Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis
Figures
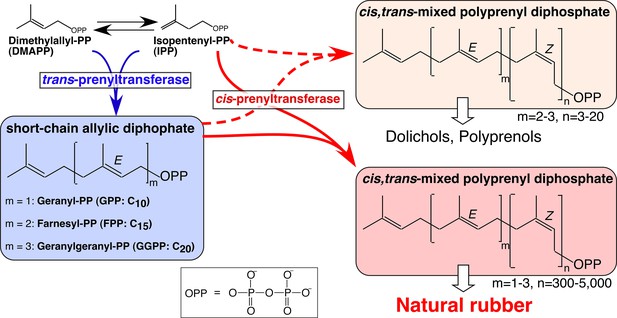
Biosynthetic pathways of backbone structures of cis,trans-mixed polyisoprenoids and natural rubber, catalyzed by trans- and cis-prenyltransferases.
https://doi.org/10.7554/eLife.19022.002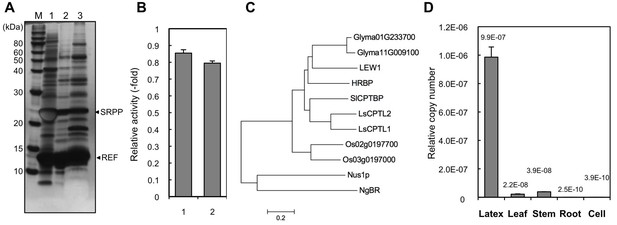
Identification of a Hevea NgBR homologue.
(A) Solubilization of proteins from RPs. Proteins on RPs were solubilized by stepwise treatment with buffers containing 4 mM CHAPS (lane 1), 16 mM CHAPS (lane 2), and 7 M urea, 4 M thiourea, and 6.5 mM CHAPS (lane 3), analyzed using SDS-PAGE, and then visualized by silver staining. (B) Relative RTase activities of the RPs, with RTase activity of non-detergent-washed RPs set at 1.0. 1: Sustained activity on RPs after the second wash with 16 mM CHAPS; 2: activity of the second-washed RPs co-incubated with the proteins released by the two-step washes, corresponding to the mixture of samples shown in Figure 2A, lanes 1 and 2, after dialysis removal of the detergent. RTase activities were assayed using the standard conditions (see Materials and methods). Results are presented as the mean of three independent determinations ± SD. (C) Molecular phylogenetic tree of the amino acid sequences of identified NgBR family proteins. The tree was constructed using the neighbor-joining method and drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Tree members are HRBP, the NgBR homologue from H. brasiliensis (LC057267); LsCPTL1 from L. sativa (AIQ81186); LsCPTL2 from L. sativa (AIQ81187); SlCPTBP from Solanum lycopersicum (XP_004241992); LEW1 from A. thaliana (NP_001077518); Nus1p from S. cerevisiae (NP_010088); NgBR (AAI50655), and non-functionally identified NgBR-like proteins in Oryza sativa (Os02g0197700: NP_001046201, Os03g0197000: NP_001049268) and Glycine max (Glyma01G233700: NP_001242452, Glyma11G009100: XP_003537718). (D) Tissue specificity of the expression levels of the NgBR homologue from H. brasiliensis. Transcript levels were determined as copy number of HRBP, normalized with those of the 18S rRNA quantified from the total RNA extracted from latex, leaves, stems, roots and suspension-cultured cells of H. brasiliensis. Results are presented as the means of three independent determinations ± SD. RP, rubber particle.
-
Figure 2—source data 1
Proteins related to the natural rubber biosynthesis (upper list) and vesicular trafficking (lower list) identified in the RP proteomics.
RP, rubber particle.
- https://doi.org/10.7554/eLife.19022.004
-
Figure 2—source data 2
Proteins identified in the proteomics of the detergent-washed RPs.
RPs, rubber particles.
- https://doi.org/10.7554/eLife.19022.005
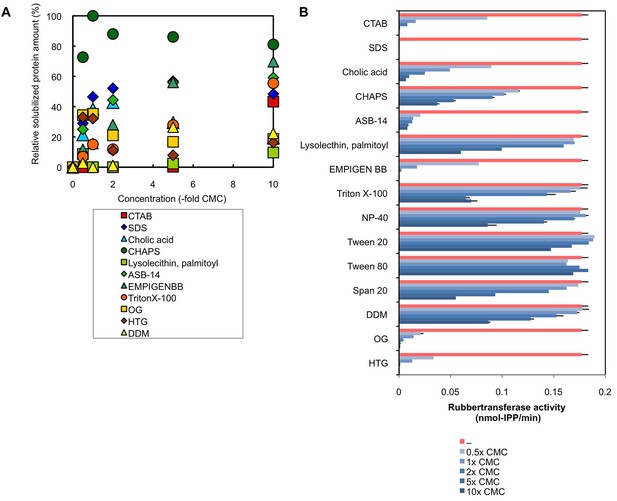
Treatments of RPs with various detergents.
(A) Solubilization of proteins on RPs by the treatments with various detergents. RPs (50kRP) was incubated overnight with the TD buffer (see Materials and methods) containing various concentrations of detergents and then ultracentrifuged at 50,000 ×g and 4°C for 45 min to separate rubber and soluble fractions. Solubilized protein amounts in the soluble fractions were determined using Protein Assay Bicinchoninate Kit (nacalai tesque, Kyoto, Japan), expressed as relative values, with the highest values taken to be 100%. Critical micelle concentrations (CMCs) for each detergent are; cetyltrimethylammonium bromide (CTAB): 0.92 mM, SDS: 8.2 mM, cholic acid: 14 mM, CHAPS: 8 mM, lysolecithin, palmitoyl (1-palmitoyl-sn-glycero-3-phosphocholine): 0.2 mM, ASB-14: 8 mM, EMPIGEN BB: 2.1 mM, Triton X-100: 0.24 mM, OG (n-octyl-β-D-glucoside): 25 mM, HTG (n-heptyl-β-D-thioglucoside): 30 mM, DDM (n-dodecyl-β-D-maltoside): 0.17 mM. (B) Effects of detergents on the RTase activity of RPs. RTase activity of 50kRP in the reaction mixture (see Materials and methods) containing each detergent indicated was assayed. Results are presented as averages of three independent determinations ± standard deviations. RPs, rubber particles.

Multiple alignments of amino acid sequences.
(A) Comparison of the deduced amino acid sequences of HRT1 (BAB71776) and HRT2 (Q8W3U4). Underlined HRT2/HRT1 sequences indicate peptides detected by LC-MS/MS in the RP proteomics. (B) Comparison of the deduced amino acid sequences of the NgBR family proteins. The peptide fragments of HRBP identified in the proteomics of RP are indicated with red bars at the top of the HRBP sequence. A deduced transmembrame region predicted by Phobius (http://phobius.binf.ku.dk/) of HRBP is indicated as blue dotted line at the top of the HRBP sequence. The sequences similar to HRBP are shaded. Identical amino acid residues in greater than five of the seven sequences are boxed. RP, rubber particle.
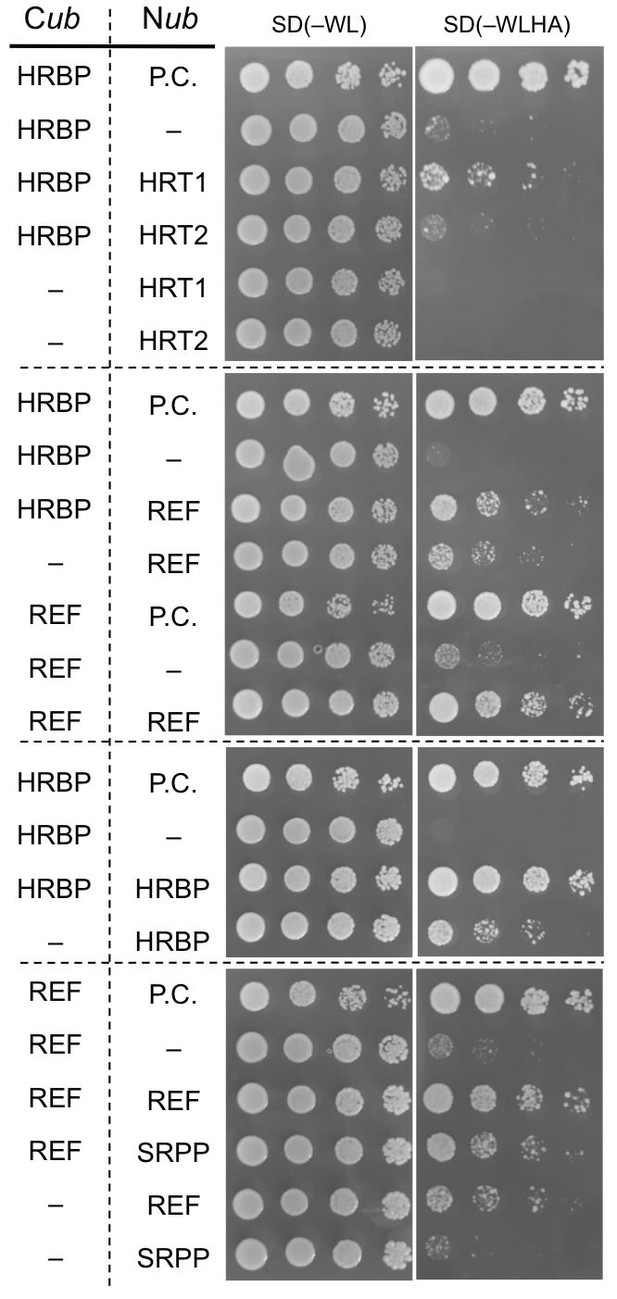
Physical interaction analyses of the proteins by a split-ubiquitin-based Y2H assay.
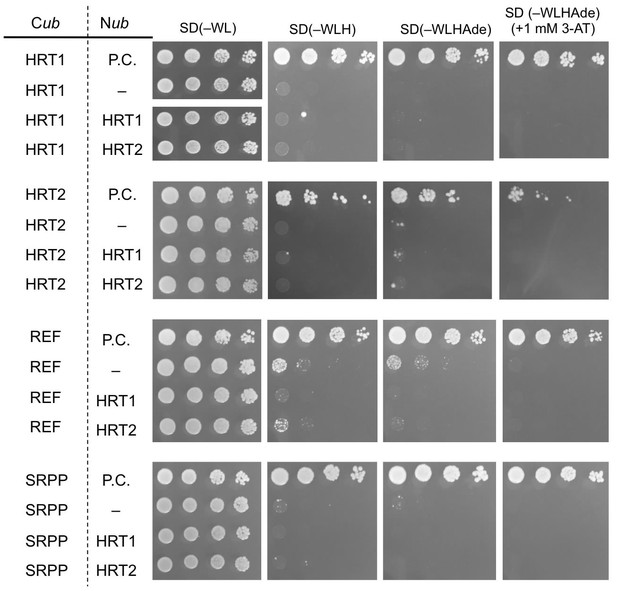
Growth of yeast strains, harbouring genes fused with N-terminal (Nub) or C-terminal (Cub) halves of split ubiquitin fragments (listed on left side), on SD(-WL) and SD(-WLHAde). In each panel, five-fold dilution series of cultures, with cell densities adjusted to be equivalent for each strain, were spotted. P.C.: the positive control plasmid harboring Ost1-Nub, which is designed to activate the reporter system independent of a protein-protein interaction. –: empty vector as negative control.
Split-ubiquitin-based Y2H assays.
Growth of yeast strains, harbouring genes fused with N-terminal (Nub) or C-terminal (Cub) halves of split ubiquitin fragments (indicated on left side), on SD media lacking Trp and Leu [SD(–WL)], those lacking Trp, Lew, and His [SD(–WLH)], those lacking Trp, Lew, His and adenine [SD(–WLHAde)] and SD(–WLHAde) supplemented with 1 mM 3-AT. In each panel, five-fold dilution series of cultures, with cell densities adjusted to be equivalent for each strain, were spotted. P.C.: positive control for ubiquitin reconstitution; –: negative control with empty vector.
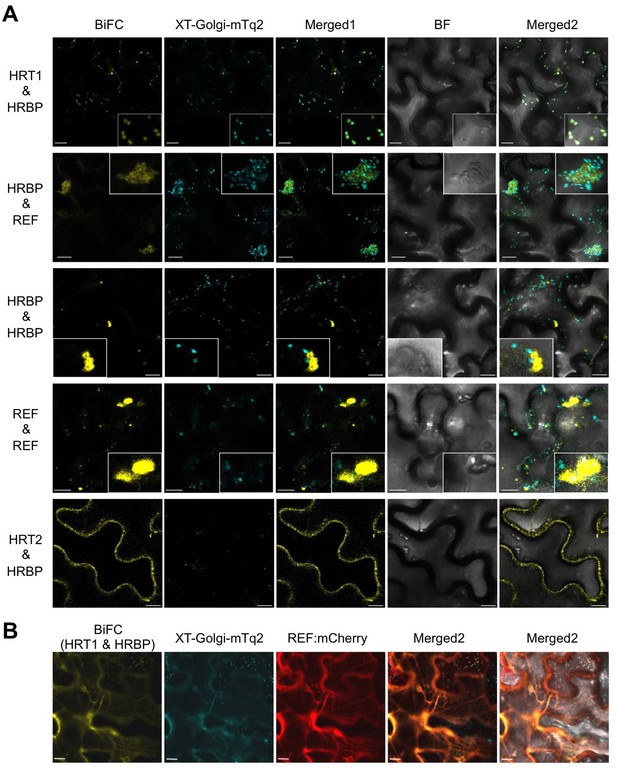
In planta interaction analyses of the proteins by monitoring BiFC.
(A) Fluorescent images of N. benthamiana leaf epidermal cells infected with Agrobacterium harbouring a binary vector constructed with pDOE-07 (HRT1–HRBP, HRT2–HRBP HRBP–REF and HRBP–HRBP) or pDOE-05 (REF–REF) were observed using a confocal laser microscope. Fluorescence signals of mVenus (BiFC) and XT-Golgi-mTq2 were obtained independently and superimposed to create merged images (Merged1), which were further superimposed (Merged2) on bright-field images (BF). (B) BiFC assay for the HRT1–HRBP interaction in N. benthamiana epidermal cells expressing a mCherry, N-terminally fused with REF. Bars, 10 µm. REF, rubber elongation factor.
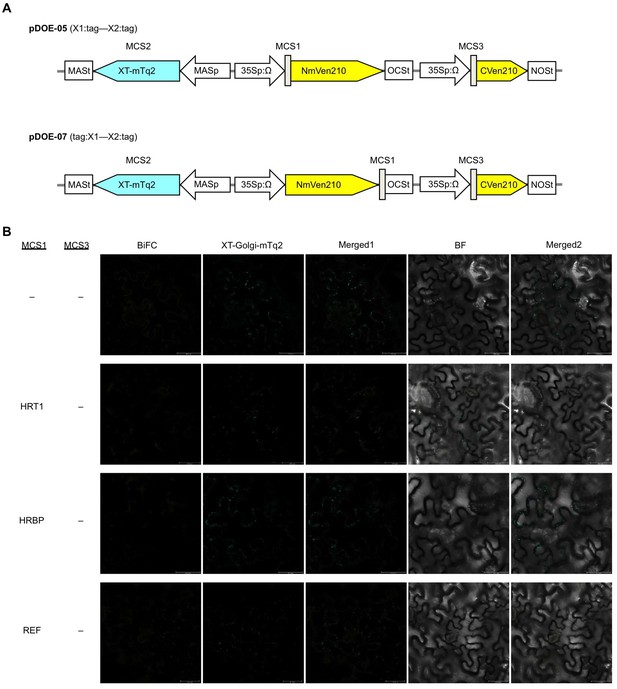
BiFC assays for in planta interactions with split mVenus protein fragments.
(A) Binary vectors pDOE-05 and pDOE-07 utilised for BiFC analyses, developed by Gookin and Assmann (Metcalfe, 1967). NmVen210: N-terminal fragment (1–210) of mVenus, which contains the A206K monomerizing mutation; CVen210: C-terminal fragment (210–238) of Venus; XT-mTq2: mTurquoise2 (mTq2) fused with the N-terminal transmembrane domain of Arabidopsis β-1,2 xylosyltransferase as a marker protein locates a medial subset of cisternae of Golgi stacks; 35Sp:∧, CaMV35S promoter linked to an Omega translation enhancer (Ω); NOSt: NOS terminator; MASp: MAS promoter; MASt: MAS terminator; OCSt: the OCS 3’ terminator; MCS1 and MCS3: multicloning sites 1and 3. (B) BiFC assays using the pDOE-07 binary vector. Fluorescent images of N. benthamiana epidermal cells infected with Agrobacterium harbouring binary vector constructed with pDOE-07 were observed using confocal laser microscope. Fluorescence signals corresponding to mVenus re-assembly (BiFC) and mTq2 fused with the Golgi marker peptide (XT-Golgi-mTq2) were obtained and superimposed to create merged images (Merged1), which were further superimposed (Merged2) on bright-field images (BF). –: no insert in corresponding multicloning site. Bars, 50 µm.
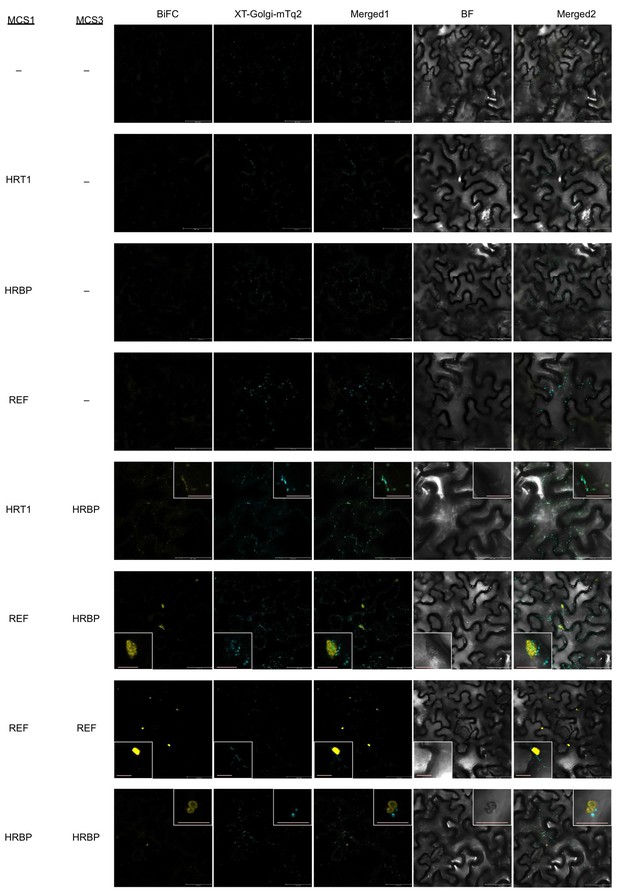
BiFC assays for in planta interactions with split mVenus protein fragments using the pDOE-05 binary vector.
Fluorescent images of N. benthamiana epidermal cells infected with Agrobacterium harbouring binary vector constructed with pDOE-05 were observed using confocal laser microscope. Fluorescence signals corresponding to mVenus re-assembly (BiFC) and mTq2 fused with the Golgi marker peptide (XT-Golgi-mTq2) were obtained and superimposed to create merged images (Merged1), which were further superimposed (Merged2) on bright-field images (BF). –: no insert in corresponding multicloning site. Bars in main panels, 50 µm. Bar in inserted panel, 10 µm.
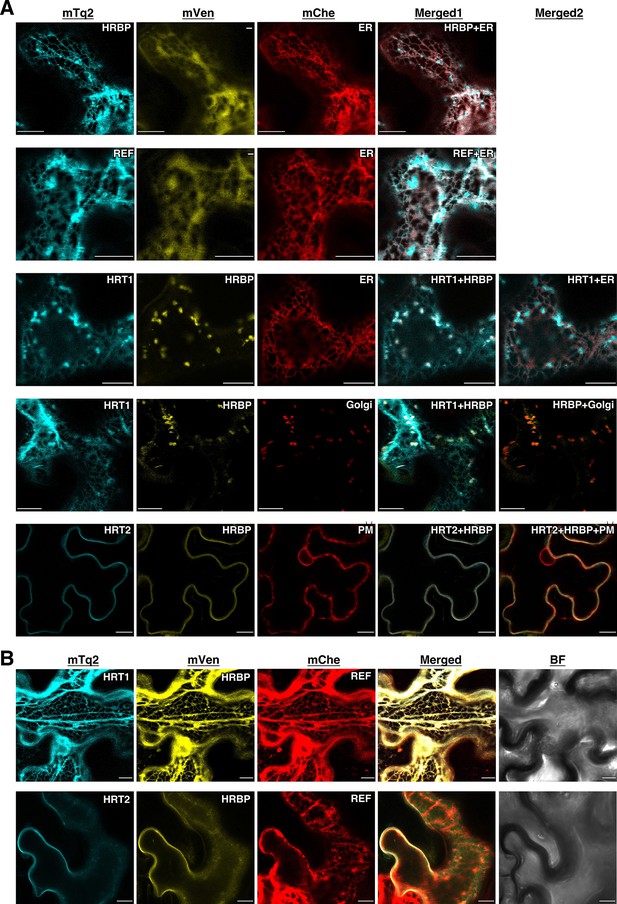
Subcellular localizations of HRT1, HRT2, HRBP and REF.
(A) Fluorescent images of N. benthamiana leaf epidermal cells expressing target proteins (indicated in each panel) fused with a fluorescent protein, mTq2, mVenus or mChe, at their C-terminus. Fluorescence signals corresponding to the fluorescent proteins were obtained using a confocal laser microscope and superimposed to create merged images (Merged1 or Merged2). ER: the ER-marker (HDEL); Golgi: the Golgi marker (N-terminal 49 amino acids of GmManI); PM: the plasma membrane marker (AtPIP2A). (B) Fluorescent images of N. benthamiana leaf epidermal cells co-expressing three proteins, HRT1:mTq2, HRBP:mVenus and REF:mChe or HRT2:mTq2, HRBP:mVenus and REF:mChe. BF: bright-field images. Bars, 10 µm. HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
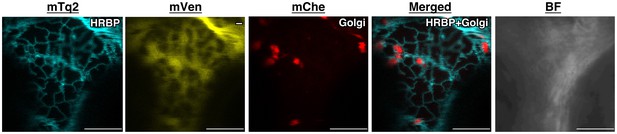
Subcellular localizations of HRBP.
Fluorescent images of N. benthamiana leaf epidermal cells expressing target proteins (indicated in each panel) fused with a fluorescent protein, mTq2, mVenus or mChe, at their C-terminus. Fluorescence signals corresponding to the fluorescent proteins were obtained using a confocal laser microscope and superimposed to create merged images. Golgi: the Golgi marker (N-terminal 49 amino acids of GmManI). BF: bright-field images.Bars, 10 µm. HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
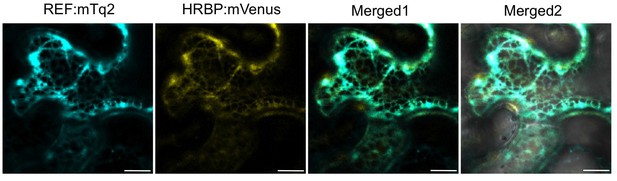
Subcellular localizations of HRBP and REF.
Fluorescent images of N. benthamiana leaf epidermal cells co-expressing REF:mTq2 and HRBP:mVenus were obtained using a confocal laser microscope and superimposed to create merged images (Merged1), which were further superimposed on bright-field images (Merged2). Bars, 10 µm. HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
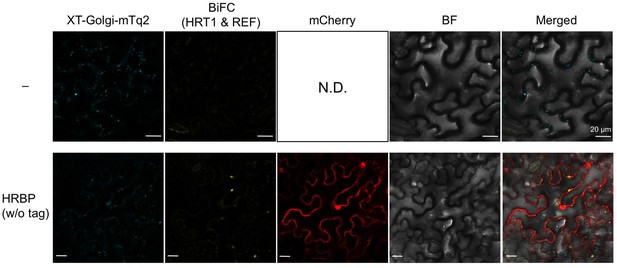
BiFC assays for interaction between HRT1 and REF in cells expressing HRBP.
N. benthamiana epidermal cells were infected with an Agrobacterium strain harbouring a BiFC binary vector constructed with pDOE-07 to assay HRT1-REF interaction (upper panels) or with mixture of two Agrobacterium strains, harbouring the BiFC binary vector for HRT1-REF interaction and a binary vector to express HRBP without protein tag and mCherry as an introduction marker, respectively (lower panels). Fluorescent images were observed using confocal laser microscope. Fluorescence signals corresponding to mVenus re-assembly (BiFC), mTq2 fused with the Golgi marker peptide (XT-Golgi-mTq2) and mCherry were obtained and superimposed to create merged images (Merged1), which were further superimposed (Merged2) on bright-field images (BF). Bars, 20 µm. HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
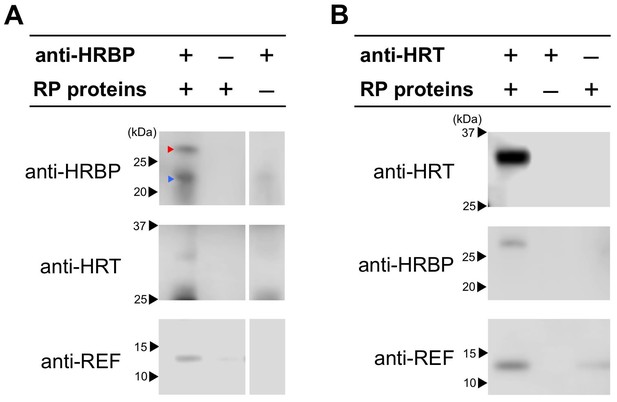
Detection of the ternary complex formation on RP by co-immunoprecipitations.
CHAPS-solubilized proteins from RPs were applied for immunoprecipitation using anti-HRBP antibody (A) or anti-HRT1/HRT2 antibody (B). The immunoprecipitation experiment without an antibody or the solubilized proteins were also conducted as negative controls. Immunoprecipitated proteins were analyzed by the immunodetections with each antibody indicated. Red and blue arrowheads on the top panel of (A) indicate HRBP and IgG light-chains of the anti-HRBP antibody, respectively. HRBP, HRT1-REF bridging protein.
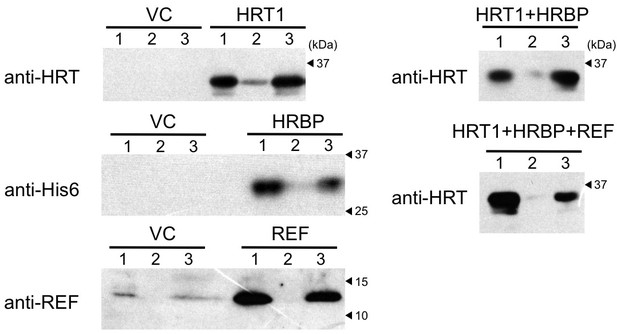
Cell-free translation-coupled protein introduction onto WRPs.
Immunodetection of HRT1, His6-tagged HRBP and REF with anti-HRT1/HRT2 antibody, anti-His6 antibody and anti-REF antibody, respectively. After in vitro translation to express the protein(s) indicated at the upper part of each lane, total proteins (lane 1) and soluble (lane 2) and RP (lane 3) fractions, recovered after ultracentrifugation of the corresponding total proteins, were analyzed. VC: vector control. HRBP, HRT1-REF bridging protein; REF, rubber elongation factor; RP, rubber particle.
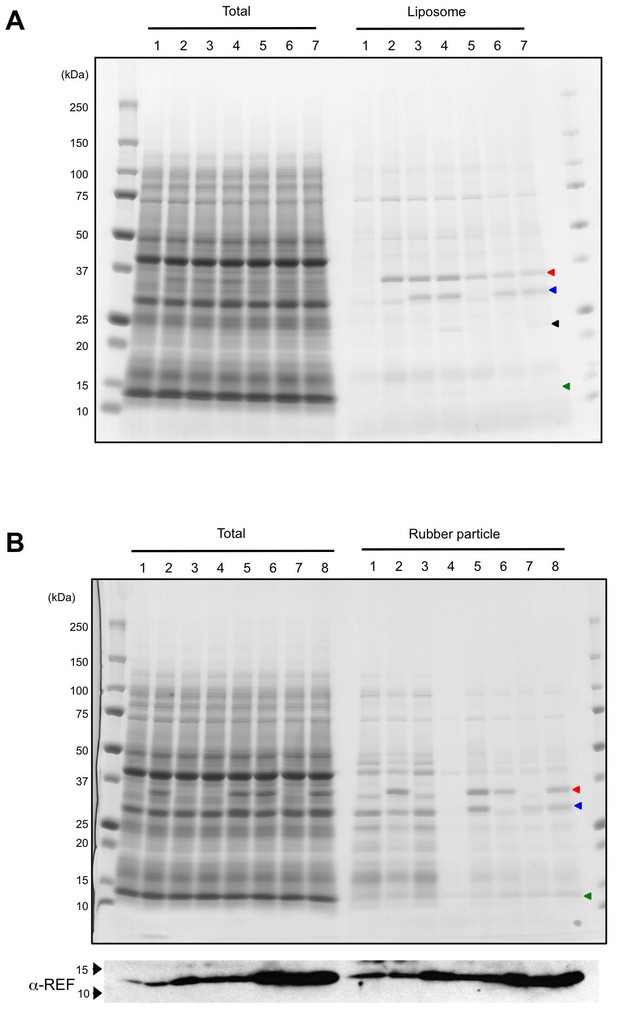
In vitro translation of HRT1, HRT2, HRBP and REF.
(A) SDS-PAGE of proteins derived from the cell-free protein expression system supplemented with liposomes. After the in vitro protein expression reaction (see Materials and methods), total crude proteins (Total) and proteins on resulting proteoliposomes separated by centrifugation (Liposome) were analyzed on 5–20% SDS-PAGE gel and stained with CBB. 1: vector control; 2: HRT1; 3: HRT1, HRBP and REF; 4: HRT1, HRBP, REF and SRPP; 5: HRT2; 6: HRT2, HRBP and REF; 7: HRT2, HRBP, REF, and SRPP. Arrowheads indicate protein band positions corresponding to HRT1/HR2 (red), HRBP (blue), SRPP (black) and REF (green). (B) SDS-PAGE of proteins derived from the cell-free protein expression system supplemented with WRPs. After the in vitrotranslation reaction (see Materials and methods), total crude proteins (Total) and proteins on RPs separated by ultracentrifugation (Rubber particle) were analyzed on 5–20% SDS-PAGE gel and stained with CBB. Lower panel indicated immunodetection of REF with an anti-REF antibody. 1: vector control; 2: HRT1; 3: HRTBP; 4: REF; 5: HRT1 and HRBP; 6: HRT1 and REF; 7: HRBP and REF; 8: HRT1, HRBP and REF. Arrowheads indicate protein band positions corresponding to HRT1/HR2 (red), HRBP (blue) and REF (green). HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
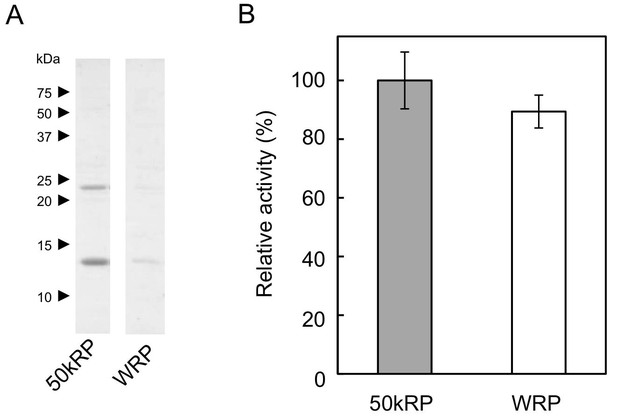
Sustained proteins and RTase activity on WRP applied for the cell-free translation.
(A) Analyses of proteins sustained on 50kRP and the WRP applied for the cell-free translation-coupled protein introduction. Proteins were solubilized from each sample with a SDS-PAGE sample buffer, analysed using SDS-PAGE, and then visualised by Coomassie Brilliant Blue staining. (B) Relative RTase activities of WRPs, with RTase activity of non-detergent-washed RPs (50kRP) set at 1.0. RTase activities were assayed using the standard conditions (see Materials and methods). Results are presented as the mean of three independent determinations ± SD. RPs, rubber particles.
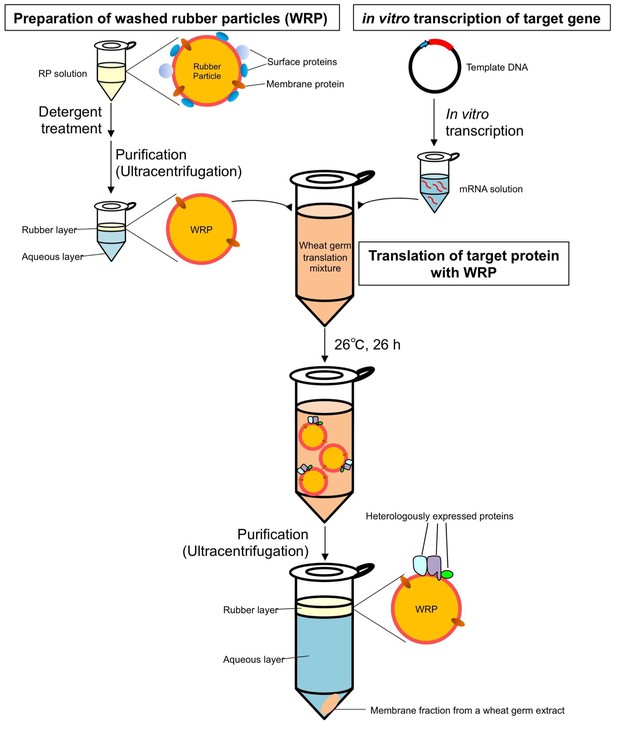
Workflow of protein expression on washed rubber particles with the wheat germ cell-free system.
https://doi.org/10.7554/eLife.19022.021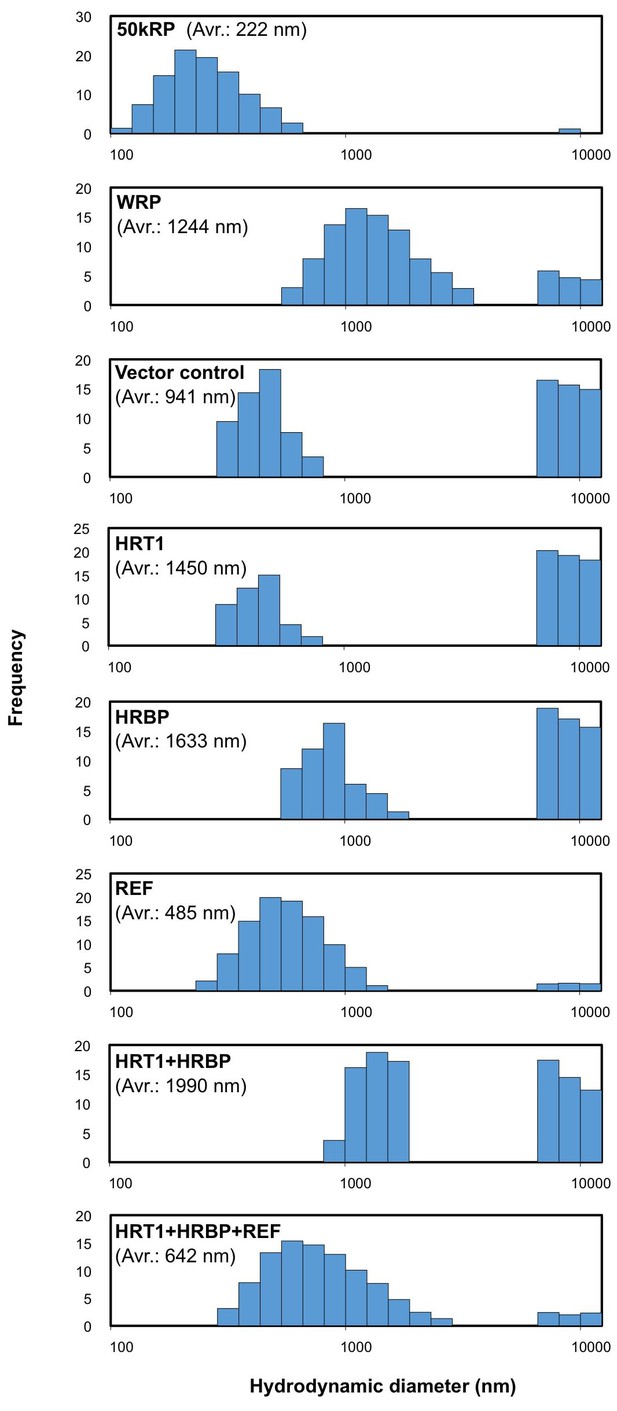
Size distributions of RPs after the in vitro translation reaction.
Typical results obtained by measurement of scattering light distributions from more than three measurements by dynamic light scattering are shown. The number in parenthesis indicates the apparent average diameter of RPs. RPs, rubber particles.
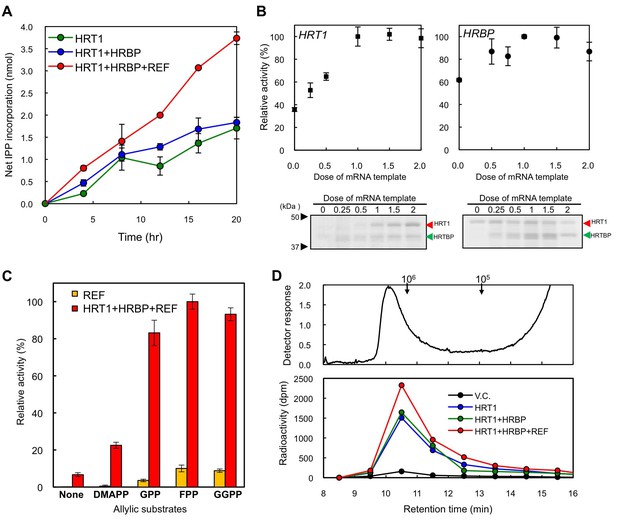
Enzymatic characterization of HRT1, HRBP and REF introduced on WRPs.
(A) In vitro RTase activities of HRT1, HRBP and REF, introduced onto 1 µg of WRPs, in the reaction with FPP and 14C-IPP as substrates. The activity is expressed as the net IPP incorporation into the resulting rubber products, calculated by subtraction of the results with the vector control WRPs from those with the WRPs carrying the protein(s) as indicated. Results are presented as the mean of three independent determinations ± SD. (B) Dependencies on HRT1 and HRBP for the RTase activity of RPs carrying HRT1, HRBP and REF. Protein levels of HRT1 (left) or HRBP (right) on WRPs were varied by changing the dose of each mRNA template in the in vitro translation reaction from the optimized standard conditions (i.e. dose 1, see Materials and methods). Protein levels on WRPs were analyzed by SDS-PAGE (lower part of each assay result). Relative activities are presented as the mean of three independent determinations ± SD. (C) Allylic substrate dependencies of RTase activities in the 8 hr reactions with RPs carrying triple components or only REF (as a control). Allylic substrates (15 mM): dimethylallyl diphosphate (DMAPP, C10), GPP, FPP and GGPP. (D) GPC analysis of reaction products from RTase assays. Molecular mass distribution of endogenous rubber molecules contained in WRPs, monitored using a refractive index detector (upper panel) and those of (Qu et al., 2015) C-labelled products synthesized in vitroby WRPs carrying protein(s) as indicated (lower panel). Elution peaks of commercially available polyisoprene standards (molecular weights 105 and 106) are indicated at the top of the upper panel. FPP, farnesyl diphosphate; GPC, gel-permeation chromatography; GPP, geranyl diphosphate; HRBP, HRT1-REF bridging protein; REF, rubber elongation factor.
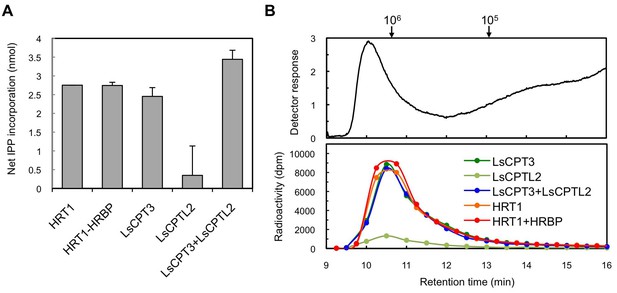
Enzymatic characterizations of LsCPT3 and LsCPT2 introduced on the Hevea WRPs.
(A) In vitro RTase activities of LsCPT3 and LsCPT2, introduced onto 1 µg of WRPs, in the reaction with FPP and 14C-IPP as substrates. The activity is expressed as the net IPP incorporation into resulting rubber products, calculated by subtraction of results with the vector control WRPs from those with the RPs carrying protein(s) as indicated. Results are presented as the mean of three independent determinations ± SD. (B) GPC analyses of reaction products from the RTase assays. Molecular mass distribution of endogenous rubber molecules contained in WRPs, monitored using a refractive index detector (upper panel) and those of 14C-labelled products synthesized in vitro by RPs carrying protein(s) as indicated. Elution peaks of commercially available polyisoprene standards (molecular weights 105 and 106) are indicated at the top of the upper panel. FPP, farnesyl diphosphate; RPs, rubber particles.
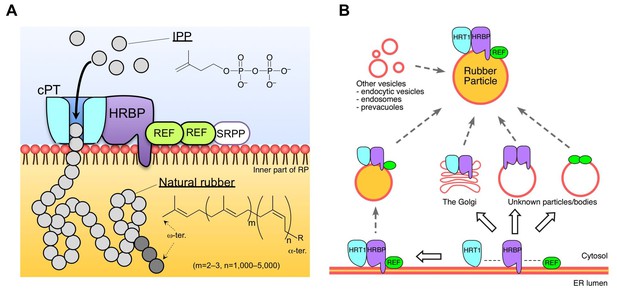
Schematic models of the rubber biosynthetic machinery on RPs (A) and RP formation correlated with the interactions of proteins (B) in the latex of H. brasiliensis. RPs, rubber particles.
https://doi.org/10.7554/eLife.19022.025Additional files
-
Supplementary file 1
List of primers used in this study.
- https://doi.org/10.7554/eLife.19022.026





