventral epidermis. N>10 animals per genotype.
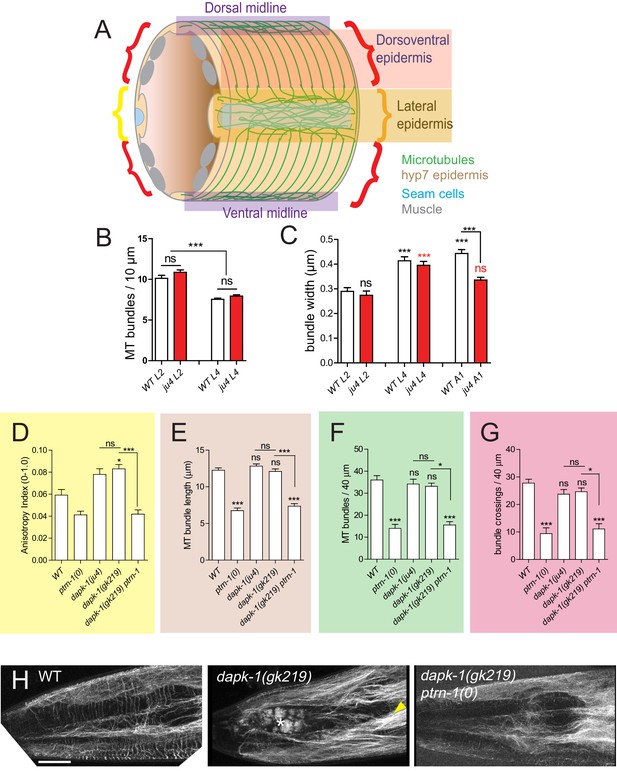
Epidermal MTs in wild type and dapk-1 mutants.
(A) Cartoon of C. elegans post-embryonic epidermis (hyp7 syncytium and lateral seam cells) showing the organization of MTs in the lateral and dorsoventral (muscle-adjacent) compartments. A large syncytium (hyp7) encloses most of the adult animal and forms two compartments: thin layers adjacent to the body wall muscle quadrants, and thickened ridges laterally and at the dorsal and ventral midlines (Figure 4—figure supplement 1A). The dorsoventral epidermis is extremely thin (~50 nm) along its apicobasal axis and is packed with arrays of hemidesmosome-like attachment structures that link muscles to the external cuticle. In contrast, the lateral epidermis is several μm thick and contains most nuclei. The two sets of MTs, in the lateral and in the dorsoventral epidermis, are seen throughout the hyp7 syncytium and in the smaller epidermal cells of the head and tail. (B) MT bundle density in WT and dapk-1(ju4) L2 and L4 animals. (C) dapk-1(ju4) adults display thinner MT bundles. (D–G) Quantitation of MT architecture in dapk-1(gk219). Graph colors refer to the ROIs in Figure 4B. All graphs: Kruskal-Wallis test and Dunn’s post-test; *p<0.05, ***p<0.001, N>8. (H) Confocal images of lateral epidermis (Pcol-19-GFP-TBB-2, juSi239). Yellow arrowhead, bundled MTs; white asterisk, Mor phenotype. Scale, 10 µm.
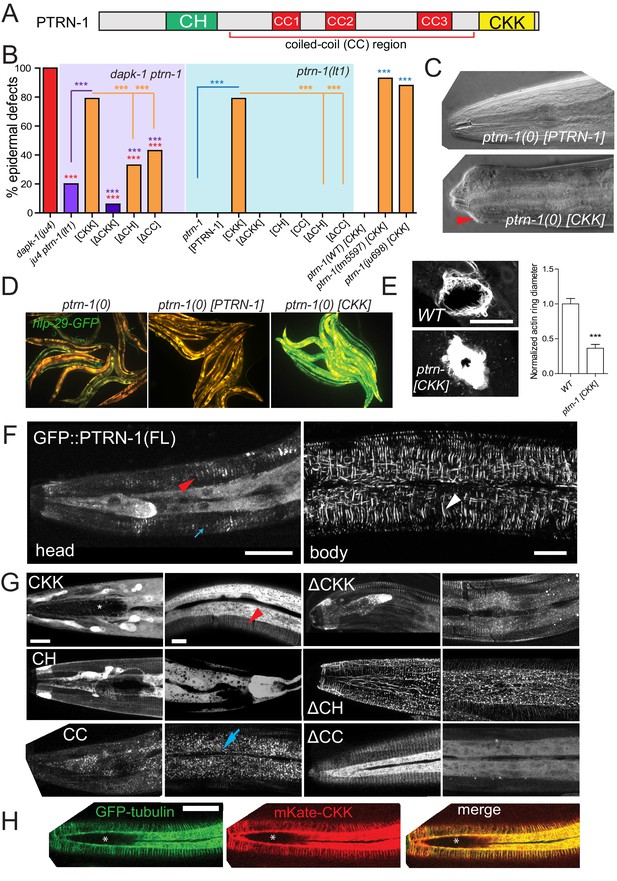
The CKK domain of PTRN-1 is required and sufficient to cause dapk-1-like defects in epidermal morphology.
(A) PTRN-1 domain organization. (B) Quantitation of epidermal defects in animals expressing different fragments of GFP::PTRN-1. N>100. Fisher’s exact test; ***p<0.001. (C) Representative DIC images of heads of animals expressing full length PTRN-1 or the CKK domain alone (dpy-7 promoter, juEx6697 and juEx6695), in ptrn-1(lt1). (D) Over-expression of the CKK domain is sufficient to induce Pnlp-29-GFP expression in ptrn-1(0) mutants (frIs7; juEx7385) and (E) speed up wound closure (juIs352; juEx6825). N>15, t-test; ***p<0.001. (F) Localization of full length GFP::PTRN-1 in the larval epidermis (juEx6697). White arrowhead points to a thick filament in the midbody lateral epidermis. PTRN-1 also localizes to puncta (blue arrow) and thin filaments (red arrowhead). All scale bars, 10 µm. (G) Localization of GFP-tagged PTRN-1 fragments in larval epidermis. (H) Colocalization of PTRN-1 CKK domain and MTs in anterior larval epidermis. Genotype: ptrn-1(lt1); juEx6825 (Pdpy-7-mKate2::CKK); ltSi570 (Pdpy-7-GFP::TBB-2). Asterisk indicates region devoid of MTs.
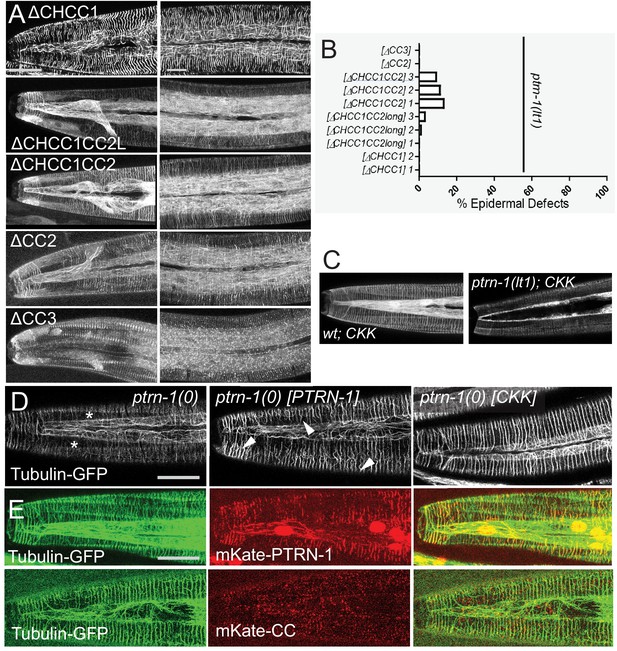
Structure-function analyses of PTRN-1.
(A) Localization of GFP-tagged fragments of the PTRN-1 coiled coil (CC) domain, in the head (left) and anterior lateral epidermis (right). Scale bars, 10 µm. (B) Quantitation of epidermal defects of transgenic animals expressing fragments of the CC domain. (C) The CKK domain localizes strongly to filaments in ptrn-1(+) but not so strongly in ptrn-1(0) backgrounds. (D) Expression of the CKK domain, but not of full-length PTRN-1, causes MT bundles to straighten (marker: ltSi570). (E) Full length PTRN-1 and the CC domain co-localize with MTs (Pdpy-7-mKate2::PTRN-1, juEx6762; Pdpy-7-mKate2::CC, juEx6822).
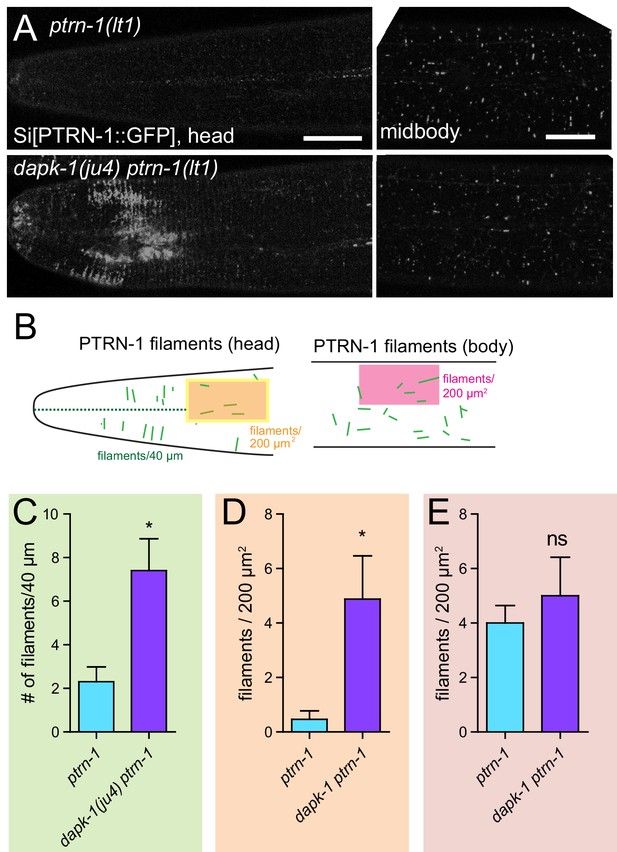
dapk-1(ju4) mutants display aberrant PTRN-1 localization.
(A) Localization of PTRN-1::GFP expressed from single-copy insertion in L4 stage anterior epidermis (left) and midbody lateral epidermis (right); transgene Pdpy-7-PTRN-1::GFP, ltSi541, in ptrn-1(0) or dapk-1(ju4) ptrn-1(0) background. Scale bars, 10 µm. (B) Schematic showing ROIs analyzed in the anterior epidermis. (C–E) Quantitation of PTRN-1 localization in ROIs indicated in B. Experiments performed at 25°C. N>8 animals per genotype. Bars indicate mean ± SEM. Student’s t-test; *p<0.05.
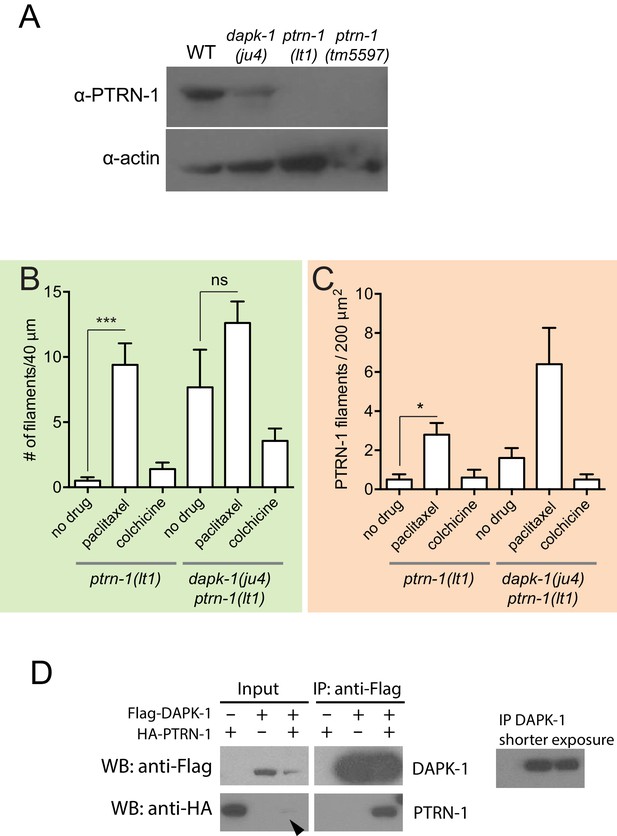
PTRN-1 localization is sensitive to MT polymerization and dapk-1.
(A) PTRN-1 is not overexpressed in dapk-1(ju4) animals; western blot using anti-CePTRN-1. Control, anti-actin. (B,C) The number of PTRN-1 filaments in the head and lateral epidermis is sensitive to MT polymerization. Paclitaxel increases the number of PTRN-1 filaments, and colchicine decreases the number of PTRN-1 filaments in dapk-1(ju4) mutants. Color code according to the ROIs in Figure 6B. Mean ± SEM, N > 8 animals per genotype. Kruskal-Wallis and Dunn’s post-test; *p<0.05; ***p<0.001. (D) Co-immunoprecipitation of DAPK-1 and PTRN-1 in HEK293T cells.
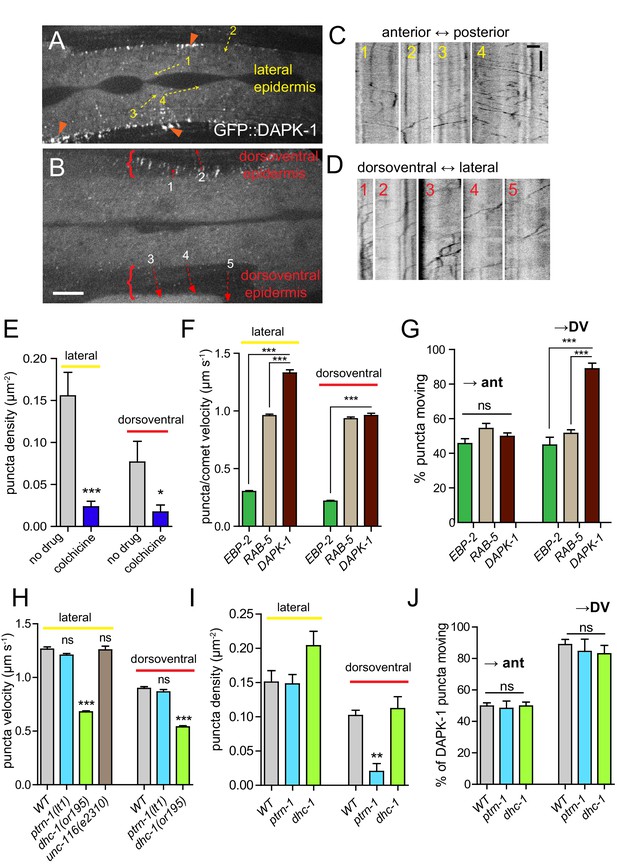
DAPK-1 undergoes MT-dependent transport in the epidermis.
(A,B) Representative images of GFP::DAPK-1 puncta in the larval epidermis (Pdpy-7-GFP::DAPK-1, juEx1774), showing lateral and dorsoventral regions, respectively. Dotted arrows indicate line scans used to generate kymographs in C,D. Orange arrowheads indicate clusters of GFP::DAPK-1 puncta at the boundary of the dorsoventral epidermis. Scale bars, 10 µm. (C) Kymographs of puncta in lateral epidermis (movie frame in A); inverted grayscale; x axis scale, 10 µm; y axis scale, 10 s. (D) Kymographs of puncta in dorsoventral epidermis (movie frame in B). (E) Density of motile GFP::DAPK-1 puncta in adult anterior lateral epidermis, with or without colchicine treatment (cat-4; Pcol-19-GFP::DAPK-1 juEx4781). N>20. (F) GFP::DAPK-1 puncta move faster than RAB-5 puncta or EBP-2 comets in the lateral epidermis (Pcol-19-GFP::DAPK-1, juEx4781). n>100. Bar charts show mean ± SEM. Statistics, Kruskal-Wallis test and Dunn’s post-test; *p<0.1, ***p<0.001. (G) Directionality of puncta or comet motion. Approximately equal numbers of RAB-5 puncta and EBP-2 comets move in each direction; only DAPK-1 displays a significant bias in the dorsoventral epidermis. (H) Reduced dynein/dhc-1 function slows GFP::DAPK-1 puncta; loss of ptrn-1 or mutation in unc-116/kinesin-1 has no effect; n>100. I. ptrn-1(0) mutants have reduced DAPK-1 puncta in the dorsoventral epidermis. J. DAPK-1 puncta directionality is unaffected in ptrn-1(0) or dhc-1 mutants.
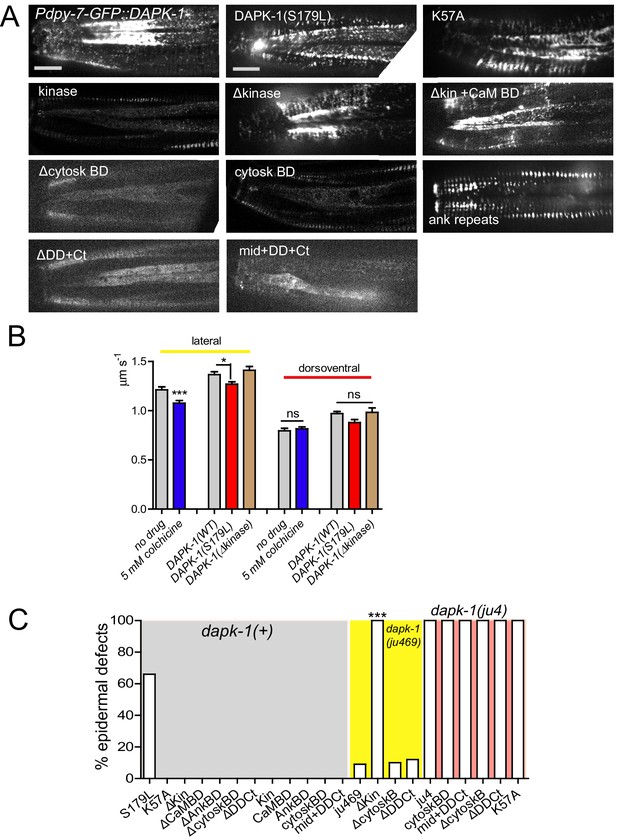
Structure function analysis of DAPK-1.
(A) Representative images of GFP-tagged DAPK-1 protein fragments and their localization in the lateral epidermis in the head. (B) Velocity of DAPK-1 puncta, with or without colchicine treatment (cat-4; Pcol-19-GFP::DAPK-1 juEx4781). n>20. Velocity of mutant DAPK-1. (C) Quantitation of Mor induction or enhancement of different DAPK-1 fragments. N>100. Fisher’s exact test; ***p<0.001.
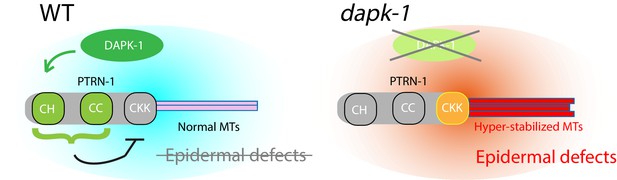
Model for DAPK-1-mediated regulation of epidermal MTs via PTRN-1.
https://doi.org/10.7554/eLife.15833.028Videos
EBP-GFP dynamics in adult lateral epidermis (Pcol-19).
https://doi.org/10.7554/eLife.15833.011EBP-GFP dynamics in adult dorsoventral epidermis.
https://doi.org/10.7554/eLife.15833.012EBP-GFP dynamics in adult lateral epidermis, dapk-1(ju4) background.
https://doi.org/10.7554/eLife.15833.013EBP-GFP dynamics in adult dorsoventral epidermis, dapk-1(ju4) background.
https://doi.org/10.7554/eLife.15833.014GFP::DAPK-1 dynamics in larval epidermis.
Lateral seam cells are in the center of the movie.
GFP::DAPK-1 dynamics in adult lateral epidermis.
Lateral seam is in center.
GFP::DAPK-1 dynamics in adult dorsoventral epidermis.
Dorsal midline is at bottom of movie.
Pdpy-7-GFP::RAB-5 dynamics.
https://doi.org/10.7554/eLife.15833.026Tables
Suppressors and enhancers of dapk-1 morphological defects.
| Gene | Alleles and sequence change | Mammalian orthologs |
|---|---|---|
| A. Suppressors (forward screen) | ||
| ptrn-1 | ju698 | CAMSAP/Patronin |
| *lt1 | ||
| tm5597 | ||
| dhc-1 | ju697 | Dynein heavy chain |
| *or195ts | ||
| dapk-1 | ju1143 | (intragenic) |
| ju1145 | ||
| B. Suppressors (candidates) | ||
| unc-116 | e2310 | Kinesin-1 |
| sma-1 | e30 | Beta-heavy spectrin |
| C. Enhancers | ||
| klp-7 | tm2143 | Kinesin-13 |
| mei-1 | or642ts | p60 katanin |
| spas-1 | tm683 | Spastin |
| cat-4 | tm773 | GTP cyclohydrolase I |
| F47G4.5 | ok2667 | p80 katanin |
| D. No interaction | ||
| ebp-1 | tm1357 | Plus-end binding protein |
| ebp-2 | gk756 | Plus-end binding protein |
| ccpp-1 | ok1821 | Cytosolic Carboxypeptidase |
| ccpp-6 | ok382 | Cytosolic Carboxypeptidase |
| mcrs-1 | tm3681 | Microspherule Protein 1 |
| efa-6 | tm3124 | EFA6 |
| ttll-5 | tm3360 | Tubulin tyrosine ligase-like |
| ttll-11 | tm4059 | Tubulin tyrosine ligase-like |
| ttll-12 | tm4957 | Tubulin tyrosine ligase-like |
| unc-70 | e524 | β-Spectrin |
| dylt-2 | gk762 | Dynein light chain |
| dnc-1 | or404ts | p150 dynactin |
| nud-1 | ok552 | NDE1/NDEL1 |
| nud-2 | ok949 | NDE1/NDEL1 |
| unc-14 | e57 | kinesin-1 adaptor |
| tbg-1 | t1465 | γ-tubulin |
| ptl-1 | ok621 | tau |
| pinn-1 | tm2235 | Pin1 |
| par-1 | zu310ts | MARK |
-
Suppressors indicated * were tested for suppression of dapk-1(ju4) and dapk-1(gk219). Enhancers were tested with dapk-1(gk219). Genes in section D were mostly tested for interaction with dapk-1(ju4). dapk-1 tbg-1 double mutants were extremely sick, and a stable strain could not be obtained; n > 100 animals scored per genotype.
PTRN-1 Structure-function analysis.
| Localization | Function | |||||
|---|---|---|---|---|---|---|
| Protein Fragment | Puncta | Thick filaments | Thin filaments | Restore Mor | Induce Mor | Co-loc with MTs |
| Full length | x | x | x | yes | no | yes |
| CH | - | - | - | no | no | no |
| CC | x | - | - | no | no | yes |
| CKK | - | - | x | yes | yes | yes |
| ΔCKK | x | - | - | no | no | yes |
| ΔCH | x | x | - | yes | no | ND |
| ΔCC | - | - | x | yes | no | ND |
| ΔCHCC1 | ? | x | - | ND | no | ND |
| ΔCHCC1CC2 | - | - | x | ND | slightly | ND |
| ΔCC2 | ? | x* | x | ND | no | ND |
| ΔCC3 | x | - | - | ND | no | ND |
-
Notes: *: Thick filaments present, but fewer compared to PTRN-1 full length or ΔCH. ?: unclear. ND: Not Determined.
Dynamics parameters in the epidermis.
| Type of Dynamics | Transgene | Velocity (mean ± SEM) μm s−1 | Directionality | ||
|---|---|---|---|---|---|
| Lateral | Dorso-ventral | Lateral | Dorso-ventral | ||
| MT plus-end growth | Pcol-19-EBP-2::GFP | 0.30 ± 0.00 | 0.22 ± 0.007 | A→P P→A | Lat→DV DV→Lat |
| Early endosome transport | Pdpy-7-GFP::RAB-5 | 0.96 ± 0.014 | 0.93 ± 0.014 | A→P P→A | Lat→DV DV→Lat |
| DAPK-1 | Pdpy-7-GFP::DAPK-1 | 1.33 ± 0.028 | 0.96 ± 0.021 | A→P P→A | Lat→DV |
| DAPK-1 | Pcol-19-GFP::DAPK-1 | 1.26 ± 0.02 | 0.90 ± 0.017 | A→P P→A | Lat→DV |
-
Notes: A→P: Anterior to Posterior. P→A: vice versa. Lat→DV: Lateral to dorsoventral epidermis. DV→Lat: vice versa.
DAPK-1 structure-function analysis.
| Protein Fragment | Puncta? | Motile? | Rescues Mor* | Induces Mor† | Enhances Mor‡ | Lethal in dapk-1(ju4) |
|---|---|---|---|---|---|---|
| Full Length | yes | yes | yes | no | no | no |
| Δ Kinase | yes | yes | no | no | yes | yes |
| Δ Kinase + CaM Bind Domain (BD) | yes | yes | no | no | yes | yes |
| Δ Cytoskeletal BD | no | no | no | no | no | no |
| Δ Death Domain + C terminus | no | no | no | no | no | no |
| Kinase only | some | no | no | no | ND | ND |
| CaM BD only | no | no | no | no | ND | ND |
| Ankyrin Domain only | some | no | no | no | no | no |
| Cytoskeletal BD only | some | no | no | no | no | no |
| Mid + DD + Ct | no | no | no | no | no | no |
| S179L dapk-1(ju4) | yes | yes | no | yes | yes | yes |
| K57A (kinase dead) | yes | yes | no | no | no | no |
-
Notes: *: In dapk-1(ju4). †: In WT background. ‡: In dapk-1(ju469).
Additional files
-
Supplementary file 1
Newly generatedstrains and plasmids.
(A) List of new strains and genotypes. (B) List of new plasmids.
- https://doi.org/10.7554/eLife.15833.029





