Crenactin forms actin-like double helical filaments regulated by arcadin-2
Figures
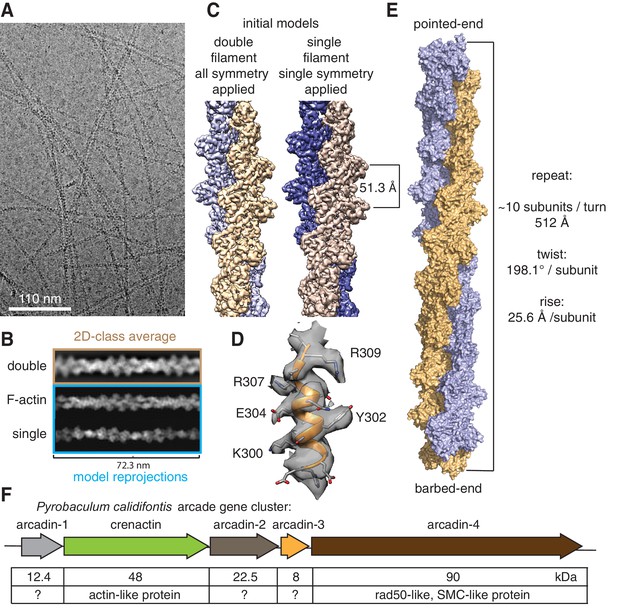
Crenactin forms double filaments.
(A) Representative cryoEM micrograph of crenactin filaments. (B) Comparison between crenactin double helical 2D class average (top), re-projections of F-actin double-stranded filament structure (middle) (von der Ecken et al., 2015) and crenactin single filament crystal structure (bottom) (Izoré et al., 2014), indicating that crenactin forms double filaments under the conditions used. (C) Comparison of cryoEM density maps obtained from double and single filament starting models. Left: crenactin double filament starting model with twist: 198.1° (equivalent to −161.9°) and rise: 25.6 Å applied during helical reconstruction. Right: crenactin single filament starting model with twist: 36.2° and rise: 51.3 Å applied. The two reconstructions converged to the same, double-stranded solution. (D) Part of the 3.8 Å cryoEM density map (resolution estimate determined through gold standard FSC 0.143 criterion, Figure 1—figure supplement 1). (E) Surface representation of crenactin double-stranded helical filament. See also Video 1. (F) Schematic showing the organisation of the arcade gene cluster, also showing similarities to proteins of known function (Ettema et al., 2011).
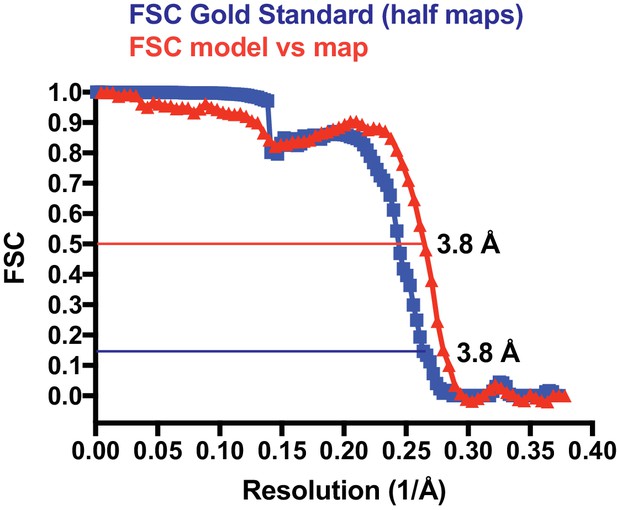
Fourier shell correlation (FSC) plot.
The blue curve has been generated by RELION after post processing, using a gold standard procedure that keep half datasets separate throughout. In order to avoid over-fitting through the presence of very similar overlapping filament segments in the two half datasets, segments were divided into half sets at the filament level. The spherical mask used covered the central 30% volume of the reconstructed map. The mask was softened at the edges with an 8 pixel raised cosine edge. The red curve describes the FSC of a map generated from the refined filament model (six central crenactin subunits) against the postprocessed map (both half maps averaged), cut around the model, but not low-pass filtered. Using the suggested resolution estimates at FSC 0.5 for the model and FSC 0.143 for the two half maps against each other (Rosenthal and Henderson, 2003), very similar values of 3.8 Å were obtained.
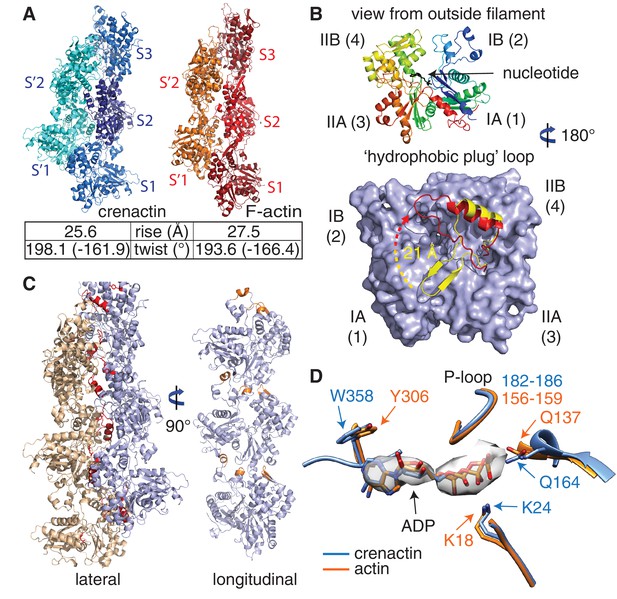
Crenactin filaments are exceptionally similar to eukaryotic F-actin.
(A) Comparison of filament architectures between crenactin (blue) and actin (orange) (von der Ecken et al., 2015) showing the exceptional similarity that is also reflected in the helical parameters (bottom). F-actin model PDB ID: 3J8A (von der Ecken et al., 2015). See also Video 2. Subunits labelled S2 were superimposed for the comparison. (B) Top: cartoon plot of crenactin in the filament conformation, showing the common subdomain nomenclature used and the position of the nucleotide. View is from the outside of the double filament, with the 'hydrophobic plug' loop positioned in the back, inside the double filament. Bottom: crenactin's hydrophobic plug loop conformational change from the single-stranded filament form (yellow) to double stranded form (red). View from inside of filament, 180° rotated from top panel. (C) Lateral (red) and longitudinal (orange) interactions in crenactin double filaments. Longitudinal interactions are represented in a single crenactin strand for clarity. For a comparison to F-actin, please see Figure 2—figure supplement 1. (D) Comparison of the ATPase active site of crenactin (blue) and actin (orange). The cryoEM density for the ADP nucleotide is superimposed.
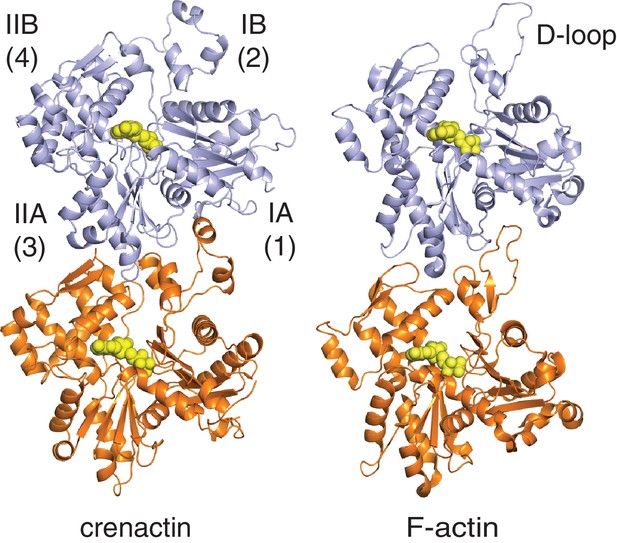
Comparison of longitudinal contacts between crenactin filaments and F-actin.
In both filaments, the tip of the top subunits (light blue) formed by subdomain IIA inserts into a cleft formed by subdomains IB (including the D-loop in F-actin, that forms a helix in crenactin) of the lower subunit (orange). Both dimers have been aligned using the orange, lower subunit, only.
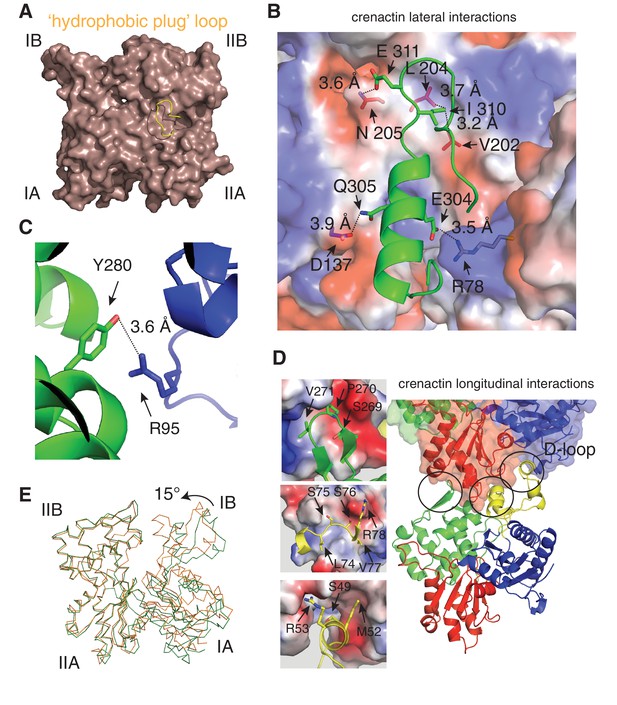
Detailed lateral and longitudinal interactions within crenactin filaments.
(A) Ribbon representation of actin's 'hydrophobic plug' loop, yellow. The core of actin is represented as a light brown surface. (B) Crenactin lateral contacts. The residues of the hydrophobic plug from one subunit (green) form an extended network of interactions with the subunit on the opposite strand (shown in surface and coloured by charge. Blue: positive charge, red: negative charge, white: non-charged). (C) Stick representation of an additional hydrophilic lateral interaction outside the hydrophobic plug interaction. (D) Crenactin longitudinal contacts. The three contact areas encircled are shown in more detail on the left, with contacts formed by the D-loop shown in the bottom panel. (E) Cα representation of crenactin monomers in single-stranded protofilaments from crystallography (this work, green) and double-stranded conformation, from this work, cryoEM (orange). Note the closure of the inter-domain cleft.
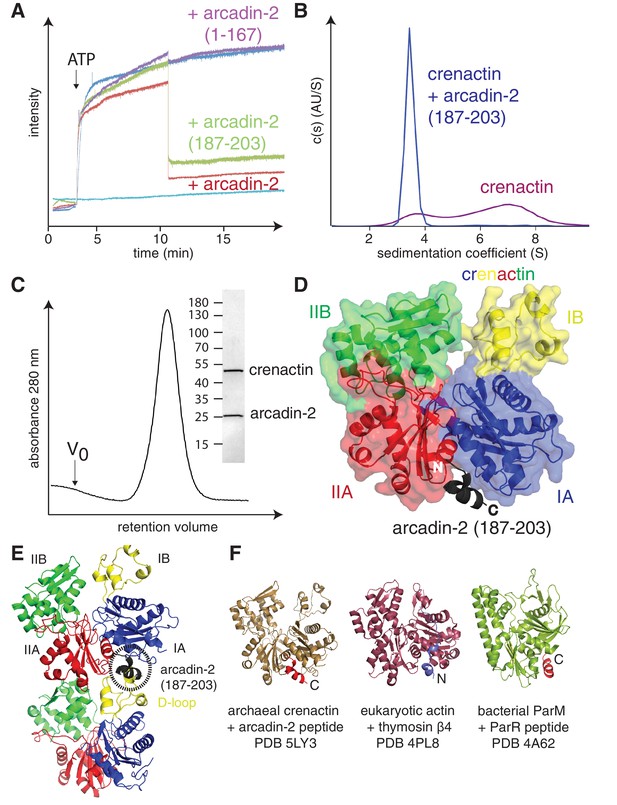
Crenactin interacts with arcadin-2.
(A) 90° light scattering assay of crenactin polymerisation. Arrow indicates ATP addition. Crenactin polymerisation is shown in dark blue (positive control). Curves representing the depolymerisation of crenactin by addition of arcadin-2 and arcadin-2 C-terminal peptide (residues 187–203) are shown in red and green, respectively. A curve following the addition of arcadin2ΔC (residues 1–167, only) is shown in purple. Crenactin and arcadin-2 premixed before the experiment is shown with the light blue curve. (B) Analytical ultracentrifugation profile of crenactin and crenactin with arcadin-2 C-terminal peptide (residues 187–203), showing monomers only for the complex sample. (C) Size exclusion chromatography profile of the crenactin:arcadin-2 complex, with corresponding Coomassie-stained SDS-PAGE. (D) Ribbon/surface representation of crenactin:arcadin-2 peptide (residues 187–203) complex crystal structure at 1.6 Å resolution, showing the binding of arcadin-2 to the hydrophobic groove, where the D-loop binds in filaments of crenactin. (E) Ribbon representation of two subunits of crenactin in the filament. The localisation of the arcadin-2 C-terminal peptide (187–203) is shown in black. Note the clash between the presence of the arcadin-2 peptide and the polymer form of crenactin, especially the D-loop. (F) Ribbon representation of archaeal, eukaryotic and bacterial actins in complex with protein domains involved in the regulation of the filaments. PDB IDs: crenactin:arcadin-2 5LY3 (this work); actin:thymosin β4 4PL8 (Xue et al., 2014); ParM:ParR 4A62 (Gayathri et al., 2012). Note that the orientation of the thymosin peptide is reversed in comparison with arcadin-2 and ParR.
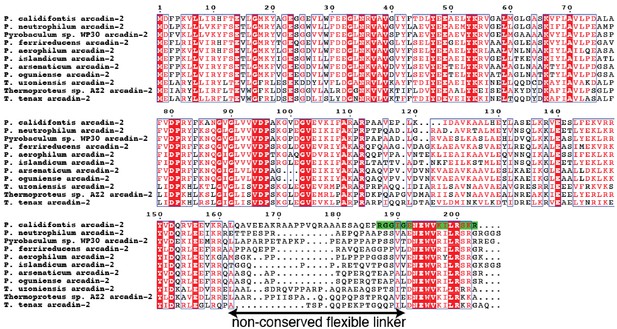
Multiple sequence alignment of arcadin-2 sequences from a BLAST search showing a small C-terminal domain separated from the body of the protein by a non-conserved, presumably flexible linker.
The C-terminal peptide sequence used in the experiments is highlighted in green (residues 187–203).
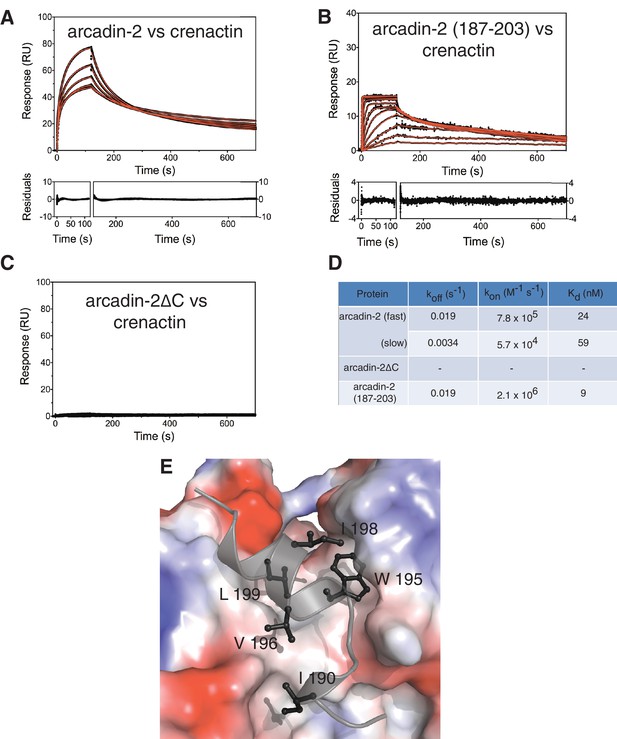
Biophysical and structural characterisation of the arcadin-2 : crenactin complex.
(A) SPR (surface plasmon resonance) data showing interaction of crenactin (immobilised) with arcadin-2. Solid red lines show the fits to the data with the resultant residuals shown underneath. (B) SPR data showing the interaction of immobilised crenactin with arcadin-2 C-terminal peptide (187–203). (C) SPR data showing lack of interaction of immobilised crenactin with arcadin-2 C-terminal truncation (arcadin-2∆C). (D) Summary of kinetic rate constants and dissociation constants for fits to data in plots A and B. The average Kd for the interaction between crenactin:arcadin-2 was 31 ± 4 nM (n = 4). (E) Surface representation of the crenactin hydrophobic groove (Dominguez, 2004) into which arcadin-2 binds. Surface of crenactin has been coloured by charge, blue: positive charge, red: negative charge, white: non-charged. Arcadin-2 peptide is shown in black/grey. For clarity only hydrophobic residues are shown.
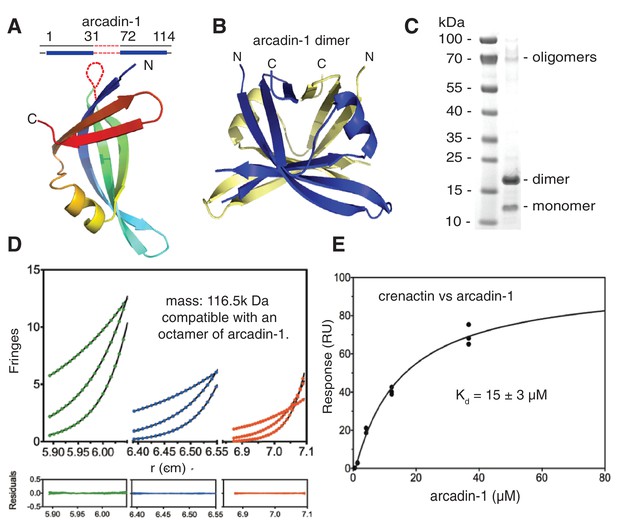
Crenactin interacts with arcadin-1.
(A) Ribbon representation of the crystal structure of arcadin-1 at 2.0 Å resolution. Note that residues 32 to 71 are presumably disordered and missing from the structure. (B) Ribbon representation of the arcadin-1 dimer present in the crystal. The two subunits of the dimer have been coloured blue and pale yellow. (C) SDS-PAGE of arcadin-1. The protein appears mainly as a dimer, although monomers and oligomers can also be seen on the Coomassie-stained gel. (D) Sedimentation equilibrium analytical ultracentrifugation of arcadin-1. Sedimentation data for 199 µM (green closed circles) 99.5 µM (blue closed circles) and 49.8 µM (red closed circles) obtained at 7800, 11800, and 15,000 rpm were fitted to an idealised single-species model (solid lines). Every tenth data point is shown for clarity. The plots below show the residuals of the fits to the data. Analysis of multiple concentrations gave a molecular weight of 116,500 ± 273 Da, close to an octamer (monomer: 14,560 Da). (E) Surface plasmon resonance (SPR) of the interaction between arcadin-1 and crenactin. Equilibrium fitting for arcadin-1 association with crenactin gave a dissociation constant of Kd = 15 ± 3 µM.
Videos
Crenactin forms F-actin-like double filaments.
The movie shows the experimental electrostatic potential density obtained from cryoEM and helical reconstruction and a ribbon representation of the refined atomic model of the filament.
Exceptional similarity between crenactin and actin.
As was previously reported, the structure of crenactin subunits very closely resembles that of eukaryotic actin, including the 'hydrophobic plug' loop. With the cryoEM filament structure we show that this similarity extends to the filament architecture, with the two filament structures being exceptionally similar. A section with two and three subunits in each strand of the double helical filaments is shown.
Tables
Crystallography and cryoEM data.
| Statistics | |||
| Sample | Pyrobaculum calidifontis crenactin: arcadin-2 peptide | Pyrobaculum aerophilum arcadin-1 | Pyrobaculum calidifontis crenactin |
| NCBI database ID | WP_011850310.1 WP_011850311.1 | NC_003364.1 | WP_011850310.1 |
| Constructs | crenactin 1-432, arcadin-2 187-203 | MGSSH6SSGLVPRGSH- 1-113 | 1-432 |
| Method Data collection | crystallography molecular replacement | crystallography SIR | cryoEM with helical reconstruction in RELION 2.0 |
| Beamline/microscope Wavelength / energy | Diamond I04-1 0.92819 Å | Diamond I04 0.97949 Å | FEI Polara / Falcon III 300 kV |
| Crystal / helical parameters | |||
| Space / point group Cell (Å/°) | P21 54.2, 70.9, 62.2, 104.21° | P6522 84.0, 84.0, 61.3, 90,90,120° | 1-start helical |
| Twist / rise | 198.1° (= −161.9°), 25.6 Å | ||
| Data | |||
| Resolution (Å) | 1.6 | 2.0 | 3.8 |
| Completeness (%)* | 97.8 (93.5) | 100.0 (99.7) | |
| Multiplicity* | 3.3 (2.8) | 19.0 (15.1) | |
| (I) / °(I)* | 14.1 (1.7) | 26.6 (2.3) | |
| Rmerge* | 0.037 (0.554) | 0.065 (1.348) | |
| Rpim* CC1/2 | 0.023 (0.378) 0.999 (0.897) | 0.015 (0.307) 1.00 (0.803) | |
| Images, pixel size Defocus range, dose Helical segments | 1474, 1.34 Å -0.8 - -3.0 µm, ~40 e/ņ 470396, 25 Å apart | ||
| Refinement | |||
| R / Rfree2† | 0.175 / 0.199 | 0.209 / 0.230 | 0.260 |
| Models | 2 chains crenactin: 4-430 arcadin-2: 188-203 ADP, 337 waters | 1 chain SH-1-32…72-113 37 waters | 6 chains refined in P1: 5-430, ADP, no waters |
| Bond length rmsd (Å) | 0.006 | 0.020 | 0.016 |
| Bond angle rmsd (°) | 0.860 | 2.12 | 1.453 |
| Favoured (%)‡ | 98.0 | 95.9 | 92.96 |
| Disallowed (%)‡ MOLPROBITY score | 0 100th percentile | 1.37 92nd percentile | 0.24 100th percentile |
| PDB/EMDB IDs | 5LY3 | 5LY5 | 5LY4, 4117 |
-
*Values in parentheses refer to the highest recorded resolution shell.
-
†5% of reflections were randomly selected before refinement.
-
‡Percentage of residues in the Ramachandran plot (PROCHECK 'most favoured' and 'additionally allowed' added together).





