sRNA-mediated activation of gene expression by inhibition of 5'-3’ exonucleolytic mRNA degradation
Figures
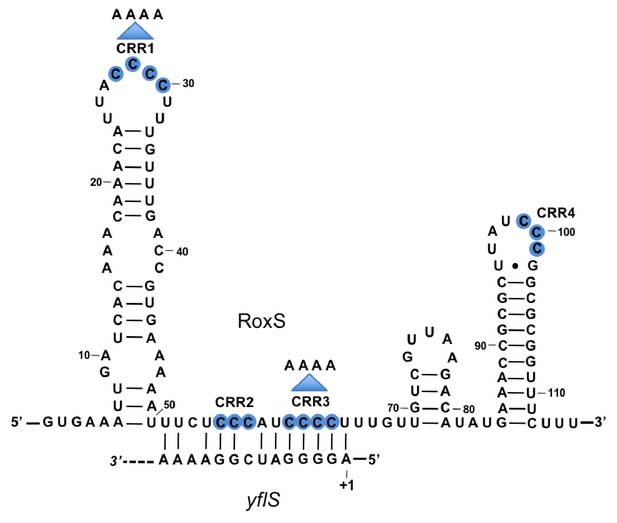
Base-pairing between RoxS and yflS mRNA predicted by the TargetRNA2 program (http://cs.wellesley.edu/~btjaden/TargetRNA2/).
The transcriptional start (+1) of the yflS mRNA is indicated. This was mapped in Figure 1—figure supplement 1 and its conservation is shown in Figure 1—figure supplement 2. The C-rich regions (CRR) of RoxS are in blue. Mutations CRR1 and CRR3 are indicated by arrows.
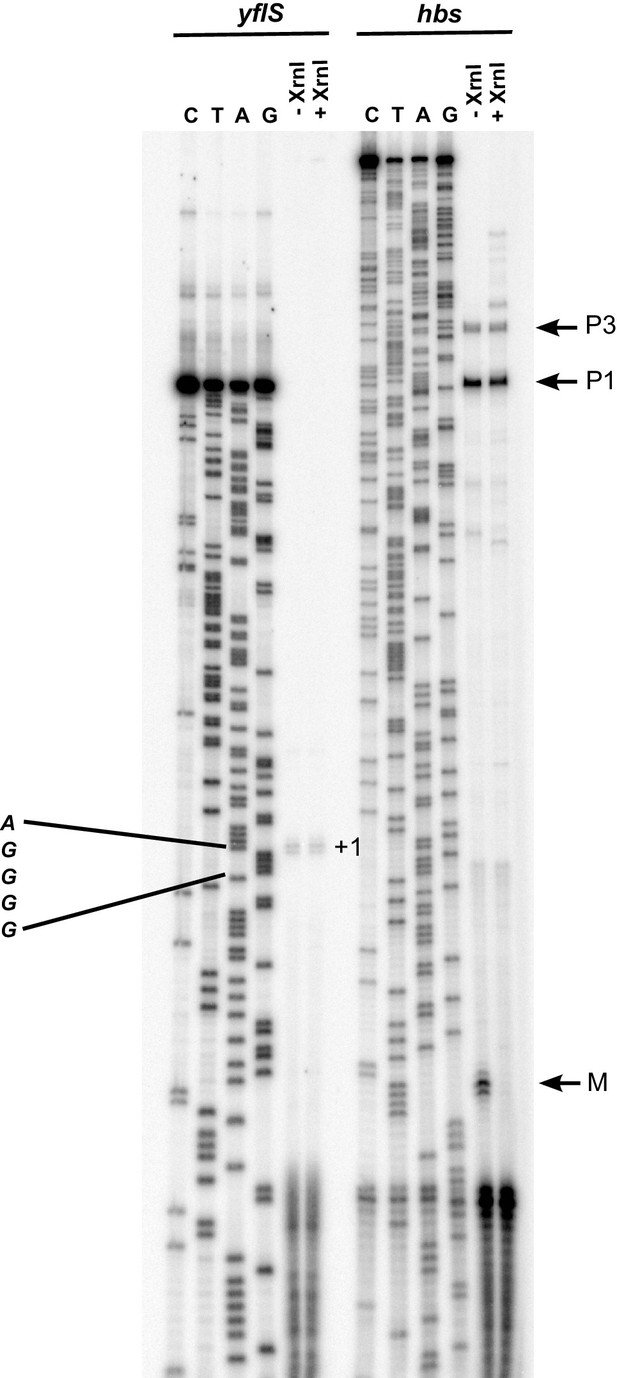
Mapping of the yflS 5’-end by primer extension.
Total RNA was extracted at mid-exponential phase (OD600 = 0.6) from the WT strain grown in the presence of 0.5% malate and treated (+) or not (−) with Xrn1 to digest 5’ monophosphorylated RNA. The sequence is labeled as its reverse complement to facilitate direct read-out. The yflS transcriptional start (+1) is indicated on the gel. As a control of the digestion of 5’ monophosphorylated RNAs with XrnI, a primer extension assay was performed with an oligonucleotide specific for the hbs mRNA. Transcripts from hbs promoters (P1 and P3, tri-P) and maturation product (M, mono-P) are indicated on the right of the gel.
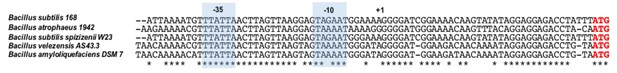
Conservation of the yflS promoter and 5'-UTR in various Bacilli.
Alignment of the yflS promoter region and 5'-UTR in Bacilli containing a clearly identifiable yflS gene through synteny. (B. subtilis and related bacteria have several di- and tri-carboxylate transporters homologous to YflS making it difficult to identify this particular transporter with certainty). The −35 and −10 regions are indicated, in addition to the transcription initiation site (+1) identified in B. subtilis. Conserved nts are indicated with an asterisk.
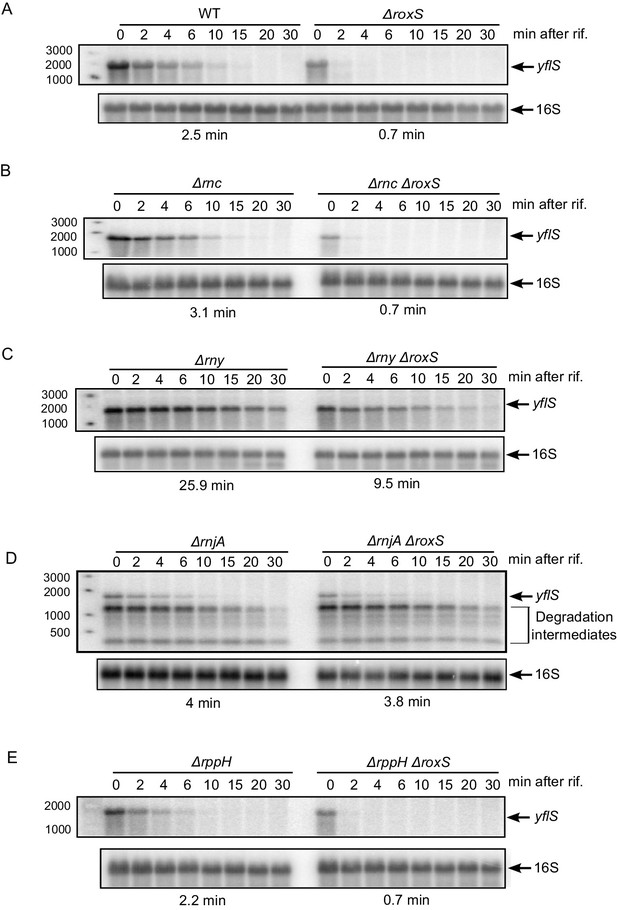
RoxS protects the full-length yflS mRNA from RNase J1 degradation in vivo.
A representative Northern blot showing total RNA isolated at times after addition of rifampicin (Rif) to (A) WT and ∆roxS strains, (B) ∆rnc and ∆rnc ∆roxS strains, (C) ∆rny and ∆rny ∆roxS strains, (D) ∆rnjA and ∆rnjA ∆roxS strains and (E) ∆rppH and ∆rppH ∆roxS strains grown in the presence of 0.5% malate. Rifampicin was added at mid-exponential phase (OD600 = 0.6). Half-lives calculated from two independent experiments (biological replicates) are given under each autoradiogram. The quantification of the Northern blots is presented in Figure 2—figure supplement 1.
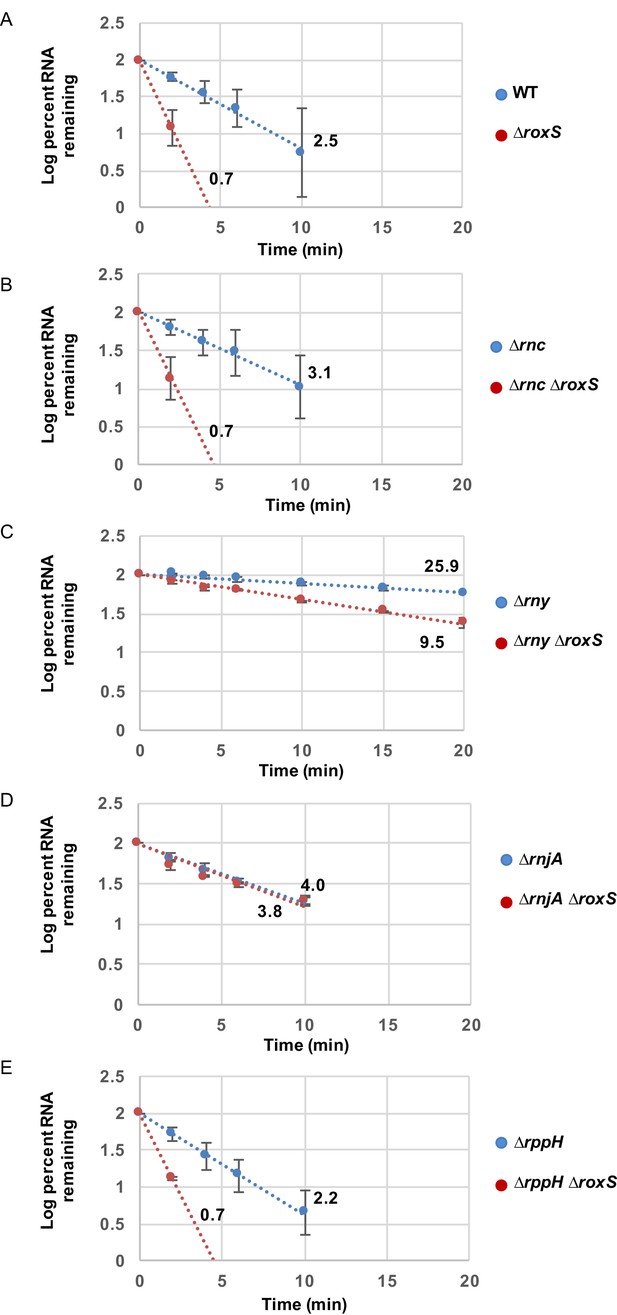
RoxS protects the full-length yflS mRNA from RNase J1 degradation in vivo.
RNA decay curves showing the log percent RNA remaining versus time after rifampicin addition to (A) WT and ∆roxS strains, (B) ∆rnc and ∆rnc ∆roxS strains, (C) ∆rny and ∆rny ∆roxS strains, (D) ∆rnjA and ∆rnjA ∆roxS strains and (E) ∆rppH and ∆rppH ∆roxS strains, grown in the presence of 0.5% malate. The data shown correspond to the quantification of two independent experiments (biological replicates). See also Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Source data for the quantification of yflS mRNA half-life in WT and Bacillus subtilis RNase mutant strains (∆rnc, ∆rnc ∆roxS, ∆rny, ∆rny ∆roxS, ∆rnjA, ∆rnjA ∆roxS, ∆rppH, ∆rppH ∆roxS).
- https://doi.org/10.7554/eLife.23602.007
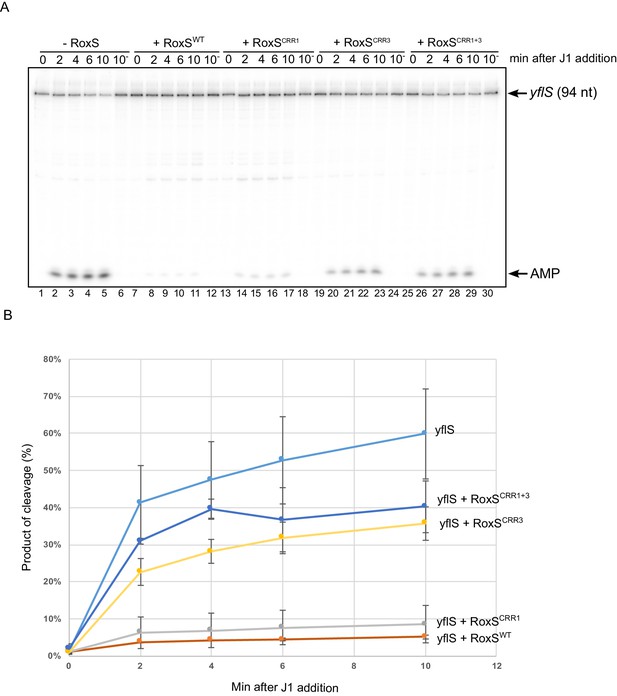
RoxS protects the yflS mRNA from RNase J1 degradation in vitro.
(A) In vitro degradation of the 5’ end of yflS RNA by RNase J1 ± RoxS sRNA or RoxS CRR mutants (see Figure 1). The yflS mRNA was in vitro transcribed, dephosphorylated with CIP and radiolabeled at its 5’-end to obtain a 5’ monophosphate extremity. Times after RNase J1 addition are indicated at the top of the gel. Reactions were done at 37°C. The migration positions of the yflS in vitro transcript (94 nt) and AMP are indicated. (B) Percent release of AMP from yflS substrate RNA by RNase J1 ± RoxS sRNA or RoxS CRR mutants as a function of time (minutes). The data shown correspond to the quantification of two independent experiments (technical replicates). See also Figure 3—source data 1.
-
Figure 3—source data 1
Source data for the quantification of yflS degradation by RNase J1 in vitro.
- https://doi.org/10.7554/eLife.23602.009
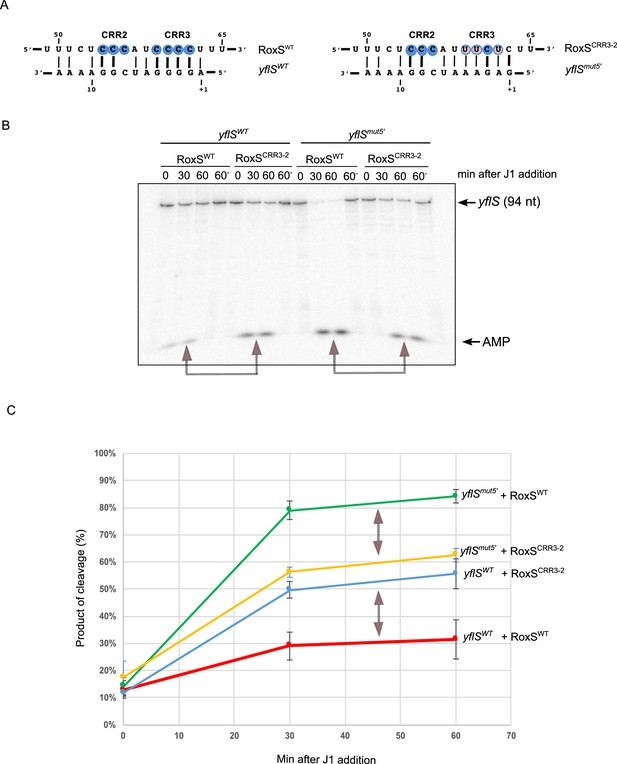
RoxS uses C-rich region 3 (CRR3) to interact with the extreme 5’-end of yflS mRNA.
(A) Base-pairing between yflS and RoxS, or yflSmut5' and RoxSCRR3-2 compensatory mutants (B) In vitro degradation of the 5’ end of yflS and yflSmut5' transcripts by RNase J1 ± RoxS or the RoxSCRR3-2 mutant. yflS and yflSmut5' mRNAs were in vitro transcribed, dephosphorylated with CIP and radiolabeled at their 5’-end to obtain a 5’ monophosphate extremities. Times after RNase J1 addition are indicated at the top of the gel. Reactions were performed at 15°C to facilitate interaction between yflSmut5' and RoxSCRR3-2 because the ∆G of the compensatory interaction is predicted to be weaker than the wt (−8.0 vs −13.7 kcal/mol). The migration positions of the yflS in vitro transcript (94 nt) and AMP are indicated. (C) Percent release of AMP from yflS and yflSmut5' substrate RNAs by RNase J1 ± RoxS or the RoxSCRR3-2 mutant as a function of time (minutes). The data shown correspond to the quantification of three independent experiments (technical replicates). Structure probing of RoxS ± yflS is shown in Figure 4—figure supplement 1. See also Figure 4—source data 1.
-
Figure 4—source data 1
Source data for the quantification of yflS and yflSmut degradation by RNase J1 in vitro.
- https://doi.org/10.7554/eLife.23602.011
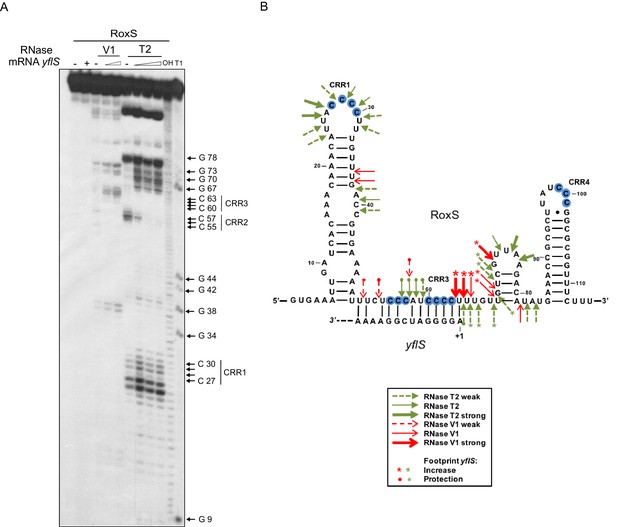
RoxS uses C-rich region 3 (CRR3) to interact with the extreme 5’-end of yflS mRNA.
(A) Enzymatic probing of the RoxS sRNA secondary structure in the presence and absence of yflS. The 5’ end-labeled RoxS sRNA was digested with RNase V1 and RNase T2 and run on an 8% polyacrylamide–urea gel. The enzymatic digestions were performed in the absence (−) or in the presence of increasing concentrations of yflS mRNA (50, 100 and 400 nM) for digestion by RNase T2, and 50 and 100 nM for the experiment with RNase V1. Incubation controls (in the absence of RNase) were done in the absence (−) and in the presence (+) of yflS (400 nM). The positions of CRR1, 2 and 3 are indicated to the right of the gel. The probing experiment was performed twice (technical replicates). (B) Secondary structure of the RoxS sRNA and effect of yflS binding. Summary of the cleavage patterns generated by RNase V1 (red arrows) and RNase T2 (green arrows). Increased cleavages upon addition of yflS are indicated by asterisks, increased protection by circles.
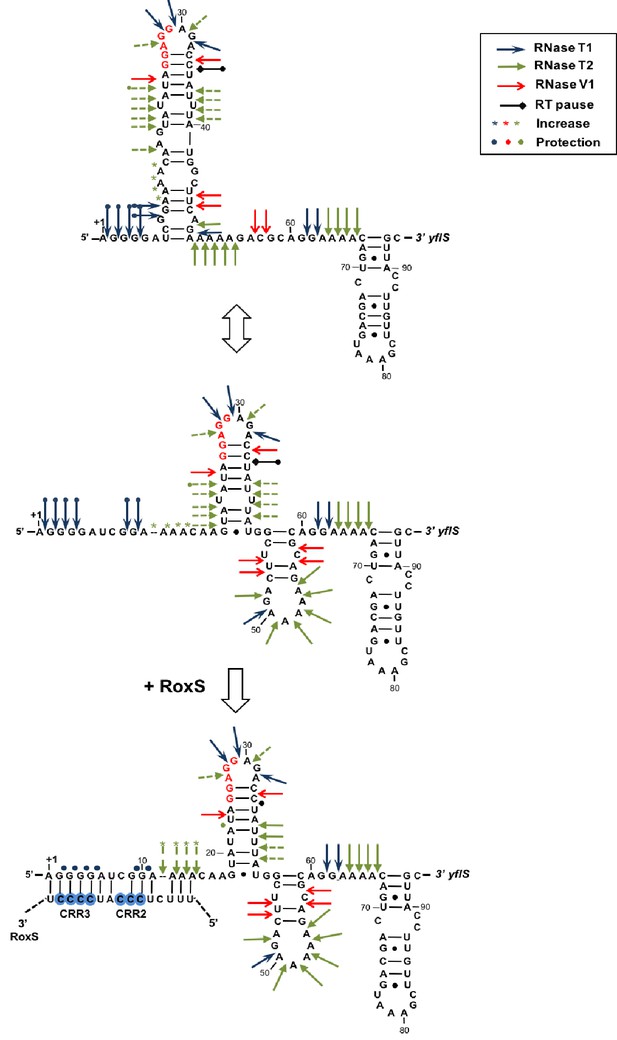
Modification of the yflS mRNA secondary structure upon RoxS binding.
Summary of the cleavage patterns (Figure 5—figure supplement 1) of yflS alone (two potential equilibrium structures) or bound to RoxS generated by RNase V1 (red arrows), RNase T1 (blue arrows) and RNase T2 (green arrows). Weaker cleavages are represented by dashed arrows. The strong RT stop observed at position U35 (−5 relative to the AUG) is also indicated (black bar). Residues affected by the RoxS binding are indicated by colored asterisks (circles for inhibition; asteriks for stimulation of cleavage). The sequencing ladder was made by reverse transcribing RNA in the presence of dideoxynucleotides. The data suggests that a structural link exists between the 5’ extremity and the downstream hairpin structure that contributes to the weak accessibility of the adenines 11 to 14 towards RNase T2 cleavages.
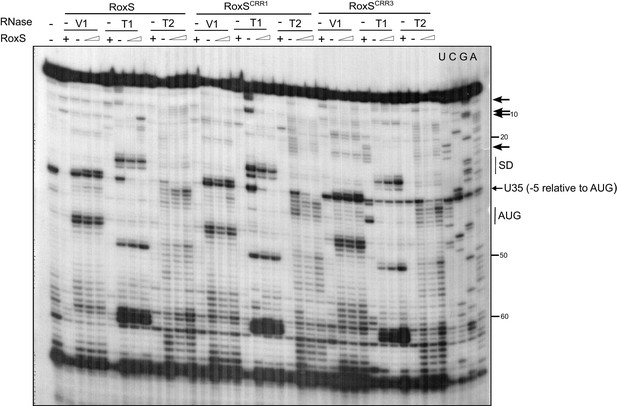
Enzymatic probing of the yflS mRNA secondary structure in the presence and absence of RoxS.
The 5′-end-labeled yflS mRNA fragment (nts + 1 to+94) was digested with RNase T1, RNase T2 and RNase V1 and run on a 10% polyacrylamide–urea gel. The enzymatic digestions were performed in the absence (−) or in the presence of increasing concentrations of RoxS (100 or 250 nM). Incubation controls (in the absence of RNase) were done in the absence (−) and in the presence (+) of RoxS (250 nM). Arrows indicate the nucleotides protected by RoxS from the attack by RNase T1 or T2. The Shine–Dalgarno sequence (SD) and the AUG codon (Met), are indicated to the right of the gel. The probing experiment was performed two times (technical replicates).
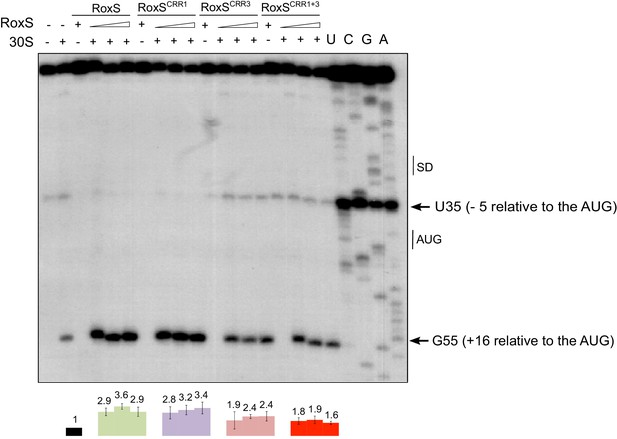
RoxS stimulates 30S binding to the yflS mRNA in vitro.
Toeprint analysis of wild-type (WT) and various mutant forms of RoxS bound to 50 nM of the yflS in vitro transcript (94 nt). The toeprint formed by the 30S ribosomal subunit is indicated at +16 relative to the first nt of the AUG start codon. An additional RT stop at position U35 (−5 relative to the AUG), observed with RoxS variants containing a mutant CRR3 is also indicated. The Shine and Dalgarno sequence is indicated by SD. Three different quantities of RoxS (250, 500 and 1000 nM) were tested. The average stimulation of the toeprint at +16 by RoxS variants, normalized to the yflS mRNA alone, from three experiments (technical replicates) is presented under each lane of the autoradiogram, with standard errors as shown.
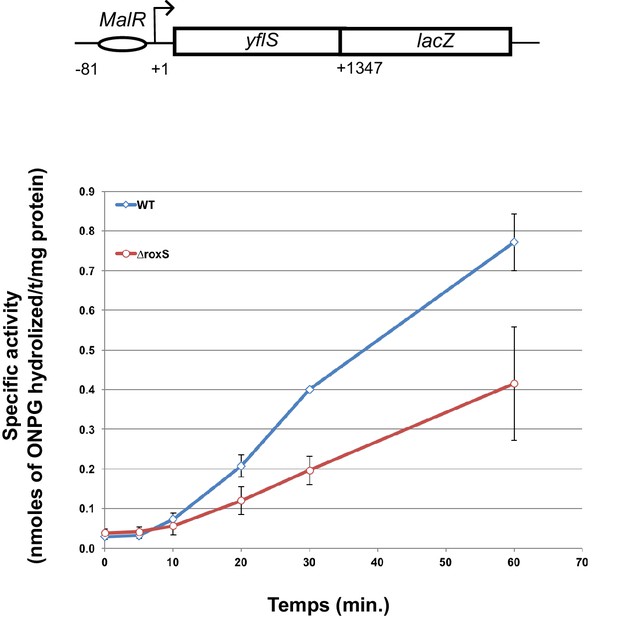
RoxS stimulates yflS translation in vivo.
ß-Galactosidase assays of yflS–lacZ translational fusions in wild-type (WT) and ∆roxS strains. The reporter is under control of the yflS promoter and regulatory sequences, and consists of 436 amino acids of yflS coding sequence fused to lacZ. Malate (0.5%) was added at mid-exponential growth phase to induce the expression of the fusion. The levels of ß-galactosidase activity were assayed between 0 and 60 min after malate addition. The specific activity, in nmoles of ONPG hydrolyzed/mg protein/min, was determined in three independent experiments. Error bars indicate the standard deviation for each point. See also Figure 7—source data 1.
-
Figure 7—source data 1
Source data for the quantification of ß-galactosidase activity of the yflS-lacZ fusion.
- https://doi.org/10.7554/eLife.23602.017
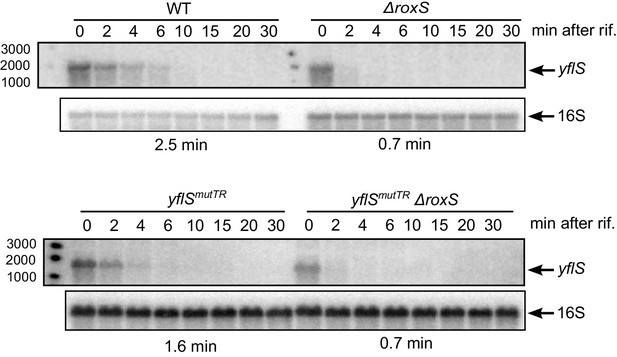
RoxS stabilizes a non-translated yflS mRNA variant in vivo.
A representative Northern blot showing total RNA isolated at times after addition of rifampicin (Rif) to WT and ∆roxS strains, and yflSmutTR and yflSmutTR ∆roxS strains grown in the presence of 0.5% malate. Rifampicin was added at mid-exponential phase (OD600 = 0.6). Half-lives calculated from two independent experiments (biological replicates) are given under each autoradiogram. The quantification of the Northern blots is presented in Figure 8—figure supplement 1.
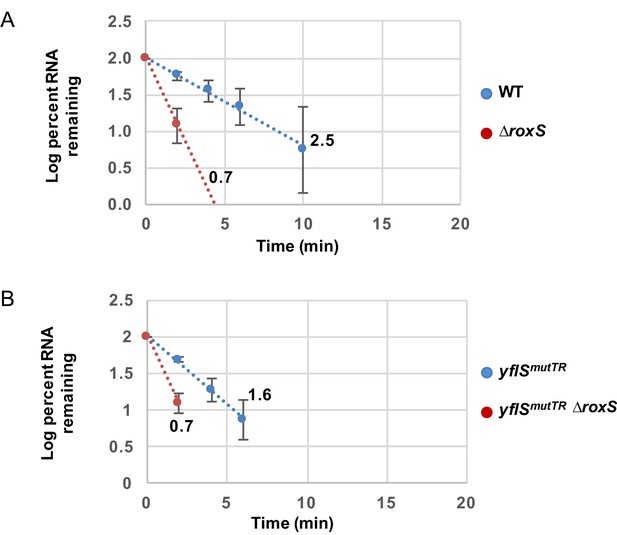
RoxS stabilizes a non-translated yflS mRNA variant in vivo.
RNA decay curves showing the log percent RNA remaining versus time after rifampicin addition to WT and ∆roxS strains, and yflSmutSD and yflSmutSD ∆roxS grown in the presence of 0.5% malate. The data shown correspond to the quantification of two independent experiments (biological replicates). See also Figure 8—figure supplement 1—source data 1.
-
Figure 8—figure supplement 1—source data 1
Source data for the quantification of yflSmutTR mRNA half-life.
- https://doi.org/10.7554/eLife.23602.020
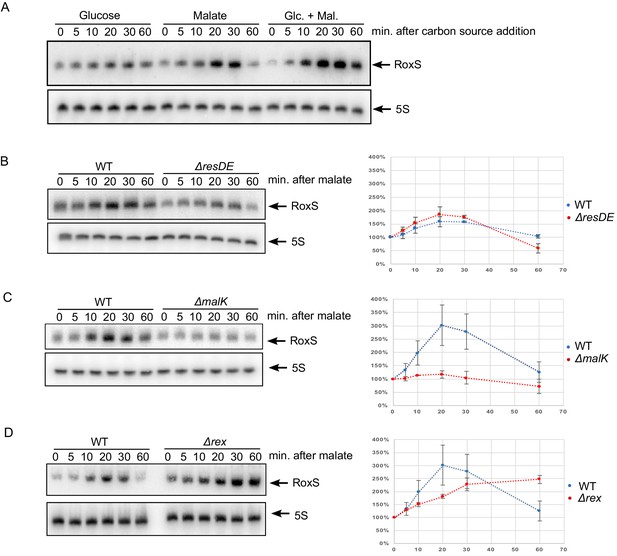
Derepression of RoxS expression in strains lacking Rex.
(A) Northern blot of total RNA extracted from B. subtilis W168 (wild-type) before (time 0) and 5, 10, 20, 30 and 60 min after addition of 0.2% malate, 0.2% glucose, or both. (B) Northern blot of total RNA extracted from wild-type and ∆resDE strains before (time 0) and 5, 10, 20, 30 and 60 min after addition of 0.5% malate. (C) Same legend as panel B using wild-type and ∆malK strains. (D) Same legend as panel B using wild-type and ∆rex strains. Quantifications of the Northern blots are from two independent experiments (biological replicates). MalR binding assays are shown in Figure 9—figure supplement 1. Conservation of the Rex binding site in different Staphylococci and Bacilli RoxS promoters is presented Figure 9—figure supplement 2. See also Figure 9—source data 1.
-
Figure 9—source data 1
Source data for the quantification of RoxS derepression by malate in Bacillus subtilis mutant strains (∆resDE, ∆malK, ∆rex).
- https://doi.org/10.7554/eLife.23602.022
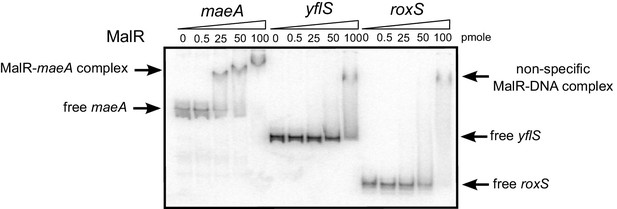
MalR does not bind the RoxS promoter.
EMSA of 32P-labelled double-stranded DNA fragments (0.5 pmol) containing the maeA MalR operator, the RoxS promoter or an internal portion of the yflS coding sequence. The amounts of MalR in the binding reaction are indicated (in pmoles) above each lane. EMSAs were performed twice (technical replicates).
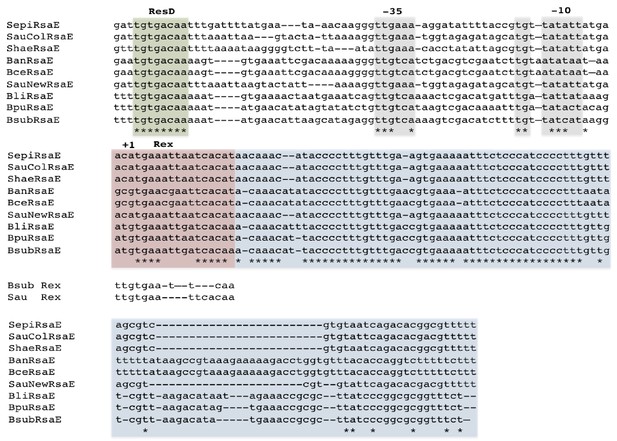
Alignment of RsaE/RoxS sequences from different Staphylococci and Bacilli adapted from Durand et al. (2015a).
The RoxS sequences are boxed in blue. Likely −35 and extended −10 sequences are boxed in mauve and the ResD and Rex binding sites are boxed in green and pink, respectively. The consensus binding sequences for B. subtilis and S. aureus Rex are indicated (Härtig and Jahn, 2012; Pagels et al., 2010). Positions showing 100% conservation are indicated by asterisks. Abbreviations are as follows: Sepi, S. epidermis; SauCol, S. aureus Colindale strain; Shae, S. haemophilus; Ban, B. anthracis; SauNew, S. aureus Newman strain; Bli, B. lichiniformis; Bpu, B. pumilus; Bsub, B. subtilis.
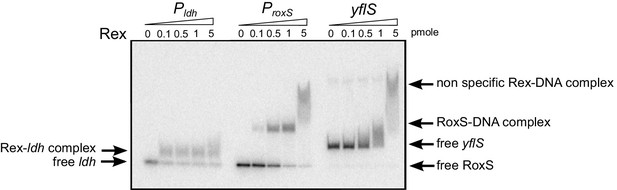
Rex binds the RoxS promoter region in vitro.
Electrophoretic mobility shift assays (EMSA) of 32P-labelled double-stranded DNA fragments (0.1 pmol) containing the ldh Rex operator, the RoxS promoter or an internal sequence of yflS coding sequence. The quantities of Rex in the binding reaction are indicated (in pmoles) above each lane. EMSAs were performed three times (technical replicates).
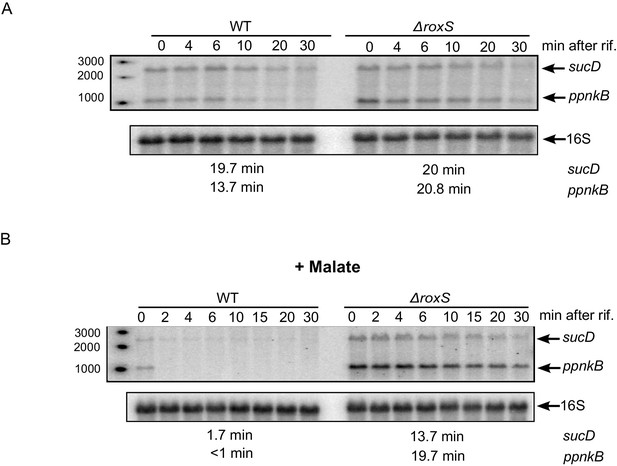
The effect of RoxS on ppnKB and sucCD mRNA stability is amplified in the presence of malate.
(A) Northern blot of total RNA isolated at times after addition of rifampicin (rif) to WT and ∆roxS strains. Rifampicin was added at mid-exponential phase (OD600 = 0.6). The half-lives of the ppnkB and sucCD mRNAs are indicated below the blot. (B) Same legend as panel A but with 0.5% malate in the medium. Northerns were performed twice (biological replicates).
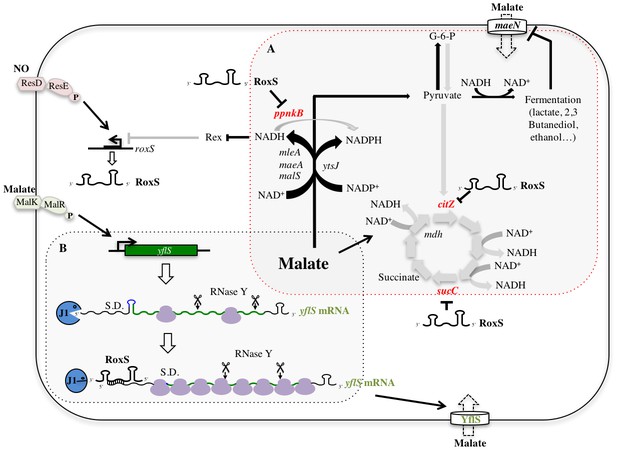
Model of various roles of RoxS upon addition of malate to culture medium.
For details see text. Panel A shows reactions affected by RoxS that impact NAD(P)+/NAD(P)H balance. Panel B shows the role of RoxS in yflS mRNA degradation. Targets negatively regulated by RoxS are in red; targets positively regulated by RoxS are in green.
Additional files
-
Supplementary file 1
Strains used in this study.
- https://doi.org/10.7554/eLife.23602.028
-
Supplementary file 2
Oligos used in this study.
- https://doi.org/10.7554/eLife.23602.029





