Discovery of novel determinants of endothelial lineage using chimeric heterokaryons
Figures
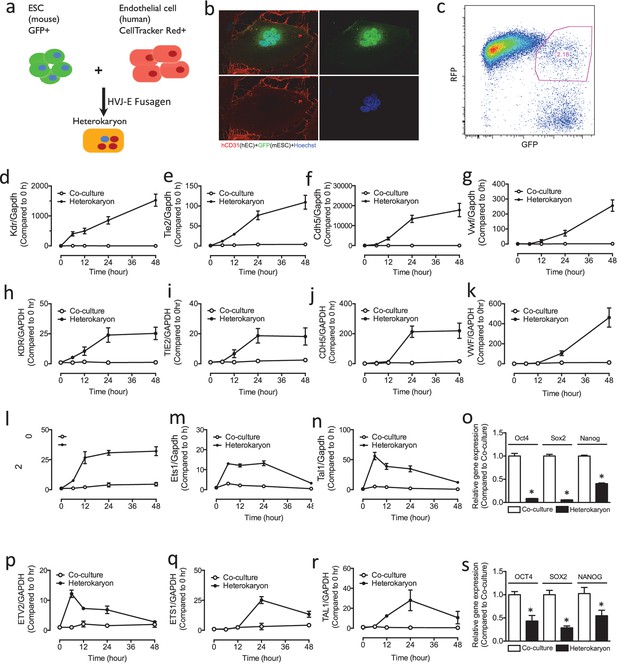
Heterokaryon recapitulates gene expression of endothelial ontogeny.
(a) Scheme for heterokaryon generation. GFP-labeled murine ESCs (mESCs) were fused with Cell Tracker Red labeled human ECs (hECs) by HVJ-enveloped fusagen to induce multinucleated heterokaryons. (b) Representative image of non-dividing multinucleated heterokaryons labeled with CD31 (Red) and GFP (Green), Hoechst (Blue) dye were used to label nuclei. (c) Representative FACS plots for heterokaryons. (d–g) Up-regulation of murine EC genes including Kdr, Tie2, Cdh5 and Vwf in heterokaryons consisting of mESC and hEC compared to co-culture control. (h–k) Up-regulation of human EC genes including Kdr, Tie2, Cdh5 and Vwf in heterokaryons consisting of human iPSC (hiPSC) and murine EC (mEC) compared to Co-culture control. (l–n) Increased expression of transcription factors involved in endothelial development such as Etv2, Ets1 and Tal1 during cell fusion of mESC with hEC. (p–r) Increased expression of transcription factors involved in endothelial development such as Etv2, Ets1 and Tal1 during cell fusion of hiPSC with mEC. (o and s) Down-regulation of genes encoding pluripotent factors (Oct4, Sox2 and Nanog) in heterokaryons compared to Co-culture control. All data represented as mean ± S.E.M. (n = 3). p<0.05 vs Co-culture control.
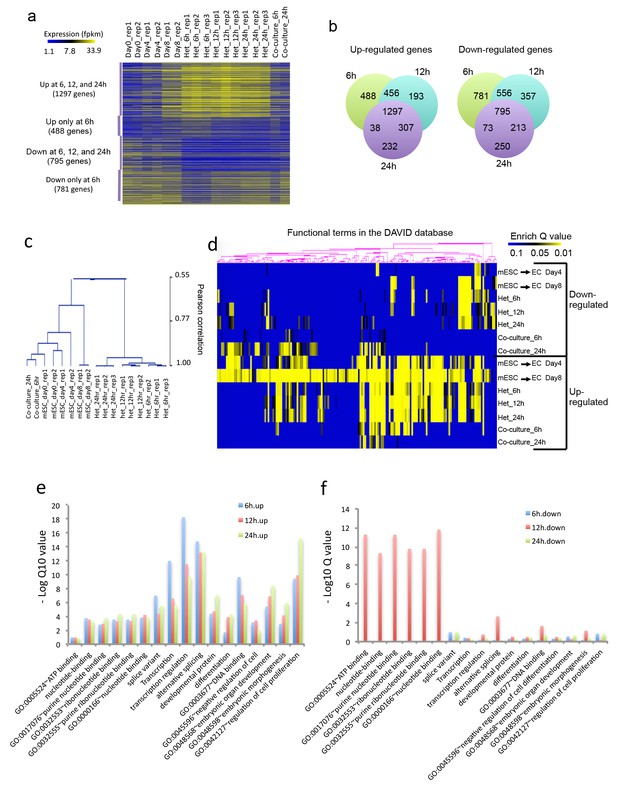
Heterokaryon bi-species RNA-seq reveals transcriptome reprogramming during differentiation of pluripotent stem cells into endothelial lineages.
(a) Heat map to show expression level of genes that are up- or down-regulated either uniquely in the Heterokaryon (Het) at 6 hr after fusion or commonly in all het samples relative to the mESC samples. mESCs were exposed to a standard endothelial differentiation protocol for 4 or 8 days. Het_6 hr, Het_12 hr, and Het_24 hr indicate mouse stem cells (mESC) fused with human endothelial cells (hEC) for 6, 12 or 24 hr. Co-Culture_6 hr and Co-Culture_24 hr indicate mESC co-cultured but not fused with hEC. (b) Venn diagram to show overlap of upregulated (left) or downregulated (right) mouse genes in heterokaryon at 6, 12 and 24 hr after fusion relative to mESC. (c) Unbiased hierarchical clustering of all samples based on all genes that are differentially expressed in mESC samples relative to at least one of the other samples. (d) Heat map displaying upregulated and downregulated genes in heterokaryons and in differentiating mESC at different time points. Genes differentially expressed were clustered into groups for functional analysis and presented as a heat map based on their enrichment Q value.( e–f) Bar plot showing enrichment Q values of 17 functional terms in genes upregulated (left) or downregulated (right) in mESCs within heterokaryons relative to the mESCs. Upregulated or downregulated genes were defined based on EdgeR FDR cutoff 1e-5. Overlap p value in pie chart was calculated based on Fisher’s Exact test. N = 3 for Het, n = 2 for mESCs, n = 1 for co-culture.
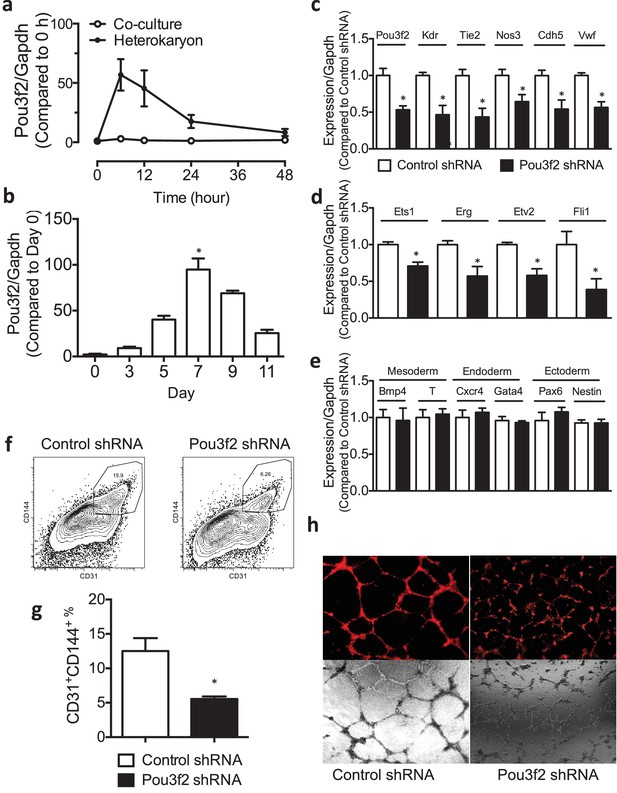
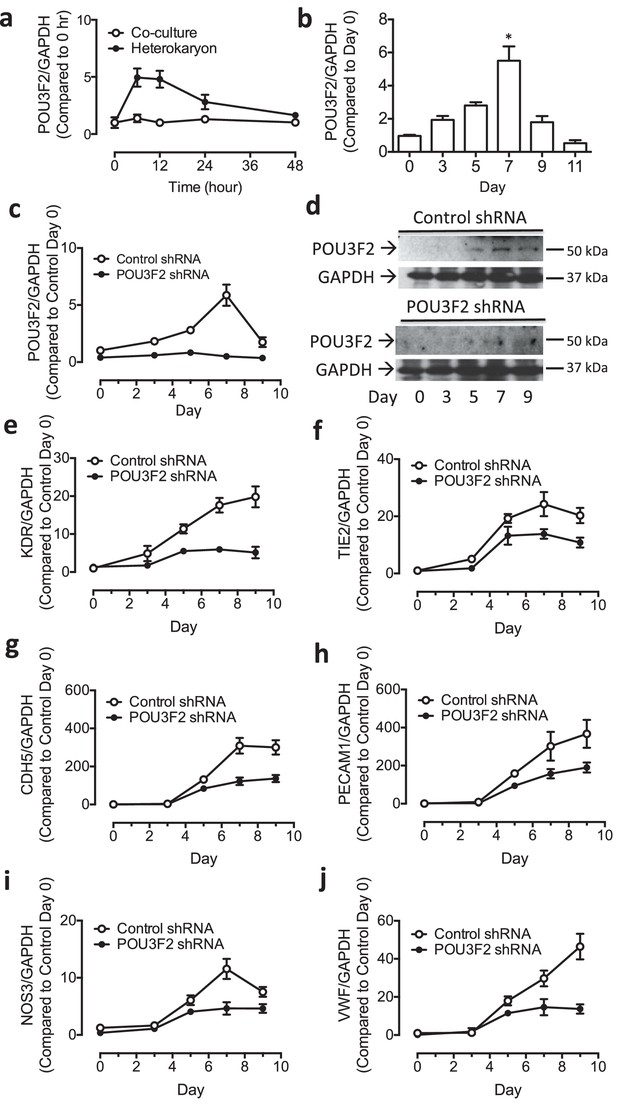
Role of Pou3f2 as a novel transcription factor in endothelial differentiation from murine embryonic stem cells.
(a) Gene expression pattern of Pou3f2 in heterokaryons consisting of mESC and hEC compared to Co-culture control. (b) Validation of expression of Pou3f2 during differentiation of mESC into endothelial lineage. (c) Lentiviral mediated shRNA KD of Pou3f2 reduced the gene expressions of endothelial markers including Kdr, Tie2, Nos3, Cdh5 and Vwf at Day 8 following endothelial differentiation from mESC. (d) KD of Pou3f2 reduced the gene expressions of transcription factors involved in endothelial differentiation such as Ets1, Erg, Etv2 and Fli1 in mESC differentiated to endothelial lineage at Day 8. (e) No differences were found in the expressions of mesodermal (Bmp4, T), endodermal (Cxcr4 and Gata4) and ectodermal (Pax6 and Nestin) in Pou3f2 shRNA treated mESC following endothelial differentiation at Day 8. (f and g) Representative FACS plots and summarized diagram showing that Pou3f2 KD reduces the yield of mESC-derived CD31+ and CD144+ cells at Day 10 of endothelial differentiation protocol. (h) Representative images showing that Pou3f2 KD mESC-derived ECs manifest an impaired ability to form endothelial networks on matrigel. All data represented as mean ± S.E.M. (n = 3). p<0.05 vs control shRNA.
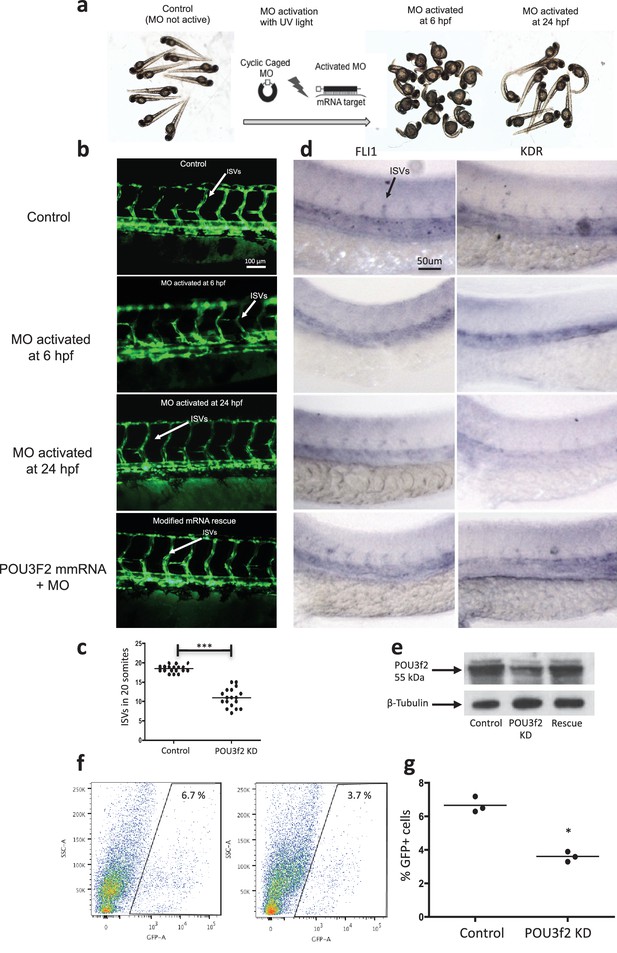
Pou3f2 knockdown in the tg(fli1:EGFP)y1 zebrafish embryo.
(a) Bright-field images of embryos injected with caged morpholino against Pou3f2 translation start site in the absence of photoactivation (control), or with photoactivation with UV light at 6 or 24 hpf. (b) Fluorescence images of embryos at 48 hpf. Experimental groups were injected with caged morpholino against Pou3f2 in the absence of photoactivation (control), or with photoactivation with UV light at 6 or 24 hpf, or with photoactivation at 6 hr in the presence of rescue mRNA encoding Pou3f2. (c) Quantitation of the number of intersegmental vessels in 20 somites in embryos at 48 hpf. (d) In situ hybridization with antisense RNA probes specific for Kdr and Fli1 in whole zebrafish embryos 28 hpf. (e) Western blotting showing the reduction level of Pou3f2 following morpholino injection and rescue by mRNA encoding Pou3f2. β-Tubulin was used as loading control. ISV – Intersegmental Vessels; hpf – hour post fertilization. (f and g). Representative FACS plot and scatter plot showing a significant reduction of GFP+ cells in Pou3f2 KD embryos. GFP+ cells were sorted following isolation by enzymatic digestion from tg(fli1:EGFP)y1 zebrafish embryos at 24 hpf. All data represented as mean ± S.E.M. N = 3. Student t-test, *p=0.01; ***p=0.001.
-
Figure 4—source data 1
Intersegmental vessel analysis in zebrafish embryos following Pou3f2 knockdown.
- https://doi.org/10.7554/eLife.23588.007
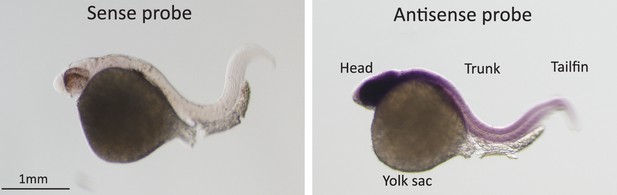
In-situ hybridization for Pou3f2 in zebrafish embryos.
Sense and Pou3f2-specific antisense RNA probe shows high expression of Pou3f2 in the head region, including the eye, hindbrain, midbrain and forebrain. Sense RNA probe was used as negative control.
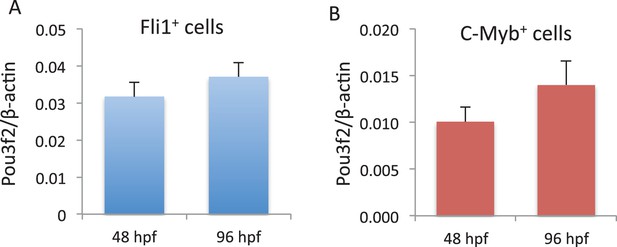
Pou3f2 gene expression in endothelial and hematopoietic lineages.
Endothelial (A) and hematopoietic (B) cells were FACS purified from Tg(Fli1:EGFP cells) and Tg(C-myb:EGFP) larvae at 48 and 96 hpf, respectively. Total RNA was extracted and real-time PCR performed for Pou3f2 (β-actin was used as housekeeping gene). All data represented as mean ± S.E.M. N = 3.
-
Figure 4—figure supplement 2—source data 1
Real time PCR analysis of Pou3f2 in zebrafish embryo Fli1+ and Cmyb+ cells.
- https://doi.org/10.7554/eLife.23588.010
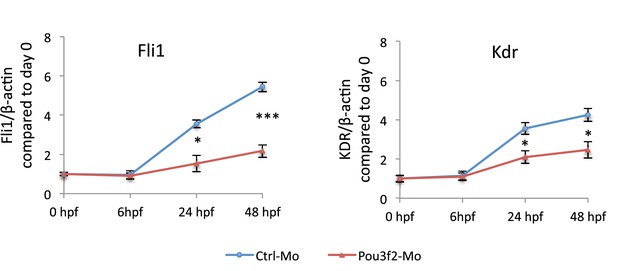
Gene expression of endothelial markers following Pou3f2 knockdown.
Total RNA was isolated from whole embryos injected with Pou3f2-targeted morpholino (Pou3f2-Mo) or a mismatch (Ctrl-Mo). Real-time PCR showed that Pou3f2 KD impaired significantly the expression of Fli1 and Kdr. All data represented as mean ± S.E.M. N = 3. Student t-test, *p=0.01; **p=0.001.
Role of Pou3f2 as a novel transcription factor in endothelial differentiation from human-induced pluripotent stem cells.
(a) Gene expression pattern of Pou3f2 in heterokaryons consisting of hiPSC and mEC compared to co-culture control. (b) Validation of expression of Pou3f2 during differentiation of hiPSC into endothelial lineage. (c) Expression of Pou3f2 in lentiviral mediated shRNA KD of Pou3f2 in hiPSC following differentiation into endothelial phenotype compared to Control shRNA group. (d) Representative images of Western blots showing the KD effects of Pou3f2 in hiPSC during endothelial differentiation, the same results were obtained at least three times. (e–j) Pou3f2 KD reduced the gene expression of endothelial markers including Kdr, Tie2, Cdh5, Pecam1, Nos3 and Vwf following endothelial differentiation of hiPSC. All data represented as mean ± S.E.M. (n = 3). p<0.05 vs co-culture control or control shRNA group.
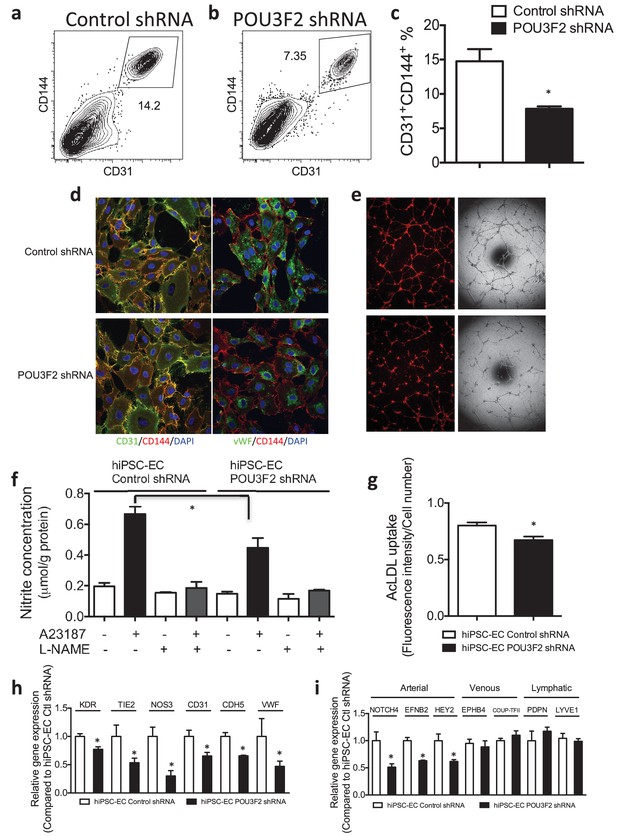
Functional assays further reveal the importance of Pou3f2 during differentiation of human iPSC into endothelial phenotype.
(a–c) Representative FACS plots and summarized diagram showing Pou3f2 KD reduced iPSC-EC generation compared to scrambled control. (d) Representative immunofluorescence images revealed lower expression of CD31, CD144 and Vwf in Pou3f2 KD iPSC-ECs. (e) The iPSC-ECs generated from Pou3f2 KD cells manifested poor formation of networks of tubular structures on matrigel. (f) The ability of Pou3f2 KD iPSC-ECs to produce nitric oxide in response to calcium ionophore A23187 was significantly reduced compared to scrambled control iPSC-ECs. (g) Reduced capacity in taking up AcLDL in Pou3f2 KD iPSC-ECs compared to scrambled control iPSC-ECs. (h) Reduced gene expression of endothelial markers including Kdr, Tie2, Nos3, CD31, Cdh5 and Vwf in Pou3f2 KD human iPSC-ECs. (i) Arterial markers (Notch4, Efnb2, Hey2) but not venous (Ephb4 and Coup-TFII) nor lymphatic markers (Pdpn and Lyve1) were affected in Pou3f2 KD iPSC-ECs compared to scrambled control iPSC-ECs. All data represented as mean ± S.E.M. (n = 3). p<0.05 vs Control shRNA group.
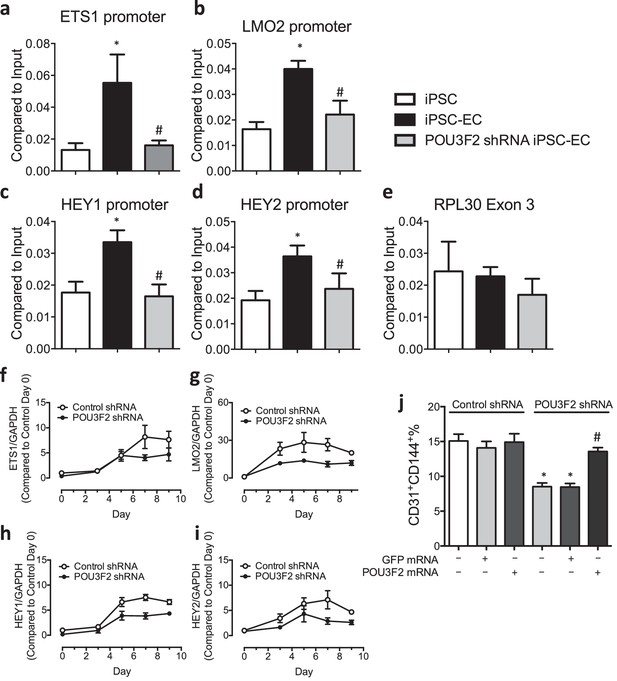
Chromatin immunoprecipitation (ChIP) and qPCR analysis reveal Pou3f2 binds to the promoters of endothelial-related transcription factors.
(a–d) Binding of Pou3f2 to the promoters of endothelial-related transcription factors including Ets1, Lmo2, Hey1 and Hey2 was significantly inhibited in Pou3f2 KD cells compared to scrambled control at Day 8 of endothelial differentiation protocol, without affecting the control promoter RPL30 Exon 3 (e). (f–i) Downregulation of gene expression of Ets1, Lmo2, Hey1 and Hey2 in Pou3f2 KD cells during differentiation into endothelial lineage. (j) Rescue experiments with modified mRNA encoding Pou3f2 improved CD31+CD144+ cell generation from Pou3f2 KD cells. All data represented as mean ± S.E.M. (n = 3). p<0.05 vs co-culture control or control shRNA group.





