Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii
Figures
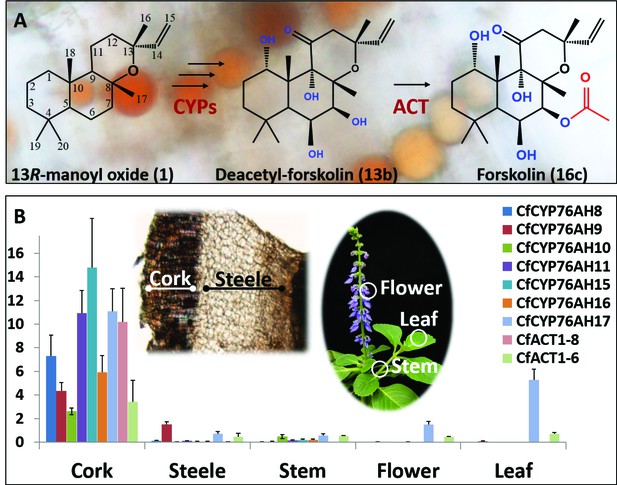
Biosynthesis of forskolin in the root cork cells of C. forskohlii.
(A) Scheme showing the structures of the diterpene precursor 13R-manoyl oxide, deacetylforskolin and forskolin on a background of root cork cells with forskolin containing oil bodies. (B) Transcript profiles of biosynthetic candidate genes in selected tissues of C. forskohlii as shown on the illustrations.
-
Figure 1—source data 1
cDNAs identified in the C. forskohlii root cork transcriptome and cloned during this work, with the GeneBank accession numbers.
- https://doi.org/10.7554/eLife.23001.004
-
Figure 1—source data 2
Table of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values of the first 20 most abundant cDNAs identified in the root cork transcriptome library.
cDNAs involved in terpenoid metabolism are marked in bold.
- https://doi.org/10.7554/eLife.23001.005
-
Figure 1—source data 3
Table of primers used in this study.
Construction of plasmids for expression of CfTPS2, CfTPS3, CfTPS1 is described in Andersen-Ranberg et al. (2016). U (uracil, marked in bold), represents the cleavage site, used in the USER cloning (Nour-Eldin et al., 2006).
- https://doi.org/10.7554/eLife.23001.006
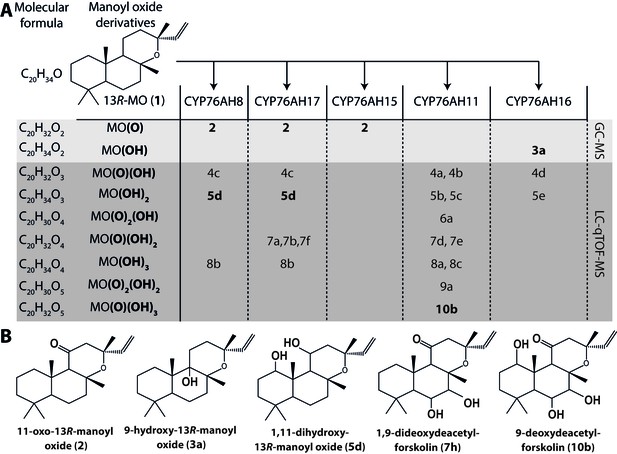
13R-manoyl oxide oxide-derived hydroxylated products formed following transient expression of CfCYP76AHs in N.
benthamiana leaves. (A) Molecular formulas of the hydroxylated products obtained using different CfCYP76AHs. The number of hydroxylations of each compound was deduced from its accurate molecular mass (<5 ppm, Supplementary file 1) as determined by LC-qTOF-MS or NMR. Each different compound is marked by a number. (B) Chemical structures of the compounds marked with numbers in bold in A as determined by NMR (Tables 1 and 2). MO: manoyl oxide
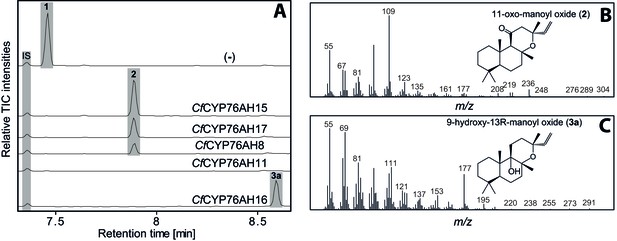
GC-MS analysis of 13R-manoyl oxide (1) derived diterpenoids obtained by transient expression of CYP76AHs from C.
forskohlii in N. benthamiana leaves. (A) GC-MS total ion chromatograms (TIC) of extracts from N. benthamiana transiently expressing CfCXS, CfGGPPS, CfTPS2 and CfTPS3 (13R-manoyl oxide biosynthesis) genes in combination with water (-), CfCYP76AH15, CfCYP76AH17, CfCYP76AH8, CfCYP76AH11 or CfCYP76AH16. 1-eicosene was used as internal standard (IS). 13R-manoyl oxide (1) was identified only in (-), indicating that it is further metabolized by the CfCYP76AH15, CfCYP76AH17, CfCYP76AH8, CfCYP76AH11 and CfCYP76AH16 enzymes. (B) m/z spectrum of 11-oxo-13R-manoyl oxide (2). (C) m/z spectrum of 9-hydroxy-13R-manoyl oxide (3a). The structure of both compounds was verified by NMR analysis (Tables 1 and 2). The compounds have been identified previously in C. forskohlii as putative intermediates in the in planta biosynthesis of forskolin (Asada et al., 2012). For each combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
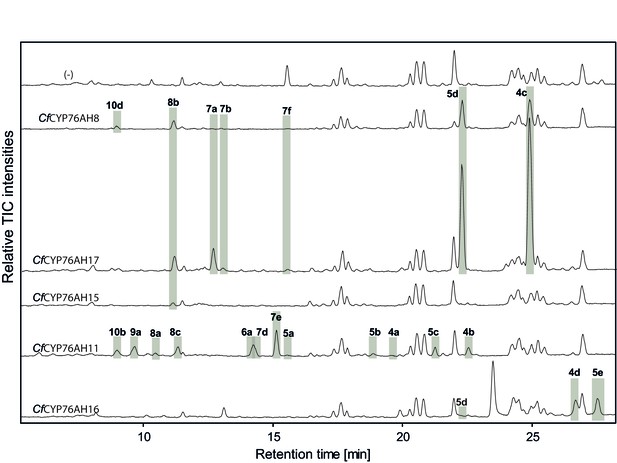
LC-qTOF-MS analysis of 13R-manoyl oxide-derived diterpenoids obtained by transient expression of C. forskohlii CYP76AH encoding genes in N. benthamiana leaves.
Total ion chromatograms (TIC) of extracts expressing the 13R-manoyl oxide biosynthesis genes (CfCXS, CfGGPPS, CfTPS2, CfTPS3) in combination with water (-), CfCYP76AH8, CfCYP76AH17, CfCYP76AH15, CfCYP76AH11 or CfCYP76AH16 are shown. 13R-manoyl oxide-derived oxygenated compounds formed (marked with grey bars) and their identity including their molecular formulas was confirmed by their accurate mass (5 ppm tolerance, Supplementary file 1). The identity of 1,11-dihydroxy-13R-manoyl oxide (5d) and 9-deoxydeactylforskolin (10b) was confirmed by NMR analysis (Figure 4 and Tables 1 and 2). No 13R-manoyl oxide-derived diterpenoids were detected in the water control (-). For each combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
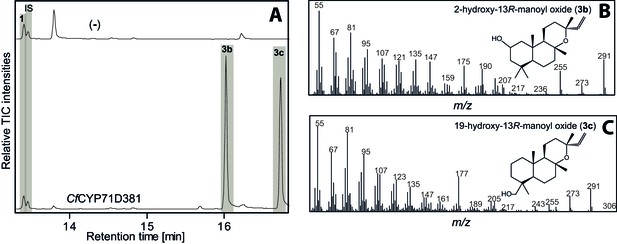
GC-MS analysis of 13R-manoyl oxide-derived diterpenoids following transient expression in N. benthamiana leaves of the C. forskohlii gene encoding CfCYP71D281 together with genes encoding the required enzymes for biosynthesis of 13R-manoyl oxide (CfCXS, CfGGPPS, CfTPS2, CfTPS3).
(A) GC-MS total ion chromatograms (TIC) of extracts from N. benthamiana transiently expressing 13R-manoyl oxide biosynthesis genes in combination with water (-) or CfCYP71D381, respectively. 1-Eicosene was used as internal standard (IS) and 13R-manoyl oxide (1) was identified in both (-) and the CfCYP71D381 samples. Compounds 3b and 3c were identified in extracts from N. benthamiana leaves expressing CfCYP71D381 together with the genes in 13R-manoyl oxide biosynthesis. CfCYP71D381 efficiently converted compound 1 to a mixture of two mono-hydroxylated 13R-manoyl oxide derivatives (3b and 3c). Structural elucidation by NMR (Figure 4 and Tables 1 and 2) showed hydroxylation of 1 at positions C-2 (3b) and C-19 (3c). These hydroxylation positions do not coincide with those found in forskolin and to our knowledge have not been observed in other diterpenoids known from C. forskohlii. (B) m/z spectrum of 2-hydroxy-13R-manoyl oxide (3b). (C) m/z spectrum of 19-hydroxy-13R-manoyl oxide (3c). For each combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
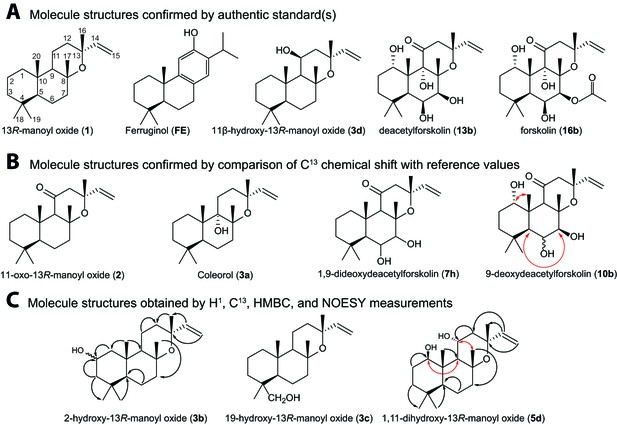
Structures of key compounds presented in this work.
(A) Compounds confirmed using authentic standards. (B) Compounds which structure was confirmed/identified by comparison of 13C NMR data with existing literature. (C) Compounds which structure was confirmed/identified by HMBC and NOE correlations for assigning position of OH-groups (marked in red), whereas couplings identified in the previously uncharacterized compounds 3b, 3c and 5d are marked in black. All other molecular structures were confirmed by 13C chemical shifts in comparisons to reference values (Table 1, Figure 4—source data 1).
-
Figure 4—source data 1
NMR spectra’s of selected 13R-manoyl oxide derived molecules.
- https://doi.org/10.7554/eLife.23001.012
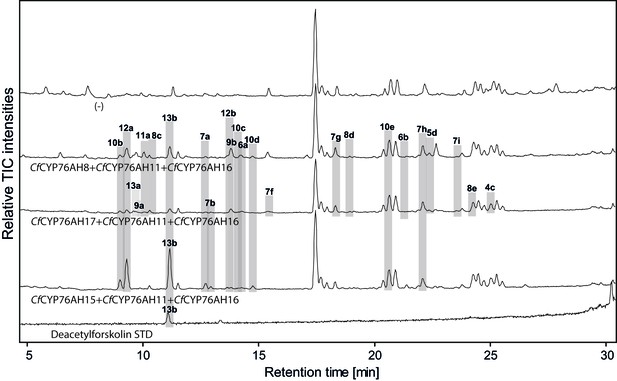
LC-qTOF-MS analysis of 13R-manoyl oxide-derived diterpenoids obtained by transient expression of combinations of C. forskohlii CYP encoding genes, together with genes encoding the required enzymes for biosynthesis of 13R-manoyl oxide in N. benthamiana leaves.
Total ion chromatograms (TIC) of extracts expressing the 13R-manoyl oxide biosynthesis genes (CfCXS, CfGGPPS, CfTPS2, CfTPS3), in combination with (from the top) water (-), CfCYP76AH8 + CfCYP76AH11 + CfCYP76AH16, CfCYP76AH17 + CfCYP76AH11 + CfCYP76AH16, and CfCYP76AH15 + CfCYP76AH11 + CfCYP76AH16 are shown. Hydroxylated 13R-manoyl oxide-derived diterpenoids (marked with grey bars) and their identity including their molecular formulas were confirmed by accurate mass (5 ppm tolerance, Supplementary file 1). Compounds present in trace amounts are not marked. The identity of 1,11-dihydroxy-13R-manoyl oxide (5d), 9-deoxydeacetylforskolin (10b) and 1,9-dideoxydeacetylforskolin (7h) was confirmed by NMR analysis (Figure 4 and Tables 1 and 2), whereas the identity of deacetylforskolin (13b) was confirmed by comparison to an authentic chemically synthesized standard. No 13R-manoyl oxide-derived diterpenoids were identified in the water control (-). For each combinaton, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
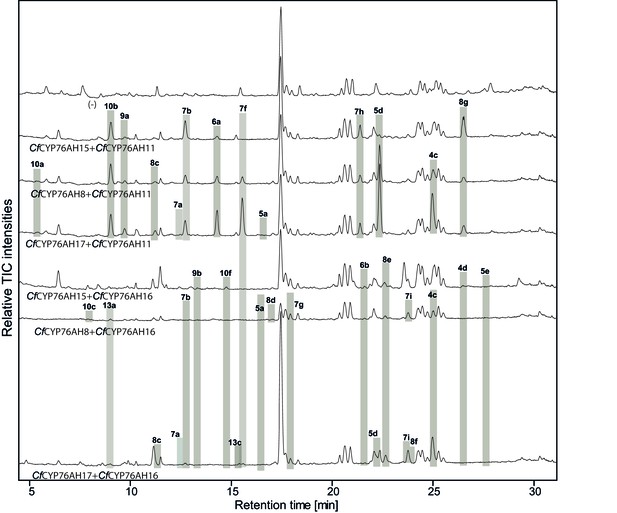
LC-qTOF-MS analysis of 13R-manoyl oxide-derived diterpenoids obtained by transient expression of combinations of C. forskohlii CYP76AH encoding genes in N. benthamiana leaves.
Total ion chromatograms (TIC) of extracts expressing the 13R-manoyl oxide biosynthesis genes (CfCXS, CfGGPPS, CfTPS2, CfTPS3) in combination with (from the top) water (-), CfCYP76AH15 + CfCYP76AH11, CfCYP76AH8 + CfCYP76AH11, CfCYP76AH17 + CfCYP76AH11, CfCYP76AH15 + CfCYP76AH16, CfCYP76AH8 + CfCYP76AH16 and CfCYP76AH17 + CfCYP76AH16 are shown. Oxygenated 13R-manoyl oxide-derived diterpenoids (marked with grey bars) and their identity including their molecular formulas were confirmed by their accurate mass (5 ppm tolerance, Supplementary file 1). Compounds present in trace amounts are not marked. The identity of 1,11-dihydroxy-13R-manoyl oxide (5d), 9-deoxydeacetylforskolin (10b), 1,9-dideoxydeacetylforskolin (7h) was confirmed by NMR analysis (Figure 4 and Tables 1 and 2). No 13R-manoyl oxide-derived diterpenoids were detected in the water control (-). For each combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
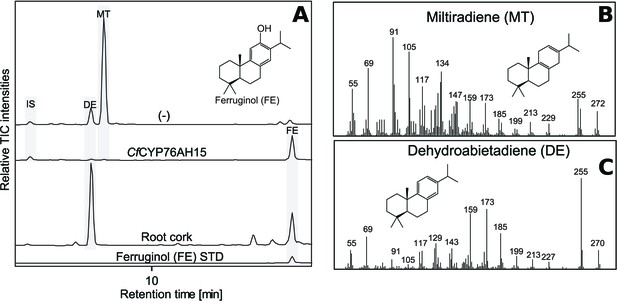
GC-MS analysis of miltiradiene-derived diterpenoids obtained by transient expression of CfCYP76AH15 in N. benthamiana leaves.
(A) Total ion chromatograms (TIC) of extracts transiently expressing CfCXS, CfGGPPS, CfTPS1 and CfTPS3 (miltiradiene biosynthesis genes) in combination with water (-) or CfCYP76AH15. Dehydroabietadiene (DE) and miltiradiene (MT) were observed in the (-) extract, whereas ferruginol was observed in extracts from tissue expressing the miltiradiene biosynthesis genes together with CfCYP76AH15. In root cork extracts, ferruginol was detected together with dehydroabietadiene. Presence of ferruginol was confirmed by comparison to an authentic standard (Ignea et al., 2016a), while presence of miltiradiene (B) and dehydroabietadiene (C) were confirmed by comparison of m/z spectra with previously characterized compounds (Andersen-Ranberg et al., 2016). For every combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
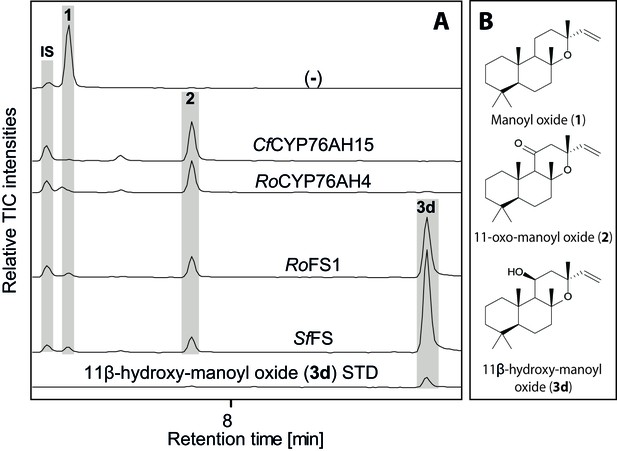
GC-MS analysis of 13R-manoyl oxide-derived diterpenoids obtained by transient expression of CYP76AHs in N. benthamiana leaves.
(A) Total ion chromatograms (TIC) of extracts transiently expressing CfCXS, CfGGPPS, CfTPS2 and CfTPS3 (13R-manoyl oxide biosynthesis genes) in combination with water (-), CfCYP76AH15, RoCYP76AH4, RoFS1 and SpFS are shown. 13R-manoyl oxide was observed in the (-) extracts, while 11-oxo-13R-manoyl oxide (2) was observed in the CfCYP76AH15, RoCYP76AH4, RoFS1 and SfFS extracts. 11-Hydroxy-13R-manoyl oxide (3d) is observed only in extracts expressing the RoFS1 and SfFS1 genes. Presence of 11-hydroxy-13R-manoyl oxide was verified by comparison to an authentic standard (Ignea et al., 2016b) while identification of 11-oxo-13R-manoyl oxide was confirmed by comparison of m/z spectra with a previously characterized compound (2). The results show RoCYP76AH4 has an activity similar to CfCYP76AH15, able to convert efficiently and specifically 13R-manoyl oxide to 2. RoFS1, as well as SfFS, can also convert 13R-manoyl oxide to 2 but they catalyze the synthesis of an additional product, 11-hydroxy-13R-manoyl oxide (3d). For every combination, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
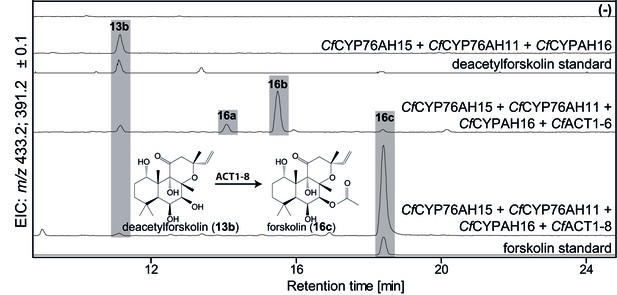
De novo biosynthesis of forskolin by transient expression of C. forskohlii genes in N. benthamiana as monitored by LC-MS-based extracted ion chromatograms (EIC).
To monitor both deacetylforskolin (13b) and forskolin (16c), the EIC were selected as the sum of m/z 433.2 ± 0.1 and m/z 391.2 ± 0.1. Chromatograms represent LC-MS analysis of extracts from leaves expressing the 13R-manoyl oxide biosynthesis genes (CfDXS, CfGGPPS, CfTP2 and CfTPS3) in combination with (from the top): water (-); CfCYP76AH15, CfCYP76AH11 and CfCYPAH16; CfCYP76AH15, CfCYP76AH11, CfCYPAH16 and CfACT1-6; CfCYP76AH15, CfCYP76AH11, CfCYPAH16 and CfACT1-8, shown together with authentic standards (deacetylforskolin and forskolin). Forskolin (16c) was identified together with two other acetylated compounds (e.g. 16a, 16b) with the same molecular mass in leaves expressing CfACT1-6 together with forskolin-specific CYPs (Supplementary file 1). When CfACT1-8 was expressed instead of CfACT1-6, a predominant accumulation of forskolin was observed, with a drastic reduction of non-specific acetylated products. For all combinations, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
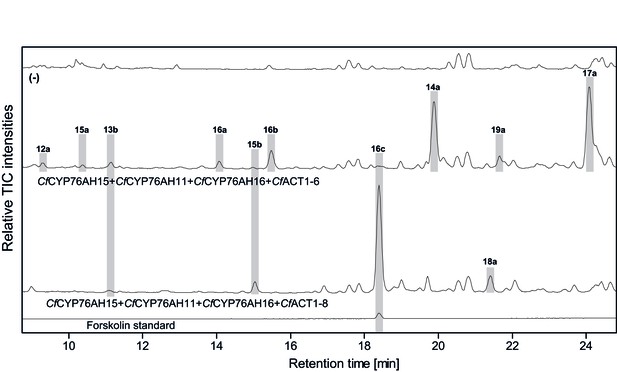
LC-qTOF-MS analysis of 13R-manoyl oxide-derived diterpenoids obtained by transient expression of combinations of C.forskohlii CYP and ACT encoding genes in N. benthamiana leaves.
Total ion chromatograms (TIC) from extracts expressing the 13R-manoyl oxide biosynthesis genes (CfCXS, CfGGPPS, CfTPS2, CfTPS3) in combination with (from the top) water (-), CfCYP76AH15 + CfCYP76AH11+ CfCYP76AH16 + CfACT1-6, and CfCYP76AH15 + CfCYP76AH11 + CfCYP76AH16 + CfACT1-8 are shown. Hydroxylated and acetylated 13R-manoyl oxide-derived diterpenoids (marked with grey bars) and their identity, including their molecular formulas, was confirmed by their accurate mass (5 ppm tolerance, Supplementary file 1). Compounds present in trace amounts are not marked. The identity of deacetylforskolin (13b) and forskolin (16c) was confirmed by comparison to authentic standards. No 13R-manoyl oxide-derived diterpenoids were detected in the water control (−). For all combinations, extracts from leaves of three different N. benthamiana plants have been analyzed and representative chromatograms are shown.
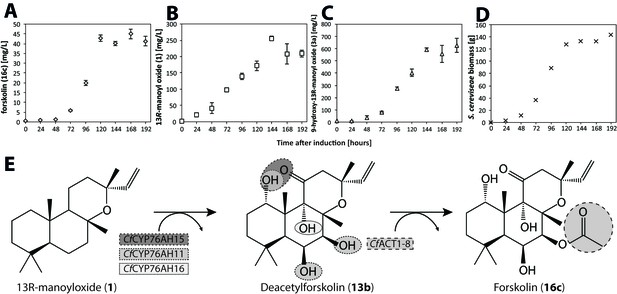
Forskolin production in S. cerevisiae following stable genomic integration of codon-optimized C.forskohlii genes.
(A) Forskolin (16c) accumulation in a fermenter batch using the EVST21543 strain (expressing CfCYP76AH15, CfCYP76HA11, CfCYP76AH16 and CfACT1-8 encoding genes in the EFSC4498 S. cerevisiae strain, optimized for the production of 13R-manoyl oxide [Andersen-Ranberg et al., 2016]). (B) 13R-manoyl oxide (1) accumulation in EVST21543 strain. (C) 9-Hydroxy-13R-manoyl oxide (3a) accumulation in EVST21543 strain. (D) EVST21543 strain biomass monitored during the fermentation process. (E) The biosynthetic pathway used for the production of forskolin in yeast. The fermentation event occurred once, and a triplicate of samples were analysed from each time course.
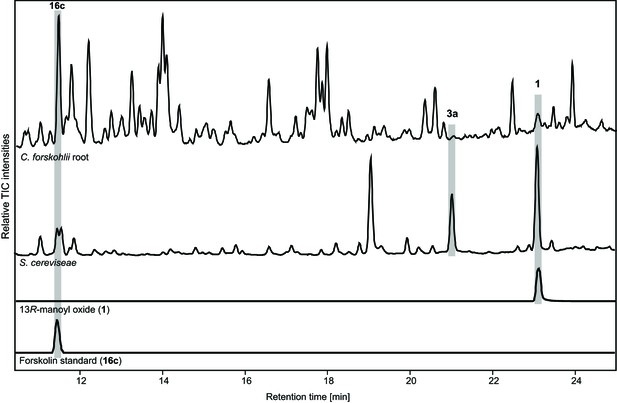
Comparison of metabolite profiles between fermenter grown yeast culture of the EVST21543 strain and C.forskohlii root extract analyzed by LC-MS.
Forskolin (16c) and 13R-manoyl oxide (1) were identified based on co-elution with standards and 9-hydroxy-13R-manoyl oxide (3a) was identified based on the presence of the [M+Na]+ ion, 329.2457 (C20H34O2Na+, Δ 0.7 ppm).
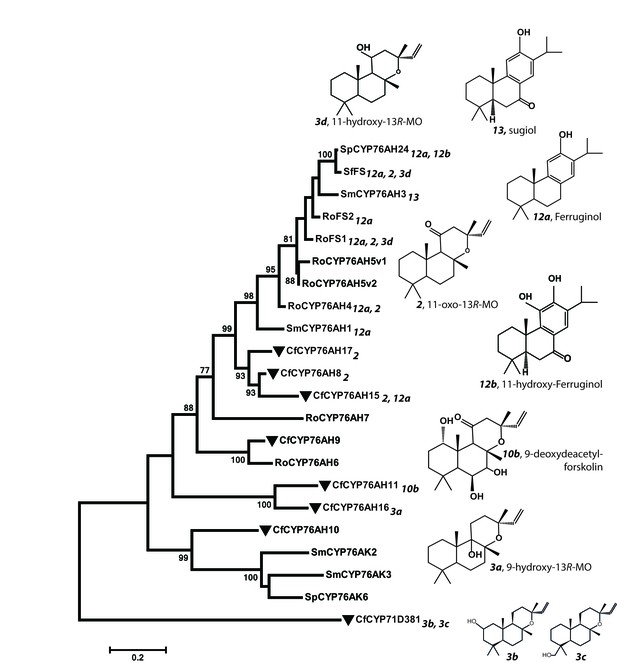
Phylogeny of known full-length CYP76AHs.
The enzymes used are listed below with their accession numbers or source of publication: CfCYP76AH15, KT382358; CfCYP76AH17, KT382360; CfCYP76AH8, KT382348; CfCYP76AH11, KT382349; CfCYP76AH16, KT382359; CfCYP76AH9, KT382347; CfCYP76AH10, KT382346; CfCYP71D381, KT382342; RoFS1, AJQ30187 (Božić et al., 2015); SmCYP76AH3, KR140168 (Guo et al., 2016); RoFS2, AJQ30188 (Božić et al., 2015); SfFS, AJQ30186 (Božić et al., 2015); RoCYP76AH4, (Zi and Peters, 2013); RoCYP76AH5v1, (Zi and Peters, 2013); RoCYP76AH5v2, (Zi and Peters, 2013); RoCYP76AH6, (Zi and Peters, 2013); RoCYP76AH7, (Zi and Peters, 2013); SmCYP76AH1, AGN04215 (Guo et al., 2013); SpCYP76AH24, ALM25796 (Ignea et al., 2016a). Coleus forskohlii enzymes are indicated by a solid black triangle. CfCYP71D381 was chosen as a root because it can accept 13R-manoyl oxide as a substrate, but does not catalyze the synthesis of forskolin-related products. The number subscripts indicated at each enzyme refer to their respective enzymatic products, the structures of which are given on the right. Only the main products of each enzymes are mentioned. MO stands for manoyl oxide.
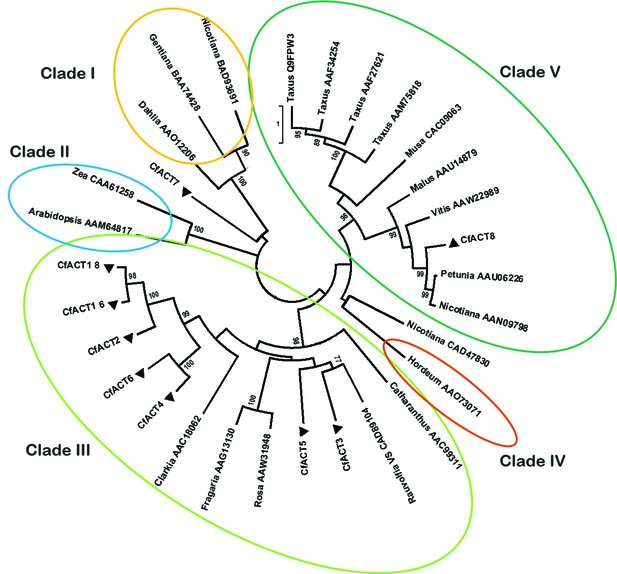
Phylogenetic tree of CfACT encoding candidate genes together with BAHD family acyltransferase representatives from all clades according to D'Auria (2006).
Accession numbers of the non-Coleus forskohlii selected protein sequences are shown next to the tree taxon names, while C. forskohlii peptide accession numbers are provided in Figure 1—source data 1. The analysis only includes functionally characterized members. Coleus forskohlii enzymes are indicated by a solid black triangle. The majority of the selected CfACTs belong to Clade III, which includes mainly members which accept a diverse range of hydroxylated substrates and use acetyl-CoA as the main acyl donor (D'Auria, 2006). Interestingly, the ACTs known to be involved in Taxol biosynthesis belong to Clade V.
Tables
1H-NMR and 13C-NMR chemical shifts (Figure 4—source data 1) of novel oxygenated 13R-(+)-manoyl oxide-derived diterpenoids formed following transient expression of CYP encoding genes from C. forskohlii.
| 19-hydroxy- 13R-manoyl oxide (3c)* | 2-hydroxy- 13R-manoyl oxide (3b)* | 1,11-dihydroxy- 13R-manoyl oxide (5d)* | ||||
|---|---|---|---|---|---|---|
| Pos. | 1H (nH; m; J(Hz)) | 13C | 1H (nH; m; J(Hz)) | 13C | 1H (nH; m; J(Hz)) | 13C |
| 1 | 0.89 (1H;m) 1.63 (1H; m) | 39.1 | 1.10 (1H; t(br); 11.9, 11.9) 1.77 (1H; m) | 51.3 | 3.49 (1H; dd;11.1, 4.5) | 79.0 |
| 2 | 1.44 (1H; m) 1.56 (1H; m) | 18.1 | 3.92 (1H; m) | 65.3 | 1.75 (1H; td; 13.5, 11.1, 3.9) 1.60 (1H; m) | 29.0 |
| 3 | 0.95 (1H; m) 1.78 (1H; m) | 35.8 | 0.76 (1H; t(br); 11.9, 11.9) 1.99 (1H; d(br); 11.9) | 48.2 | 1.47 (1H; dd; 13.6, 3.9) 1.39 (1H; td; 13.5, 3.6) | 39.6 |
| 4 | 38.5 | 34.9 | 33.4 | |||
| 5 | 1.10 (1H; dd; 2.3, 12.6) | 56.9 | 0.95 (1H; dd; 2.2, 12.4) | 55.9 | 0.84 (1H; dd; 11.3, 2.0) | 55.6 |
| 6 | 1.36 (1H; dd; 3.6, 12.6) | 20.1 | 1.68 (1H; m) | 19.7 | 1.47 (1H; m) | 20.2 |
| 1.75 (1H; m) | 1.27 (1H; m) | 1.64 (1H; m) | ||||
| 7 | 1.42 (1H; m) 1.83 (1H; dt; 3.3, 12.2) | 43.6 | 1.45 (1H; dd(br); 3.6, 12.5) 1.85 (1H; dt(br); 2.9, 12.5) | 43.2 | 1.48 (1H; m) 1.85 (1H; m) | 44.0 |
| 8 | 75.1 | 75.1 | 75.3 | |||
| 9 | 1.35 (1H; dd; 4.3, 12.0) | 55.7 | 1.40 (1H; dd; 4.2, 11.9) | 55.4 | 1.54 (1H; d; 5.8) | 55.8 |
| 10 | 37.3 | 38.7 | 43.8 | |||
| 11 | 1.48 (1H; m) 1.58 (1H; m) | 15.4 | 1.53 (1H; m) 1.61 (1H; m) | 15.6 | 4.38 (1H; br q; ≈8.6) | 65.6 |
| 12 | 1.78 (1H; m) 1.64 (1H; m) | 35.7 | 1.78 (1H; m) 1.66 (1H; m) | 35.5 | 2.02 (1H; dd; 14.3, 8.7) 2.27 (1H; dd; 14.3, 8.7) | 35.8 |
| 13 | 73.4 | 73.4 | 72.8 | |||
| 14 | 5.87 (1H; dd; 10.8, 17.4) | 147.7 | 5.87 (1H; dd; 10.8, 17.4) | 147.7 | 5.90 (1H; dd; 17.4, 10.8) | 147.1 |
| 15 | 4.92 (1H; dd; 1.5, 10.8) 5.14 (1H; dd; 1.5, 17.4) | 110.2 | 4.92 (1H; d; 10.8) 5.14 (1H; d; 17.4) | 110.3 | 4.94 (1H; dd; 10.7, 1.5) 5.17 (1H; dd; 17.4, 1.5) | 111.2 |
| 16 | 1.27 (3H; s) | 28.5 | 1.27 (3H; s) | 28.7 | 1.27 (3H; s) | 32.1 |
| 17 | 1.28 (3H; s) | 25.3 | 1.29 (3H; s) | 25.7 | 1.49 (3H; s) | 27.8 |
| 18 | 0.97 (3H; s) | 26.8 | 0.93 (3H; s) | 33.5 | 0.78 (3H; s) | 13.5 |
| 19 | 3.70 (1H; d; 10.9) 3.46 (1H; d; 10.9) | 65.4 | 0.85 (3H; s) | 22.2 | 0.85 (3H; s) | 32.8 |
| 20 | 0.78 (3H; s) | 15.7 | 0.84 (3H; s) | 16.5 | 0.79 (3H; s) | 21.1 |
-
* 1H and 13C NMR data acquired at 600 and 150 MHz, respectively, in methanol-d4, at 300 K. s = singlet, d = doublet, t = triplet, m = multiplet, br = broad
Structural identification of four oxygenated 13R-manoyl oxide-derived diterpenoids formed following transient expression of CYP encoding genes from C. forskohlii based on comparison of their 1H-NMR and 13C-NMR (Figure 4—source data 1) chemical shifts to literature data. Chemical shifts for reference compounds marked with * have not been assigned to a specific carbon. The 13C chemical shifts of 9-deoxyforskolin (Gabetta et al., 1989) were used as reference for 6,7-dihydroxy-11-oxo-13R-manoyl oxide (7h).
| 9-Deoxydeacetylforskolin (10b)† | 1,9-Dideoxydeacetylforskolin (7h)† | 11-oxo-13R-manoyl oxide (2)† | Coleorol (3a)† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos. | 1H (nH; m; J(Hz)) | 13C | (Gabetta et al., 1989) | 1H (nH; m; J(Hz)) | 13C | (Gabetta et al., 1989) | 13C | (Gabetta et al., 1989) | 13C | (Asada et al., 2012) |
| 1 | 4.38 (1H; t; 2.8) | 71.6 | 71.2 | 2.45 (1H, d(br); 13.1) 0.78 (H; m) | 41.5 | 43.1 | 42.1 | 41.9 | 31.7 | 31.6 |
| 2 | 1.47 (1H; m) 2.14 (1H; m) | 25.8 | 25.6 | 1.78 (H; m) 1.40 (H; m) | 18.7 | 18.4 | 18.5 | 18.4 | 18.6 | 18.4 |
| 3 | 1.12 (1H; dt; 3.4, 13.2) 1.62 (1H; dt; 3.5, 13.5) | 36.4 | 36.3 | 1.36 (H; m) 1.15 (H; m) | 43.8 | 43.7 | 43.4 | 43.3 | 41.9 | 41.8 |
| 4 | 34.2 | 34.1 | 34.4 | 34.1 | 33.4 | 33.2 | 33.3 | 33.2 | ||
| 5 | 1.34 (1H; d; 2.1) | 47.5 | 47.4 | n.d. | 55.7 | 55.2 | 56.0 | 55.8 | 45.7 | 45.5 |
| 6 | 4.44 (1H; t; 2.6) | 70.8 | 70.2 | 4.39 (1H; m) | 70.4 | 70.2 | 19.8 | 19.7 | 19.5 | 19.4 |
| 7 | 3.68 (1H; d; 3.6) | 80.7 | 81.1 | 3.71 (1H; d; 3.8) | 81.0 | 80.7 | 39.6 | 39.4 | 36.6 | 36.4 |
| 8 | 80.0 | 78.5 | 80.1 | 79.9 | 77.5 | 77.2 | 78.0 | 77.8 | ||
| 9 | 3.32 (1H; s) | 58.0 | 58.2 | 2.59 (1H; s) | 65.5 | 65.4 | 66.9 | 66.7 | 75.3 | 75.2 |
| 10 | 42.2 | 41.7 | 38.0 | 37.8 | 37.3 | 37.1 | 41.1 | 40.9 | ||
| 11 | 207.7 | 207.6 | 206.3 | 205.7 | 207.7 | 207.1 | 21.1 | 21.0 | ||
| 12 | 2.63 (1H; d; 18.0) 2.69 (1H; d; 18.0) | 49.8 | 49.9 | 2.60 (1H; d; 18.1) 2.66 (1H; d; 18.1) | 50.0 | 49.8 | 50.4 | 50.2 | 31.6 | 31.5 |
| 13 | 75.1 | 74.8 | 75.1 | 75.1 | 75.1 | 74.4 | 72.9 | 72.8 | ||
| 14 | 5.94 (1H; dd; 10.8, 17.4) | 146.2 | 145.8 | 5.95 (1H; dd; 10.7, 17.4) | 146.9 | 146.4 | 146.9 | 146.0 | 147.4 | 147.3 |
| 15 | 5.04 (1H; d; 10.8) 5.14 (1H; d; 17.4) | 112.4 | 112.7 | 5.04 (1H; d; 10.7) 5.17 (1H; d; 17.4) | 112.3 | 112.1 | 112.3 | 111.9 | 110.1 | 110.0 |
| 16 | 1.30 (3H; s) | 31.5 | 31.5* | 1.28 (3H; s) | 31.6 | 33.2* | 31.4 | 31.2* | 28.9 | 28.8 |
| 17 | 1.54 (3H; s) | 24.1 | 24.5* | 1.50 (3H; s) | 23.5 | 31.4* | 28.1 | 27.9* | 27.0 | 29.9 |
| 18 | 1.38 (3H; s) | 33.1 | 18.2* | 0.97 (3H; s) | 33.4 | 23.9* | 15.6 | 15.5* | 33.7 | 33.6 |
| 19 | 1.21 (3H; s) | 23.7 | 23.6* | 1.21 (3H; s) | 24.0 | 23.7* | 21.8 | 21.6* | 21.5 | 21.4 |
| 20 | 1.01 (3H; s) | 18.5 | 32.8* | 1.30 (3H; s) | 17.2 | 16.7* | 33.6 | 33.5* | 17.0 | 16.8 |
-
†1H and 13C NMR data acquired at 600 and 150 MHz, respectively,in methanol-d4, at 300 K. s = singlet, d = doublet, t = triplet, m = multiplet, br = broad
Additional files
-
Supplementary file 1
Overview of 13R-manoyl oxide-derived diterpenoids identified in N. benthamiana, expressing combinations of C. forskohlii genes encoding CYPs and acetyltransferases together with genes encoding the required enzymes for biosynthesis of 13R-manoyl oxide (CfDXS, CfGGPPs, CfTP2 and CfTPS3).
GC-MS and LC-qTOF-MS chromatograms of the identified diterpenoids are shown in previous figures.
- https://doi.org/10.7554/eLife.23001.025





