Auxin production in the endosperm drives seed coat development in Arabidopsis
Abstract
In flowering plants, seed development is initiated by the fusion of the maternal egg and central cells with two paternal sperm cells, leading to the formation of embryo and endosperm, respectively. The fertilization products are surrounded by the maternally derived seed coat, whose development prior to fertilization is blocked by epigenetic regulators belonging to the Polycomb Group (PcG) protein family. Here we show that fertilization of the central cell results in the production of auxin and most likely its export to the maternal tissues, which drives seed coat development by removing PcG function. We furthermore show that mutants for the MADS-box transcription factor AGL62 have an impaired transport of auxin from the endosperm to the integuments, which results in seed abortion. We propose that AGL62 regulates auxin transport from the endosperm to the integuments, leading to the removal of the PcG block on seed coat development.
https://doi.org/10.7554/eLife.20542.001eLife digest
The seeds of rice, wheat and other flowering plants store a variety of nutrients, largely in the form of sugars, proteins and oils. These stored reserves provide the main source of calories for humans and livestock all over the world, so they are of major social and economic importance.
Seed development is an intricate process. It begins after male sperm cells fuse with female gametes inside the flower. This leads to the formation of the embryo, which will develop into a new plant, and a structure called the endosperm, which nourishes the growing embryo. A protective seed coat surrounds the embryo and endosperm, which develops from certain parts of the parent flower. In order for the seed to develop successfully, these three components have to communicate so they can coordinate their growth.
Auxin is a key plant hormone that is needed for plants to grow and develop properly and is necessary for the endosperm to form. Previous research has shown that the endosperm is also required to trigger the formation of the seed coat, but the signal that triggers this process has not yet been identified. Figueiredo et al. now address this question in a small flowering plant called Arabidopsis thaliana.
The experiments show that the endosperm produces auxin, which acts as a molecular signal for the seed coat to start forming. Exposing unfertilized flowers to auxin caused a seed coat to form even though the endosperm was absent. This suggests that this hormone alone is sufficient to trigger the formation of the seed coat without any other signals. Further analysis revealed that a protein called AGL62 regulates the movement of auxin to the parts of the flower that give rise to the seed coat. In the absence of AGL62, the hormone remains trapped in the endosperm and the seed coat fails to develop. The next step following on from this work is to understand how auxin moves from the endosperm to the parts of the flower that form the seed coat.
https://doi.org/10.7554/eLife.20542.002Introduction
In flowering plants, fertilization of the two female gametes, egg cell and central cell by the two male sperm cells results in the development of the embryo and the endosperm, a nourishing tissue which supports embryo growth. These two fertilization products are surrounded by the seed coat, a sporophytic tissue of purely maternal origin. While the embryo is the only component of the seed that will form the next generation, it contributes little to seed growth. Instead, the interplay between the endosperm and seed coat determines the final size of the seed. Impaired endosperm proliferation has a negative effect on seed coat development, as is the case in the Arabidopsis haiku or miniseed mutants (Garcia et al., 2003; Luo et al., 2005) or in transgenic lines expressing diphtheria toxin A in the endosperm (Weijers et al., 2003). And, conversely, mutations that affect seed coat expansion limit endosperm growth, such as transparent testa glabra 2 (Garcia et al., 2005), while mutants with increased integument cell proliferation like megaintegumenta/auxin responsive factor 2, result in enlarged seeds with more abundant endosperm (Schruff et al., 2006). Furthermore, the absence of the endothelium integument layer results in seed abortion, highlighting the importance of the developing seed coat for the establishment of a viable seed (Mizzotti et al., 2012).
Endosperm initiation in Arabidopsis is coupled to the production of auxin in the central cell following fertilization (Figueiredo et al., 2015); however, the factor initiating seed coat development is yet to be described. Seed coat initiation is dependent on the fertilization of the female gametophyte, but mutants for sporophytically-acting Polycomb Group (PcG) proteins develop a seed coat without fertilization in a dosage-sensitive manner (Roszak and Köhler, 2011). PcG proteins assemble into multimeric complexes, of which the Polycomb Repressive Complex 2 (PRC2) represses target loci through the deposition of trimethyl groups on lysine 27 of histone H3 (H3K27me3) (Mozgova et al., 2015). Therefore, the initiation of seed coat development prior to fertilization is inhibited by sporophytically active PRC2 complexes and this block is relieved following fertilization through a signal derived from the fertilization products (Figueiredo and Köhler, 2014; Mozgova et al., 2015). Seed coat development is dependent on the development of the sexual endosperm (Weijers et al., 2003; Ingouff et al., 2006; Roszak and Köhler, 2011), strongly suggesting that the seed coat-initiation signal is generated in the fertilized central cell/endosperm.
Here, we show that seed coat development requires production of auxin in the fertilized central cell/endosperm and most likely the transport of auxin to the ovule integuments in an AGL62-dependent manner. We demonstrate that application of auxin is sufficient to drive seed coat development and that AGL62 regulates the expression of P-GLYCOPROTEIN 10 (PGP10), in the endosperm, which likely functions as an auxin transporter to the sporophytic tissues. Finally, we show that auxin is sufficient to remove the PRC2 block on seed coat development.
Results
Auxin and gibberellin signaling are active in the developing seed coat
Seed coat initiation is dependent on the fertilization of the central cell by one of the paternally-contributed sperm cells, but this requirement can be bypassed in mutants of sporophytic PRC2 components that initiate the autonomous development of the seed coat (Roszak and Köhler, 2011). In order to identify the signaling pathways that could be involved in seed coat initiation, we generated transcriptome data from non-fertilized wild-type (WT) ovules and vrn2/- emf2/+ ovules at four days after emasculation (4 DAE), and WT seeds at two days after pollination (2 DAP). In contrast to WT ovules, non-fertilized vrn2/- emf2/+ ovules initiate autonomous seed coat development (Roszak and Köhler, 2011), correlating with the activation of genes involved in auxin and gibberellin (GA) response or signaling that became also activated in fertilized seeds (Table 1 and Table 1—source data 1).
Significantly enriched biological processes for genes commonly upregulated in fertilized WT and autonomous vrn2 emf2/+ seeds, compared to unfertilized WT ovules (p-value<0.05).
GO-term | p-value | Number of genes | Description |
|---|---|---|---|
GO:0016043 | 2,12E-10 | 63 | cell organization and biogenesis |
GO:0006412 | 2,17E-09 | 66 | translation |
GO:0009058 | 1,46E-07 | 96 | biosynthetic process |
GO:0042545 | 1,04E-04 | 10 | cell wall modification |
GO:0009753 | 6,58E-04 | 10 | response to jasmonic acid |
GO:0009739 | 1,80E-03 | 8 | response to gibberellin |
GO:0009605 | 1,94E-03 | 16 | response to external stimulus |
GO:0008361 | 1,98E-03 | 10 | regulation of cell size |
GO:0009861 | 2,09E-03 | 10 | jasmonic acid and ethylene-dependent systemic resistance |
GO:0009611 | 2,60E-03 | 12 | response to wounding |
GO:0016049 | 5,22E-03 | 9 | cell growth |
GO:0006694 | 7,68E-03 | 4 | steroid biosynthetic process |
GO:0007155 | 8,49E-03 | 4 | cell adhesion |
GO:0009723 | 1,02E-02 | 8 | response to ethylene |
GO:0009733 | 1,46E-02 | 12 | response to auxin |
GO:0007267 | 1,81E-02 | 3 | cell-cell signaling |
GO:0007276 | 1,96E-02 | 6 | gamete generation |
GO:0009698 | 2,51E-02 | 7 | phenylpropanoid metabolic process |
GO:0009813 | 2,60E-02 | 4 | flavonoid biosynthetic process |
GO:0009812 | 3,53E-02 | 4 | flavonoid metabolic process |
-
Table 1—source data 1
Seed and ovule transcriptome data.
This table includes absolute and relative gene expression values for the microarray data of WT vs. vrn2/- emf2/+ ovules and seeds, and for the mRNAseq data of WT vs. agl62/+ seeds.
- https://doi.org/10.7554/eLife.20542.004
Post-fertilization activation of both auxin and GA signaling was previously shown in Arabidopsis seeds (Dorcey et al., 2009). In order to test whether this activation was specific to the seed coat we investigated the behavior of auxin and GA reporter lines before and after fertilization (Figure 1). To monitor auxin signaling, we investigated expression of the DR5v2::VENUS reporter (Liao et al., 2015) before and after fertilization. There was no DR5v2::VENUS reporter activity in unfertilized ovules (Figure 1A), with the exception of a few cells neighboring the vascular bundle and occasionally near the antipodal cells. However, shortly after fertilization VENUS activity was detectable in the sporophytic tissues, where it persisted throughout seed development, with particular incidence in the funiculus and the micropylar region (Figure 1B–C). We further substantiated these observations with the use of the R2D2 auxin sensor system (Liao et al., 2015). Before fertilization there was a strong DII:VENUS expression in the integuments, indicating very low or absent auxin signaling (Figure 1D). However, this signal was quickly depleted following fertilization, as early as the first division of the central cell (Figure 1E). Conversely, the auxin-insensitive mDII:Tdtomato remained stable in the seed coat, indicating a stable expression of the RPS5a promoter, that drives both reporter genes. These observations indicate a rapid accumulation of auxin in the sporophytic tissues following fertilization of the maternal gametes.
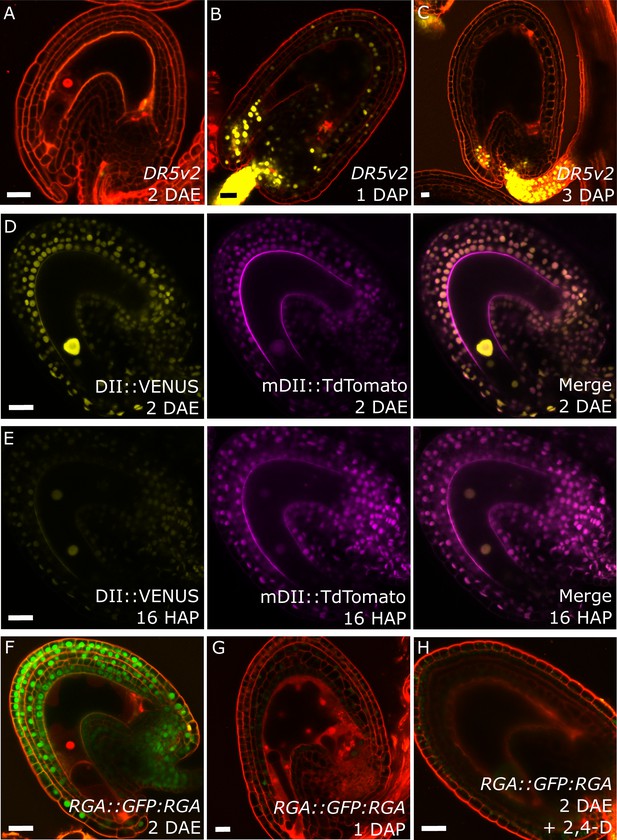
Fertilization activates auxin and GA signaling in the seed coat.
(A–C) DR5v2::VENUS expression before (A) and one and three days after pollination (B–C). (D–E) R2D2 auxin sensor before (D) and 16 hr after pollination (HAP) (E). (F–H) RGA::GFP:RGA reporter before (F), one day after pollination (G) and after 2,4-D treatment without pollination (H). Bars indicate 20 µm. Red staining is propidium iodide (PI).
We further analyzed plants expressing the DELLA reporter RGA::GFP:RGA, as a marker for active GA signaling (Dorcey et al., 2009). Before fertilization we observed a strong GFP signal in the integuments which quickly diminished after fertilization (Figure 1F–G), indicating an activation of GA signaling, similarly to what we observed with the auxin reporters.
To test whether auxin and GA act in the same pathway during seed coat development, we treated unpollinated pistils of plants expressing the RGA reporter with the synthetic auxin 2,4-Dichlorophenoxyacetic acid (2,4-D) and observed a removal of the RGA protein, similarly to what happens after fertilization or application of gibberellic acid (GA3; Figure 1H and Figure 1—figure supplement 1). Conversely, the application of GA3 did not have an influence on the expression of the auxin reporters DR5v2 or R2D2 (Figure 1—figure supplement 1). These observations reveal that auxin acts upstream of GA during seed coat development.
Auxin and GA trigger autonomous seed coat development
Seed coat growth initiating after fertilization is a process driven by cell elongation rather than cell division, since the integument cell number did not change after fertilization (Figure 1—figure supplement 2). Given that both auxin and gibberellin signaling is active in the developing seed coat, and that both hormones are known for having a role in cell growth and expansion (Rayle and Cleland, 1992; Cowling and Harberd, 1999), we tested whether exogenous application of either auxin or GA3 was sufficient to drive seed coat development. We treated unpollinated pistils with either GA3 or 2,4-D and investigated autonomous seed coat development after three days (Figure 2A–B). We used vanillin staining as a marker for the seed coat development, which stains the proanthocyanidins produced in the endothelium after fertilization (Debeaujon et al., 2000). Mock-treated ovules were either not or only weakly stained with vanillin, while treatments with GA3 or 2,4-D resulted in 30% to 40% of ovules showing strong or very strong staining. Furthermore, the ovules treated with either GA3 or 2,4-D were significantly larger than the mock-treated ones (Figure 2C).
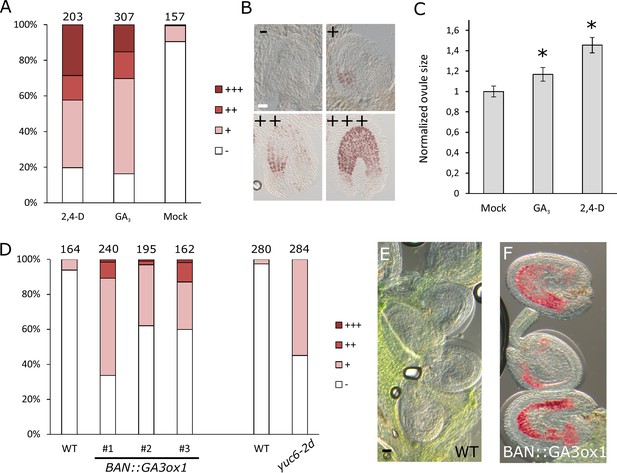
GA and auxin are sufficient to drive seed coat development.
(A–B) Scoring of vanillin staining of ovules after 2,4-D, GA3 and mock treatments, as categorized in (B). Numbers on top indicate total ovules counted. Bar indicates 20 µm. (C) Ovule area three days after 2,4-D, GA3 and mock treatments. Ten ovules were measured per treatment. The size is normalized to the mock-treated ovules. Error bars indicate standard deviation. * Differences are significant for p<0.00001(T-Test). (D) Scoring of vanillin staining in ovules of WT, auxin and GA-overexpressing lines, as categorized in (B). Numbers on top indicate total ovules counted. (E–F) Vanillin-stained WT (E) and BAN::GA3ox1 (F) ovules at 5 DAE. Bar indicates 50 µm.
We further tested whether over-production of either one of these hormones specifically in the integuments of unfertilized ovules would trigger seed coat development. We raised transgenic plants expressing a GA biosynthesis gene (GA3ox1) under the control of the BANYULS (BAN) promoter, which is active in the innermost integument/seed coat layer (Figure 2—figure supplement 1) (Debeaujon et al., 2003). We emasculated BAN::GA3ox1 transgenic plants and performed vanillin staining at 5 DAE to test for initiation of seed coat development. As with the GA3 treatments, we observed an increased number of ovules stained in the transgenic lines when compared to WT plants (Figure 2D–F). In order to test whether increased auxin production in the integuments triggers autonomous seed coat development, we analyzed the gain-of-function yuc6-2D mutant, in which the auxin biosynthesis gene YUCCA6 (YUC6) is ectopically expressed due to the insertion of a Cauliflower Mosaic Virus 35S (CaMV35S) enhancer sequence in its promotor region (Kim et al., 2007).Activation of the YUC6 gene in yuc6-2D is expected to be restricted to sporophytic tissues, as the CaMV35S enhancer is not active in the female gametophyte (Roszak and Köhler, 2011).We therefore tested whether increased auxin production in the integuments of yuc6-2D plants resulted in the autonomous development of the seed coat, as scored by vanillin staining. Indeed, up to 50% of yuc6-2D ovules stained with vanillin, while less than 5% of WT ovules were stained (Figure 2D).These observations reveal that both auxin and GA3 are sufficient to trigger seed coat development without fertilization.
Auxin production is necessary for seed coat development
Based on our findings that ectopic auxin and GA3 are sufficient to trigger the seed coat development and that auxin acts upstream of GA, we hypothesized that auxin may be the post-fertilization signal that triggers seed coat development. This hypothesis is in agreement with our previous observations that auxin is produced in the endosperm after fertilization (Figueiredo et al., 2015). In order to test whether auxin produced in the endosperm is necessary for seed coat development, we analyzed mutants deficient either in auxin signaling (axr1/+ axl-1/-) or auxin biosynthesis (wei8/- tar1/- tar2-1/+ and wei8/- tar1/- tar2-2/+). The homozygous axr1/- axl-1/- mutant is zygotic lethal, thus 25% of seeds from an axr1/+ axl-1/- plant will abort (Dharmasiri et al., 2007). Nevertheless, those seeds did not differ in size compared to WT seeds at 3 DAP and were only slightly smaller compared to their WT counterparts at 5 DAP, indicating that seed coat development is not impaired (Figure 3A and Figure 3—figure supplement 1). We thus conclude that auxin signaling in the fertilization products (endosperm and embryo) is not necessary to initiate seed coat development. In contrast, wei/tar mutants affected in auxin biosynthesis showed seeds aborting at different sizes (Figure 3B and Figure 3—figure supplement 1). Namely, around 2.5% of seeds completely failed to develop a seed coat and another 20–30% of seeds did not reach full WT size (Figure 3C–E). No differences were observed in the integument cell number between the mutant and WT ovules (Figure 1—figure supplement 2), indicating that cell expansion following fertilization is affected in the wei/tar mutant. These observations strongly support the idea that post-fertilization production of auxin in the fertilization products is necessary for the early stages of seed coat development in Arabidopsis, but that auxin signalling does not seem to be required for full seed expansion until later developmental stages.
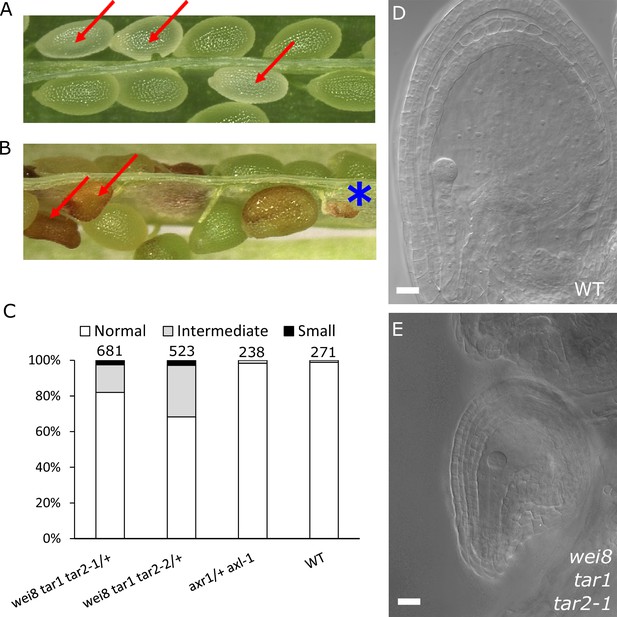
Auxin is necessary for seed coat development.
(A) Opened silique of an axr1 axl-1/+ mutant showing aborting white seeds (red arrows) that undergo full seed coat development. (B) Opened silique of a wei8 tar1 tar2-1/+ mutant showing seeds that do not develop a seed coat (asterisk) and seeds aborting at an intermediate stage compared to WT (red arrows). (C) Determination of final seed size in auxin biosynthesis and signaling mutants and WT. Numbers on top indicate total seeds counted. (D) WT seed at 2 DAP, showing a developed seed coat. (E) wei8 tar1 tar2-1 seed at 2 DAP that failed to develop seed coat. Bars indicate 20 µm.
Previous work had revealed that the endosperm, rather than the embryo, is solely responsible for triggering the seed coat development in Arabidopsis (Weijers et al., 2003; Ingouff et al., 2006; Roszak and Köhler, 2011). Thus, our data showing that auxin biosynthesis after fertilization is necessary for seed coat development, strongly supports our hypothesis that auxin needs to be produced in the endosperm to initiate seed coat development. Additionally, we previously showed that seeds mutant for axr1 axl-1 have endosperm proliferation defects (Figueiredo et al., 2015). Our observations reported here that these seeds develop a seed coat similar to that of WT seeds indicates that the proliferation of the endosperm per se is not required for seed coat initiation. This is in line with previous observations that mutants with severe endosperm proliferation defects, such as the titan mutants, still develop a seed coat (Liu and Meinke, 1998).
Production of auxin in the central cell drives seed coat development
To test whether production of auxin in the central cell is sufficient to trigger seed coat development, we expressed the auxin biosynthesis genes TAA1 and YUC6 under control of the central cell and early endosperm specific promoter DD65 (Steffen et al., 2007; Figueiredo et al., 2015) and investigated the autonomous seed coat development at 5 DAE. Indeed, DD65::TAA1; DD65::YUC6 expressing ovules initiated seed coat development, as evidenced by a significant increase in size (Figure 4A–B) and the production of protoanthocyanidins (Figure 4C–D). The integument cell number was not affected in the transgenic lines (Figure 1—figure supplement 2). The increased ovule size in DD65::TAA1; DD65::YUC6 expressing lines correlated with an increased auxin signaling in sporophytic tissues, revealed by the activation of DR5v2 in the integuments as well as the removal of the DII:VENUS signal in R2D2 expressing lines (Figure 4—figure supplement 1). Ovules expressing DD65::TAA1; DD65::YUC6 initiate central cell division in the absence of fertilization (Figueiredo et al., 2015). However, the autonomous central cell division in these ovules only occurs at 6 DAE, while at 5 DAE seed coat development did clearly initiate. This data reveal that seed coat development precedes replication of the central cell, further indicating that endosperm development per se is not required for the initiation of seed coat development.
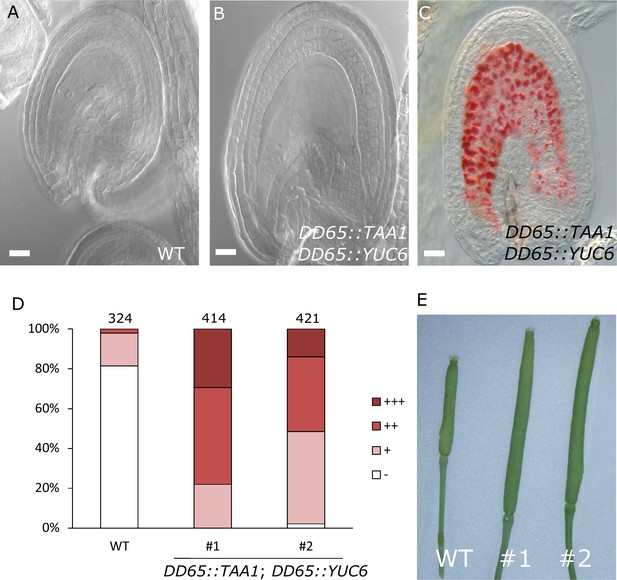
Auxin production in the central cell triggers seed coat development.
(A–B) Ovule development at 5 DAE: (A) WT and (B) DD65::TAA1; DD65::YUC6. (C) Vanillin-stained ovules of DD65::TAA1; DD65::YUC6 at 5 DAE. Bars indicate 20 µm. (D) Scoring of vanillin staining in WT and two independent transgenic lines expressing DD65::TAA1; DD65::YUC6. The scoring was done as indicated in Figure 2B. Numbers on top indicate total ovules counted. (E) Pistil size in WT and the two transgenic lines expressing DD65::TAA1; DD65::YUC6 at 5 DAE.
Our observations demonstrate that ectopic production of auxin in the central cell is sufficient to drive seed coat development without fertilization. Additionally, the transgenic lines showed pronounced parthenocarpic growth of the gynoecium (Figure 4E), supporting the hypothesis that endosperm-produced auxin likely acts as a signal to the maternal tissues, inducing fruit development (Dorcey et al., 2009).
agl62 seeds show retention of auxin in the endosperm
Our data strongly suggest that production of auxin in the developing endosperm is likely to be the trigger for seed coat development. To further challenge this hypothesis, we investigated plants mutant for the MADS-box transcription factor AGL62. agl62 seeds abort early after fertilization (around 3–4 DAP), correlating with early endosperm cellularization (Kang et al., 2008) and failure to develop a seed coat (Roszak and Köhler, 2011). The agl62 mutant did not show defects in integument cell number, implying compromised cell expansion following fertilization (Figure 1—figure supplement 2). To test whether the early cellularization of agl62 was the cause for the defects in seed coat development, we crossed plants mutant for agl62/+ with a mutant for HALLIMASCH (hal/+), whose seeds fail to undergo endosperm cellularization (Mayer et al., 1999). In agl62 seeds the endosperm cellularizes at around 3 DAP, but in double mutant agl62 hal seeds this early cellularization phenotype is reverted (Figure 5A–C). However, similarly to the single agl62 mutant, agl62 hal seeds do not develop a seed coat and remain small. We thus propose that early cellularization of agl62 endosperm is a consequence, rather than a cause of the failure to develop seed coat. Given that AGL62 is solely expressed in the endosperm (Kang et al., 2008; Roszak and Köhler, 2011), we hypothesized whether this transcription factor could be involved in regulating the pathways driving seed coat development. Consistently, GA signaling was not activated in agl62 integuments, as evidenced by the maintained GFP signal of the RGA DELLA reporter (RGA::GFP:RGA) in agl62 seeds (Figures 1G and 5D). Importantly, also auxin signaling was impaired in agl62 integuments, which failed to express the DR5::VENUS reporter. Strikingly however, while no VENUS signal was observed in agl62 integuments, we could clearly observe VENUS expression in the endosperm of agl62 seeds (Figure 5E–F), contrasting the absence of VENUS activity in WT seeds. These results strongly suggest that in WT seeds, auxin is produced in the endosperm after fertilization and quickly exported into the integuments. However, in the absence of AGL62, auxin export is impaired and it accumulates in the endosperm at sufficiently high levels to activate DR5 expression.
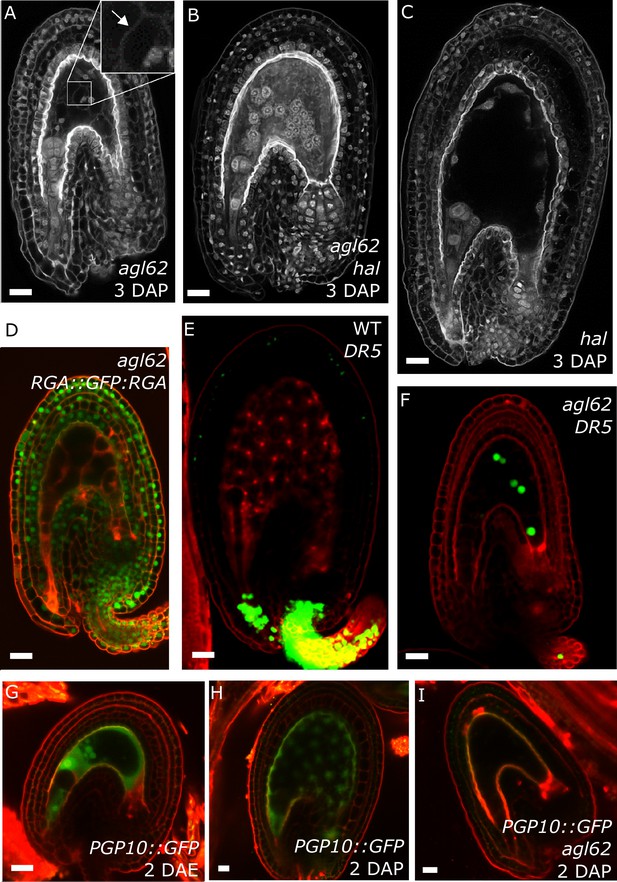
agl62 seeds fail to develop a seed coat.
(A–C) Endosperm cellularization as determined by Feulgen staining in agl62 (A), agl62 hal (B) and hal (C) seeds at 3 DAP. Inlay in (A) shows cellularized endosperm (arrow indicates cell wall). (D) Expression of RGA::GFP:RGA in agl62 mutant seeds at 1 DAP. See Figure 1 for expression of the same reporter in WT seeds. (E–F) DR5::VENUS expression in WT (E) and agl62 (F) seeds at 3 DAP. (G–I) PGP10::GFP expression in ovules (G) and in WT (H) and agl62 (I) seeds at 2 DAP. Bars indicate 20 µm. Red staining is PI.
Our observation that auxin is trapped in the endosperm of agl62 seeds suggests that AGL62 regulates the expression or activity of auxin transporters in the endosperm. We therefore investigated the transcriptome of agl62 seeds at 30 hr after manual pollination and searched for genes encoding putative auxin transporters that were downregulated in this mutant compared to WT seeds. We found that PGP10, a gene coding for an ABCB-type transporter was significantly downregulated in agl62 seeds (log2FC = −3.27, p-value=−2.83E-41; See also Table 1—source data 1). PGP-type transporters are active auxin transporters (Geisler et al., 2005; Lin and Wang, 2005), raising the possibility that lack of PGP10 expression could account for the accumulation of auxin in the endosperm of agl62 seeds.
In order to test whether PGP10 was expressed in the endosperm, we developed a PGP10::GFP reporter and investigated its expression in ovules and seeds of Arabidopsis. Consistently, PGP10 was expressed before fertilization in the central cell and its expression was maintained in the endosperm after fertilization (Figure 5G–H). The expression of PGP10 required AGL62 function, as PGP10 expression was not detectable in agl62 seeds at 2 DAP (Figure 5I), in agreement with the agl62 transcriptome data. Nevertheless, the PGP10 expression in agl62 under control of the AGL62 promoter was not sufficient to induce seed coat development (Figure 5—figure supplement 1), suggesting that additional factors are required to activate PGP10 function, similarly as previously reported for other PGPs (Bouchard et al., 2006; Wu et al., 2010; Wang et al., 2013).
Auxin and sporophytic PRC2s work in the same pathway during seed coat development
Previous observations revealed that sporophytic PRC2s exert a block on seed coat development that is lifted following fertilization (Roszak and Köhler, 2011). Lack of the core PRC2 subunits VRN2 and EMF2 results in a dosage-dependent autonomous seed coat development, as visualized by staining of protoanthocyanidins using vanillin (Figure 6A–C). Thus, lack of PRC2 function and ectopic auxin are sufficient to initiate seed coat development in the absence of fertilization; we therefore asked whether auxin and PRC2 act in the same pathway initiating seed coat development. To test this hypothesis, we generated triple mutant plants that have reduced PRC2 function and produce auxin ectopically in the integuments by combining mutant alleles for yuc6-2d, vrn2/- and emf2/+. No differences in integument cell number were observed in these mutants, compared to WT, implying that autonomous ovule growth is mediated by cell expansion (Figure 1—figure supplement 2). By scoring protoanthocyanidin production as a proxy for seed coat formation we compared the number of ovules forming autonomous seed coat in this triple mutant to those formed in the yuc6-2d single mutant and the vrn2/- emf2/+ double mutant. Ovules of the triple mutant and the yuc6-2d single mutant initiated autonomous seed coat development at similar frequencies (Figure 6D), suggesting that auxin and PRC2 act in the same pathway.
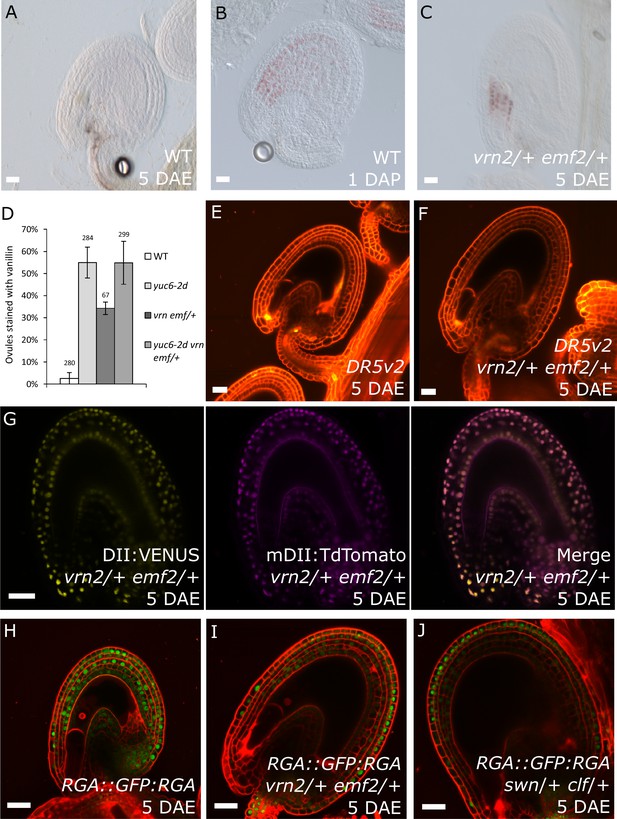
PRC2 and auxin work in the same pathway during seed coat development.
(A–C) Vanillin-stained WT ovule at 5 DAE (A), WT seed at 1 DAP (B) and vrn2/+ emf2/+ autonomous seed at 5 DAE (C). (D) Percentage of vanillin-stained ovules per silique at 6 DAE in WT and mutant lines. Bars indicate standard deviation. (E–F) DR5v2::VENUS activity in WT (E) and vrn2/+ emf2/+ (F) unfertilized ovules at 5 DAE. Red is PI. See also Figure 1 for DR5v2::VENUS activity in fertilized seeds. (G) R2D2 reporter activity in autonomous seeds of PRC2 mutant vrn2/+ emf2/+ at 5 DAE. See also Figure 1. (H–J) RGA::GFP:RGA activity at 5 DAE in WT ovules (H) or autonomous seeds of mutants for sporophytic PRC2 components: vrn2/+ emf2/+ (I) and swn/+ clf/+ (J). Red staining is PI. Bars indicate 20 µm.
Based on the results showing that auxin is the endosperm-derived seed coat initiation signal, it seems most likely that auxin acts upstream of PRC2 during seed coat development. That being the case, autonomous seed coat development in mutants for the PRC2 core components VRN2 and EMF2 should not coincide with the activation of auxin signaling in the integuments. To test this hypothesis, we crossed the DR5v2 and R2D2 auxin reporters into the vrn2 emf2 double mutant background. As predicted, despite that vrn2/+ emf2/+ ovules initiated seed coat development at 5 DAE, there was no detectable expression of DR5v2, contrasting to its expression post-fertilization (Figure 6E–F and Figure 1B–C). Similarly, we did not observe an efficient removal of the DII:VENUS signal from vrn2/- emf2/+ ovules, unlike what happens post-fertilization (Figure 6G and Figure 1E). These observations strongly support the hypothesis that auxin acts upstream of the PRC2 complex, and might have a role in removing the PRC2-block on seed coat development. To investigate how PRC2 function is depleted following either fertilization or application of auxin, we analyzed transgenic lines expressing translational reporters for the PRC2 components MSI1 and SWN, as well as for the PRC2-associated protein LHP1 (Derkacheva et al., 2013). Before fertilization there was a strong expression of all three reporters in the integuments, which markedly decreased following fertilization (Figure 7). Interestingly, a similar decrease in reporter activity was observed in unfertilized ovules following exogenous application of 2,4-D (Figure 7C,F and I), but no changes were observed in mock-treated ovules (Figure 7—figure supplement 1). These observations support the view that post-fertilization auxin transport to the integuments leads to the removal of PRC2 and LHP1 proteins. To test whether the downregulation of PRC2 components occurred at the transcriptional level, we tested the expression of genes coding for sporophytic PRC2 components. All the genes tested were strongly downregulated after fertilization (Figure 7J) as well as after exogenous application of 2,4-D in unfertilized ovules (Figure 7—figure supplement 1). This data reveals that the auxin-dependent removal of PRC2 function is likely to occur at the transcriptional level; nevertheless, an additional post-translational regulation of PRC2 components by auxin cannot be completely ruled out.
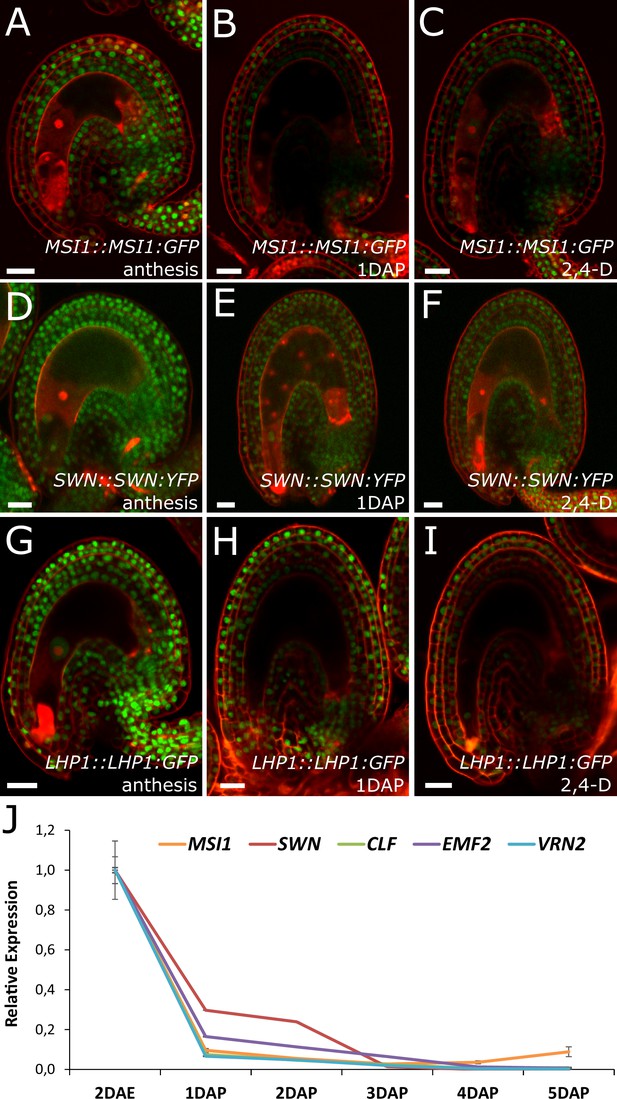
PcG activity is reduced in the developing seed coat.
(A–I) Expression of MSI1:MSI1::GFP (A–C), SWN:SWN::GFP (D–F) and LHP1:LHP1::GFP (G–I) reporters in 2 DAE ovules (A,D and G), 1 DAP seeds (B,E and G) and 2,4-D treated ovules at 3 DAE (C, F and I). For mock controls see figure supplement 1. Red staining is PI. Bars indicate 20 µm. (J) Relative expression of sporophytic PRC2 genes during seed development, as determined by RT-qPCR. Gene expression was measured at 2DAE and from 1 to 5 DAP and was normalized to the 2 DAE time-point. Error bars indicate standard deviation.
Given that GA signalling is downstream of auxin during seed development (Figure 1F–H), we asked whether the seed coat growth in mutants for sporophytic PRC2s would coincide with the ectopic activation of GA signaling in the integuments, similarly to what happens following application of auxin. Indeed, the GA biosynthesis gene GA20ox2 was upregulated in vrn2/- emf2/+ ovules while the GA catabolism genes GA2ox6 and GA2ox2 were downregulated when compared to WT (Table 1—source data 1 and Figure 6—figure supplement 1). To further test this hypothesis, we crossed the GA reporter RGA::GFP:RGA into mutants deficient for either PRC2 components VRN2 and EMF2 or SWN and CLF, and analyzed GFP expression in unfertilized ovules. Indeed, when compared to WT ovules, the ovules mutant for vrn2/+ emf2/+ or swn/+ clf/+ had a strongly reduced GFP:RGA signal, particularly in the innermost layers of the integuments (Figure 6H–J) This was observed both at 5 DAE, when the autonomous seeds were significantly larger than WT ovules, but also at 2 DAE, indicating that activation of GA signalling takes place shortly after anthesis (Figure 6—figure supplement 2). This decreased signal is most likely due to RGA protein degradation, since the RGA gene was not downregulated in the vrn2/- emf2/+ mutant (Table 1—source data 1 and Figure 6—figure supplement 1). We conclude that GA signaling is activated in the absence of fertilization, when PRC2 function is removed from the integuments.
Discussion
In this manuscript we reveal that auxin is the missing link connecting fertilization and seed coat development. As the maternal integuments do not take part in the fertilization process, the signal that coordinates the development of the fertilization products with the maternal integuments remained elusive (Figueiredo and Köhler, 2016). Based on previous research it seemed likely that the signal is generated in the endosperm, as endosperm development is required to drive seed coat initiation in Arabidopsis (Weijers et al., 2003; Ingouff et al., 2006; Roszak and Köhler, 2011). It was furthermore known that the presence of the paternal genome is necessary for the seed coat to develop, as mutants for cdka;1 and fbl17, in which one of the sperm cells fails to undergo karyogamy with the central cell, do not develop a seed coat (Gusti et al., 2009; Aw et al., 2010). These observations strongly suggest that the signal driving seed coat development should be dependent on genes that are paternally-expressed in the endosperm. Auxin biosynthesis following fertilization of the central cell is dependent on the paternally-expressed genes YUC10 and TAR1 (Figueiredo et al., 2015), raising the hypothesis that auxin could be the trigger for seed coat initiation. In this manuscript we provide multiple lines of evidence supporting this hypothesis: (i) auxin rapidly accumulates in the integuments after fertilization, (ii) impaired auxin biosynthesis but not auxin signaling in the endosperm causes defects in seed coat development, (iii) ovules of transgenic lines producing auxin ectopically in the central cell initiate seed coat development without fertilization, (iv) failure of seed coat development in the agl62 mutant correlates with failure to export auxin. Collectively, our data strongly support the hypothesis that auxin generated in the fertilized central cell/endosperm is the trigger for seed coat formation. Impaired auxin signaling in the endosperm did not impair seed coat formation, strongly supporting the hypothesis that auxin is exported to the integuments, where it initiates downstream signaling events. We could show that the ABCB transporter PGP10 is expressed in the fertilized central cell and its expression depends on the type I MADS-box transcription factor AGL62, suggesting that PGP10 may regulate the export of auxin from the endosperm to the integuments. PGPs are known to depend on additional proteins for their localization and function (Bouchard et al., 2006; Wu et al., 2010; Wang et al., 2013), likely explaining our failure to restore seed coat development in agl62 by ectopically expressing PGP10. It is furthermore possible that additional AGL62 targets are required for successful auxin export from the endosperm and are therefore lacking in the agl62 mutant. Future research will focus on identifying the remaining factors required for PGP10 function.
Post-fertilization auxin production was shown to lead to activation of GA biosynthesis in the ovules, which is then transported to the valves to promote silique growth (Dorcey et al., 2009). Our observation that auxin produced in the central cell is sufficient to drive parthenocarpic growth of the silique couples the fertilization event to fruit development.
We have previously shown that auxin drives endosperm development in Arabidopsis (Figueiredo et al., 2015). We now propose a dual role for post-fertilization auxin production, both in driving endosperm and seed coat development. Additionally, the agl62 mutant has defects both in seed coat development in fertilized ovules and endosperm development in autonomous seeds. This suggests that AGL62, like auxin, is involved in mediating both endosperm and seed coat developmental pathways. It was recently proposed that AGL62 in the endosperm also mediates non-autonomous cell signalling driving nucellus degeneration (Xu et al., 2016). This transcription factor thus seems to be a central player in modulating diverse developmental pathways during seed development.
While ectopic auxin production in the central cell was sufficient to initiate seed coat development, the autonomous seeds did not reach the final size of a fertilized seed and collapsed at around 6–7 DAE. This implies that auxin is sufficient to initiate seed coat development, but that additional signals are necessary to support its full expansion and prevent the degeneration of the ovule. Recent work has shown that the innermost layer of the outer ovule integument perceives mechanical cues from the expanding endosperm, which affects cell wall thickening and GA metabolism in that cell layer (Creff et al., 2015). It is therefore an attractive hypothesis that auxin is the initial paternally-derived signal that drives the integument-to-seed coat developmental transition, and that mechanical pressure from the syncytial endosperm sustains seed coat growth. This view is supported by observations showing that mutants affected in syncytial endosperm proliferation have smaller seeds when compared to WT plants (Garcia et al., 2003; Luo et al., 2005; Zhou et al., 2009; Wang et al., 2010). Conversely, a gain-of-function mutation in SHORT HYPOCOTYL UNDER BLUE1 results in increased proliferation of the syncytial endosperm delayed cellularization and increased seed size (Zhou et al., 2009). Additionally, it cannot be ruled out that other signalling pathways contribute to sustain seed coat growth, such as small peptide-mediated signalling (Ingram and Gutierrez-Marcos, 2015; Figueiredo and Köhler, 2016)
Mutants in sporophytic PRC2 components develop seed coat autonomously, implying that the PRC2 imposes a block on seed coat development that has to be lifted upon fertilization (Roszak and Köhler, 2011). Here, we demonstrate that ectopic auxin can bypass the PRC2 block on seed coat development, which leads to activation of GA signaling and production of protoanthocyanidins in the integuments. We propose that transport of auxin from the developing endosperm is required for the removal of PRC2 function in the integuments, allowing for the development of the seed coat by activation of GA signaling (Figure 8). Also in mammals PRC2 regulates the transition from proliferation to differentiation during organogenesis and PRC2 function has to be removed to allow differentiation of embryonic stem cells (Leeb et al., 2010; Aldiri and Vetter, 2012). We further show that exogenous auxin is sufficient to remove PRC2 function from the ovule integuments. Our gene expression analysis revealed that sporophytic PRC2-coding genes are downregulated after fertilization, likely due to increased auxin signalling in the integuments. Auxin is known to extensively modulate transcription (Weijers and Wagner, 2016), but the mechanism by which it regulates PRC2-coding genes is yet to be understood. In fact, although there are reports on the transcriptional regulation of PRC2 genes in animals (Neri et al., 2012), there is very little information on how the plant PRC2 activity is regulated. Thus, we provide the first evidence on the modulation of the PRC2 activity by a plant hormone, as a way to modulate developmental transitions.
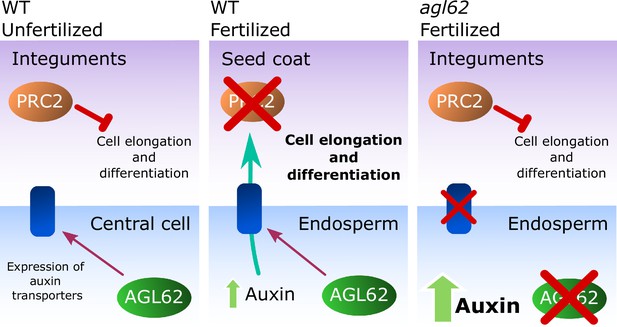
Model for the post-fertilization mechanism driving seed coat development.
Before fertilization (left panel) sporophytic PRC2 represses seed coat developmental pathways. The fertilization of the central cell (middle panel) results in the production of auxin that is exported to the integuments, lifting the PRC2 block on seed coat development and allowing for cell elongation and differentiation. However, this process is highly dependent on the activity of the transcription factor AGL62, since agl62 seeds fail to export auxin from the endosperm. This leads to auxin accumulation and consequently, failure to develop a seed coat (right panel). We propose that AGL62 activity is important for the expression, and eventually for the activity, of endosperm-specific auxin-exporters.
In conclusion, we have discovered that auxin produced in the fertilized central cell is required to induce seed coat formation after fertilization and most likely acts as mobile signal linking fertilization with the differentiation of the surrounding sporophytic tissues.
Materials and methods
Plant material, growth conditions and treatments
Request a detailed protocolThe Arabidopsis thaliana mutants and reporters used were described previously: emf2-5 (Yang et al., 1995), vrn2-1 (Chandler et al., 1996), swn-3 (Chanvivattana et al., 2004), clf-9 (Goodrich et al., 1997), wei8-1/- tar1/- tar2-1/+ and wei8-1/- tar1/- tar2-2/+ (Stepanova et al., 2008), axr1-12/+ axl-1/- (Dharmasiri et al., 2007), agl62-2/+ (Kang et al., 2008), hal/+ (Mayer et al., 1999), SWN::SWN:YFP (Wang et al., 2006), and LHP1::LHP1:GFP (Kotake et al., 2003).
Seeds were sterilized in 5% commercial bleach and 0.01% Tween-20 for 10 min and washed three times in sterile ddH2O. Sterile seeds were plated on ½ MS-medium (0.43% MS-salts, 0.8% Bacto Agar, 0.19% MES hydrate and 1% Sucrose; when necessary, the medium was supplemented with the appropriate antibiotics) and stratified at 4°C in the dark for 48 hr. Plates were then transferred to a growth chamber (16 hr light/8 hr dark; 110 μmol.s−1.m−2; 21°C; 70% humidity). After 10 days seedlings were transferred to soil and grown in a growth chamber (16 hr light/8 hr dark; 110 μmol.s−1.m−2; 21°C; 70% humidity).
The solutions used for hormone treatments contained 5% EtOH, 0.05% Tween-20 and 200 μM of 2,4-Dichlorophenoxyacetic acid (2,4-D) or 200 μM Gibberellic acid (GA3). In all experiments a mock control was run in parallel. Flowers were emasculated two days before anthesis and treated at 2 DAE by covering the whole pistils with 2 μL of the respective solution. The treated pistils were then collected at the specified time-points and processed for microscopy analyses, as described below.
Transcriptome analysis
Request a detailed protocolWT (Ler) and double mutant vrn2/- emf2/+ plants were emasculated at 1–2 days prior to anthesis. Two days after emasculation half of the WT plants were hand-pollinated. Unfertilized ovules and young seeds were dissected from the siliques at 4 DAE and 2 DAP, respectively, collected in RNAlater (Sigma-Aldrich Chemie GmbH, Switzerland) and ground frozen in a Silamat S5 for 3 times 7 s. Three biological replicates were generated per sample. RNA was extracted using the QIAGEN RNeasy KIT followed by on-column DNase treatment (QIAGEN Instruments AB, Switzerland). Labeling and hybridization to AGRONOMICS1 arrays (Affymetrix UK Ltd., United Kingdom) was done as described (Hennig et al., 2003). Signal values were derived from Affymetrix CEL files using RMA (RRID:SCR_008549) (Rehrauer et al., 2010). All data processing was done using the statistics package R (version 2.6.2, RRID:SCR_001905) that is freely available at http://www.rproject.org/. Quality control was done using the affyQCReport package in R (RRID:SCR_001318). In addition, coefficients of variation (cv) were calculated between replicates as a quantitative measure of data quality and consistency between replicates as described previously (Köhler et al., 2003). Differentially expressed genes were identified using the limma package in R (RRID:SCR_010943) (Smyth, 2004). Multiple‐testing correction was done using the q‐value method (Storey and Tibshirani, 2003). Probesets were called significantly differentially expressed when q < 0.05. The genes used in the analysis were selected by overlapping the list of significantly upregulated genes in 2 DAP seeds and in vrn2 emf2 ovules. The list of commonly upregulated genes was then used to determine enriched GO-terms using AtCOECIS. Only significantly enriched biological processes were considered (p-value<0.05). REVIGO was used to remove redundant GO terms and summarize the list. For the transcriptome analysis of agl62 seeds, we made use of homozygous agl62 mutant plants that arise at low frequency and that form small amounts of viable agl62/- seeds. agl62 plants were emasculated 1–2 days prior to anthesis, hand-pollinated and seeds were harvested 30 hr after pollination. Three biological replicates were generated per sample. Isolation of RNA and preparation of libraries was done as previously described (Wolff et al., 2015). RNA was sequenced at the Functional Genomics Center Zurich (Switzerland) on an Illumina HiSeq2000 (Ilumina, San Diego, USA) on two lanes in 100 bp paired end fashion. Sequencing reads were aligned to the TAIR 10.0 version of the Arabidopsis reference genome (Col-0) using TopHat (RRID:SCR_013035) (Trapnell et al., 2009). Only uniquely mapping reads were used for further analysis. Differentially expressed genes were identified using the DESeq package (Anders and Huber, 2010). Analysis of GO categories was performed using AtCOECiS (Vandepoele et al., 2009) and REVIGO (RRID:SCR_005825) (Supek et al., 2011). To enrich for biologically relevant changes, probesets with signal log ratio (SLR) > 0.6 were selected for the transcriptome analysis of vrn2/- emf2/+ and with SLR<−1 for the analysis of agl62. Affymetrix data and sequencing reads are deposited as CEL and fastq files, respectively, in the Gene Expression Omnibus (RRID:SCR_005012) (datasets GSE85751 and GSE85848).
Cloning and generation of transgenic plants
Request a detailed protocolTo clone the promoters of PGP10, BAN and AGL62, as well as the coding region of GA3ox1 and PGP10, WT Col-0 genomic DNA was used as a template. The primer sequences can be found in Table 2. The amplified fragments were purified from the gel, recombined into the donor vector (pDONR221) to create entry clones, and sequenced. Gateway cloning was done according to the manufacturer’s instructions (Invitrogen, Fisher Scientific, Sweden). The promoter of PGP10 was recombined into vector pB7FWG.0. The coding region of GA3ox1 and PGP10 were recombined into vector pB7WG2, where the 35 s promoter was replaced by that of BAN and AGL62, respectively, using the restriction enzymes SacI and SpeI. The MSI1 translational reporter was obtained by replacing the TAP fragment in the MSI::MSI1:TAP construct with GFP (Bouveret et al., 2006). The BAN::GFP reporter was generated by recombining the amplified promoter sequence of BAN and the coding sequence of GFP into the pBGW vector. Recombination was done using the In-Fusion enzyme (Clontech, Takara Bio Europe, France), following the manufacturer’s instructions.
Primer sequences.
Used for | Gene | Sequence 5’ - > 3’ * | |
|---|---|---|---|
RT-qPCR | AT5G58230 (MSI1) | fw | GCCCAAGTTCAGCTTCCTCT |
rv | TTTGTACCTTTCCAGTTGCACA | ||
AT4G02020 (SWN) | fw | CAACTCCTCTGGACGAATCAAG | |
rv | TCTGTTTTCCAAACCCTCGAGTC | ||
AT4G16845 (VRN2) | fw | TCATTCTCACAGAGTCCAGCC | |
rv | AGTCATCAAGCATCTGGCGAT | ||
AT5G51230 (EMF2) | fw | CGCACTTGATTTGGTGCTGG | |
rv | TGTTCATGGTTCGGGCATCA | ||
AT2G23380 (CLF) | fw | AAGTACTGCGGTTGCCCAAA | |
rv | ACATTCCCGATCTGCAGCAA | ||
Cloning | AT1G15550 (GA3ox1) | fw | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCAAGATGCCTGCTATGTTA |
rv | GGGGACCACTTTGTACAAGAAAGCTGGGTATCTAATCATTCTTCTCTGTGATTTCT | ||
AT1G61720 (promBAN) Gateway | fw | GGGGACAAGTTTGTACAAAAAAGCAGGCTGAGCTCTAACAGAACCTTACTGTAACACT | |
rv | GGGGACCACTTTGTACAAGAAAGCTGGGTACTAGTGAGTCTGGTCCATGGTTGTA | ||
AT1G61720 (promBAN) In-Fusion | fw | CCATGGCCGCGGGATATCAGATTCTTAGGTGAAGACAAG | |
rv | CGCTGAATGATTCATGATTGTACTTTTGAAATTACAG | ||
GFP | fw | ATGAATCATTCAGCGAAAACC | |
rv | CTTCACCTAAGAATCCATCTAGTAACATAGATGACA | ||
AT1G10680 (promPGP10) | fw | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGCGTTGCGTATAATCCGTT | |
rv | GGGGACCACTTTGTACAAGAAAGCTGGGTTTTCACTTTTGGATATGGAGAGA | ||
At5G60440 (promAGL26) | fw | TAAGCAGAGCTCGAATTGCATCTCGGCAATGAC | |
rv | TGCTTAACTAGTTTTTAGTGATATTTGAGAAGCT | ||
AT1G10680 (PGP10) | fw | GGGGACAAGTTTGTACAAAAAAGCAGGCTATGCAACCGTCAAATGATCCAG | |
rv | GGGGACCACTTTGTACAAGAAAGCTGGGTTTAAGGATGATGGCGCTGC | ||
-
*Primer adapters are underlined
The constructs were transformed into Agrobacterium tumefaciens strain GV3101 and Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). Transformants were selected with the appropriate antibiotics.
Histological and fluorescence analyses
Request a detailed protocolFor clearing of ovules and seeds the whole pistils/siliques were fixed with EtOH:acetic acid (9:1), washed for 10 min in 90% EtOH, 10 min in 70% EtOH and cleared over-night in chloralhydrate solution (66.7% chloralhydrate (w/w), 8.3% glycerol (w/w)). The ovules/seeds were observed under differential interference contrast (DIC) optics using a Zeiss Axioplan or Axioscope A1 microscopes (Carl Zeiss AB, Sweden). The vanillin staining was done on 5 DAE ovules in 1% (w/v) vanillin (4-hydroxy-3-methoxybenzaldehyde) in 6 N HCl. The emasculated pistils were incubated in this solution for 30 min and then the ovules were dissected out and mounted on a microscope slide. Images were recorded using a Leica DFC295 camera with a 0.63x optical adapter (Leica Microsystems, Sweden).
For fluorescence analysis seeds were mounted in 7% glucose. Where indicated, 0.1 mg/mL propidium iodide (PI) was used. Samples were analyzed under confocal microscopy on a Zeiss 780 Inverted Axio Observer with a supersensitive GaASp detector with the following settings (in nm; excitation-ex and emission-em): GFP – ex 488, em 499–525; PI – ex 488/514, em 635–719; YFP (VENUS) – ex 514, em 499–552 for DR5v2 and 525–543 for DII; tdTomato – ex 561, em 599–622. Images were acquired, analyzed and exported using Zeiss ZEN software.
For Feulgen staining of seeds, fixation, staining and embedding were performed as described (Braselton et al., 1996). Confocal imaging was performed using a Leica SP1-2, excitation wavelengths were set to 488 nm and detection to 535 nm and longer.
Ovule size and cell number measurements (Figure 1—figure supplement 2) were performed through confocal imaging of PI stained ovules. Each ovule was imaged at the median longitudinal plane, and its area was subsequently measured using the ImageJ software (RRID:SCR_002285) (Schneider et al., 2012). For each condition, the area of 10 ovules was measured. Average ovule area for each condition was normalized to the average area of mock treated ovules. The determination of integument cell number in all lines was done for the outer layer of the inner integument at 2 DAE and 10 ovules were analyzed per line. Ovule and seed area measurements for WT and auxin mutants (Figure 3—figure supplement 1) were performed through seed clearing (as described above) and measured using ImageJ software (RRID:SCR_002285) (Schneider et al., 2012). An average of 50 ovules/seeds was measured for each line per time-point.
RT-qPCR analyses
Request a detailed protocolOvules/seeds of 25 emasculated or hand-pollinated WT siliques were harvested at 2DAE, 1, 2, 3, 4 and 5 DAP in 20 μL of RNAlater solution (Invitrogen) and ground for 2 min using a TissueLyser II (Qiagen AB, Sweden). Total RNA was extracted using the Qiagen RNeasy kit, followed by DNase I treatment (Qiagen). cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Fisher Scientific, Sweden). Maxima SYBR Green qPCR Master Mix (Thermo Scientific) was used to perform the qPCR in an iQ5 qPCR system (Bio-Rad Laboratories AB, Sweden). The primers used for the RT-qPCR are described in Table 2. PP2A was used as the reference gene. Relative quantification of gene expression was performed as described (Pfaffl, 2001). Expression levels for each gene were normalized to the expression level at 2 DAE.
Data availability
-
Expression data of vrn2/- emf2/+ and wild-type ovules and seedsPublicly available at the NCBI Gene Expression Omnibus (accession no: GSE85751).
-
Expression data of agl62/- and wild-type seeds at 30 hrs after pollinationPublicly available at the NCBI Gene Expression Omnibus (accession no: GSE85847).
References
-
PRC2 during vertebrate organogenesis: a complex in transitionDevelopmental Biology 367:91–99.https://doi.org/10.1016/j.ydbio.2012.04.030
-
Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteinsJournal of Biological Chemistry 281:30603–30612.https://doi.org/10.1074/jbc.M604604200
-
Regulation of flowering time by Arabidopsis MSI1Development 133:1693–1702.https://doi.org/10.1242/dev.02340
-
Feulgen staining of intact plant tissues for confocal microscopyBiotechnic & Histochemistry 71:84–87.https://doi.org/10.3109/10520299609117139
-
Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongationJournal of Experimental Botany 50:1351–1357.https://doi.org/10.1093/jxb/50.337.1351
-
Arabidopsis MSI1 connects LHP1 to PRC2 complexesThe EMBO Journal 32:2073–2085.https://doi.org/10.1038/emboj.2013.145
-
Bridging the generation gap: communication between maternal sporophyte, female gametophyte and fertilization productsCurrent Opinion in Plant Biology 29:16–20.https://doi.org/10.1016/j.pbi.2015.10.008
-
Signalling events regulating seed coat developmentBiochemical Society Transactions 42:358–363.https://doi.org/10.1042/BST20130221
-
Arabidopsis haiku mutants reveal new controls of seed size by endospermPlant Physiology 131:1661–1670.https://doi.org/10.1104/pp.102.018762
-
Arabidopsis transcript profiling on Affymetrix GeneChip arraysPlant Molecular Biology 53:457–465.https://doi.org/10.1023/B:PLAN.0000019069.23317.97
-
Peptide signalling during angiosperm seed developmentJournal of Experimental Botany 66:5151–5159.https://doi.org/10.1093/jxb/erv336
-
Polycomb complexes act redundantly to repress genomic repeats and genesGenes & Development 24:265–276.https://doi.org/10.1101/gad.544410
-
Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cellsMolecular and Cellular Biology 32:840–851.https://doi.org/10.1128/MCB.06148-11
-
A new mathematical model for relative quantification in real-time RT-PCRNucleic Acids Research 29:e45.https://doi.org/10.1093/nar/29.9.e45
-
The Acid Growth Theory of auxin-induced cell elongation is alive and wellPlant Physiology 99:1271–1274.https://doi.org/10.1104/pp.99.4.1271
-
AGRONOMICS1: a new resource for Arabidopsis transcriptome profilingPlant Physiology 152:487–499.https://doi.org/10.1104/pp.109.150185
-
NIH Image to ImageJ: 25 years of image analysisNature Methods 9:671–675.https://doi.org/10.1038/nmeth.2089
-
Linear models and empirical bayes methods for assessing differential expression in microarray experimentsStatistical Applications in Genetics and Molecular Biology 3:Article3.https://doi.org/10.2202/1544-6115.1027
-
TopHat: discovering splice junctions with RNA-SeqBioinformatics 25:1105–1111.https://doi.org/10.1093/bioinformatics/btp120
-
Transcriptional responses to the auxin hormoneAnnual Review of Plant Biology 67:539–574.https://doi.org/10.1146/annurev-arplant-043015-112122
-
Endosperm and nucellus develop Antagonistically in Arabidopsis seedsThe Plant Cell 28:1343–1360.https://doi.org/10.1105/tpc.16.00041
-
Genetic regulation of shoot development in Arabidopsis: role of the EMF genesDevelopmental Biology 169:421–435.https://doi.org/10.1006/dbio.1995.1158
Article and author information
Author details
Funding
European Research Council
- Claudia Köhler
Vetenskapsrådet
- Claudia Köhler
Knut och Alice Wallenbergs Stiftelse
- Claudia Köhler
Olle Engkvist Byggmästare Foundation
- Claudia Köhler
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We are indebted to Dolf Weijers for providing the R2D2 system and DR5v2 reporter prior to publication, to Koji Goto for the LHP1 reporter, to Ramin Yadegari for the SWN reporter and to Miguel Blázquez for providing us with the RGA reporter. This research was supported by a European Research Council Starting Independent Researcher grant (to CK), a grant from the Swedish Science Foundation (to CK), a grant from the Olle Engkvist Byggmästare Foundation (to CK), and a grant from the Knut and Alice Wallenberg Foundation (to CK).
Copyright
© 2016, Figueiredo et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 7,575
- views
-
- 1,603
- downloads
-
- 182
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 182
- citations for umbrella DOI https://doi.org/10.7554/eLife.20542





