La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs
Figures
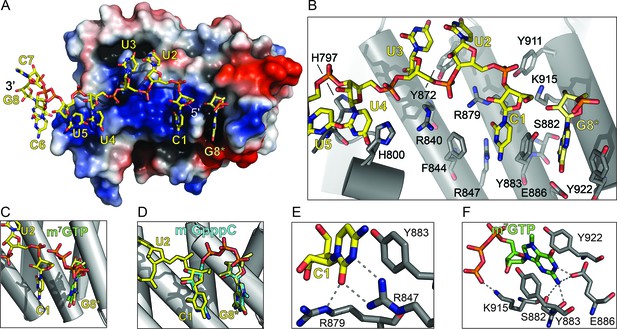
The LARP1 DM15 region recognizes the 7-methylguanosine cap and invariant 5’cytidine of TOP mRNAs.
(A) Protein surface representation is colored according to electrostatic potential (−74 kEV, red; 74 kEV, blue). (B) Zoomed view of the DM15 RNA binding site. (C) Superimposition of DM15 bound to RNA and bound to cap analog, m7GTP. (D) Superimposition of DM15 bound to RNA and bound to m7GpppC. (E–F) Zoomed views of the specific recognition of C1 (E) and m7GTP (F). Potential hydrogen bonds indicated by dotted lines.
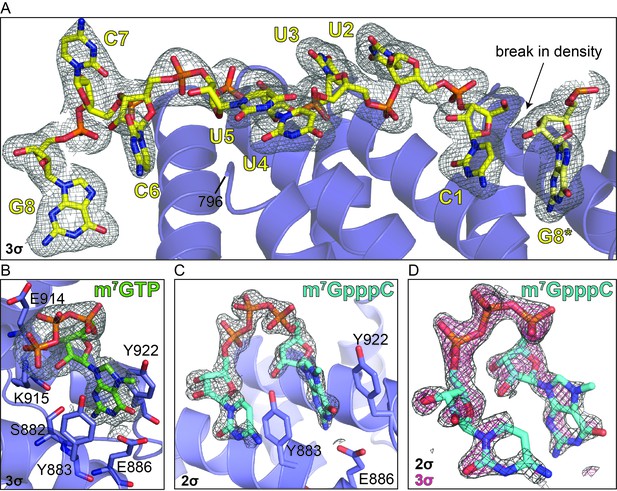
Electron density reveals RNA, cap analog, and m7GpppC bind in the same location on the conserved surface of the DM15 region of LARP1.
Composite omit maps carved around the (A) RNA (3σ), (B) m7GTP cap analog (3σ), and (C) m7GpppC dinucleotide (2σ). (D) Composite omit map carved around the m7GpppC dinucleotide at 2σ (grey) and 3σ (magenta) for comparison.
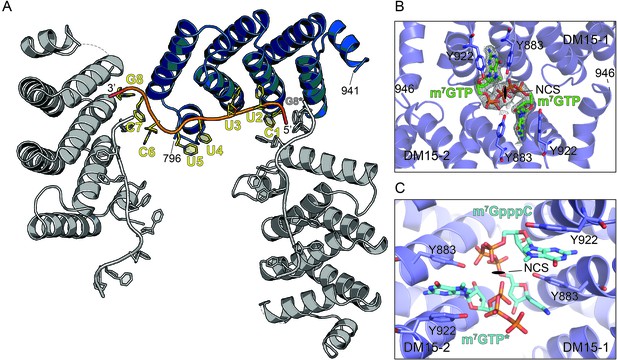
The DM15 region of LARP1 recognizes a guanosine.
(A) Three neighboring unit cells from the DM15-RNA co-crystal are shown. The protein monomer colored in blue interacts with two molecules of RNA: one from its unit cell and one originating in the unit cell on the right. This places a guanosine nucleotide 5’ to the TOP motif in the binding site of the blue DM15 region. (B) Two non-crystallographic symmetry mates from the DM15-m7GTP co-crystal show the guanine of the cap analog binds the same place as the G8* residue binds in the DM15-RNA co-crystal (A). (C) Two non-crystallographic symmetry mates from the DM15-m7GpppC co-crystal structure reveal that the m7G and C moieties bind in the same place as the G8* and C1 residues in the DM15-RNA co-crystal, respectively. In the other non-crystallographic symmetry-mate shown, only m7GTP is modeled because only one dinucleotide can bind per asymmetric unit since the inverted triphosphate linkage sits on the NCS 2-fold axis (see Figure 1—figure supplement 5).
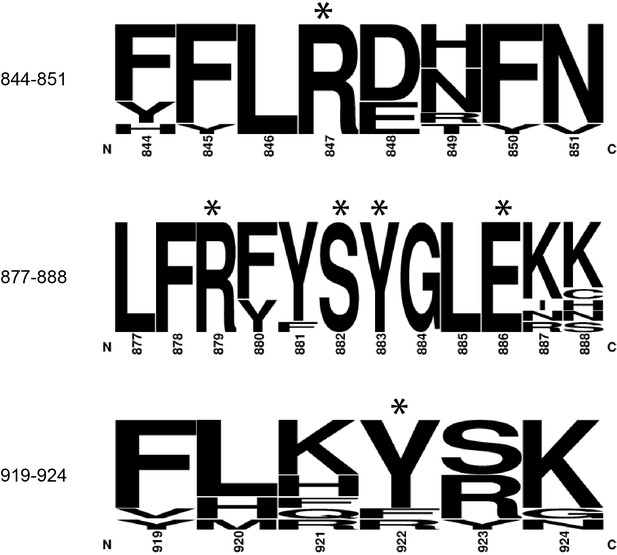
The amino acids of the DM15 region that directly bind G8*, C1, and m7GTP are nearly 100% conserved.
Frequency plots were generated from amino acid sequences of LARP1 from H. sapiens (NP_056130.2), M. musculus (NP_082727.1), D. rerio (XP_001920902.3), X. laevis (NP_001089363.1), D. melanogaster (NP_524998.1), C. elegans (NP_001040867.2), A. thaliana (NP_001190354.1), O. sativa (XP_015621184.1), B. dendrobatidis (XP_006676827.1), and M. brevicollis (XP_001744631.1). Asterisks indicate amino acids shown in Figure 1. Numbering is based on the DM15 region construct cloned from LARP1 isoform 2 (NP_056130.2).
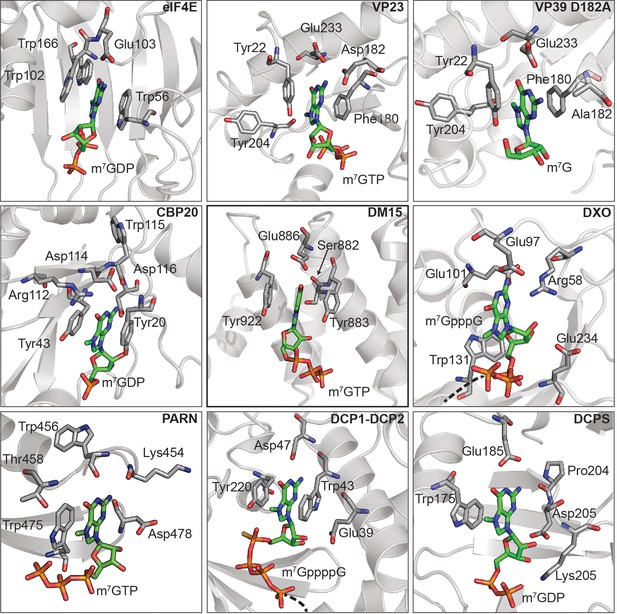
The DM15 region of LARP1 has evolved a unique fold to support a canonical cap-binding pocket.
The amino acids responsible for cap recognition in each of the proteins are shown as white sticks and the cap analog is shown as green sticks. PDBs used to generate each panel: eIF4E (1EJ1), VP23 (1AV6), VP39 D182A (4DCG), CBP20 (1H2T), LARP1 DM15 (5V4R), DXO (4J7N), PARN (3CTR), DCP1-DCP2 (5KQ4), DCPS (1XMM).
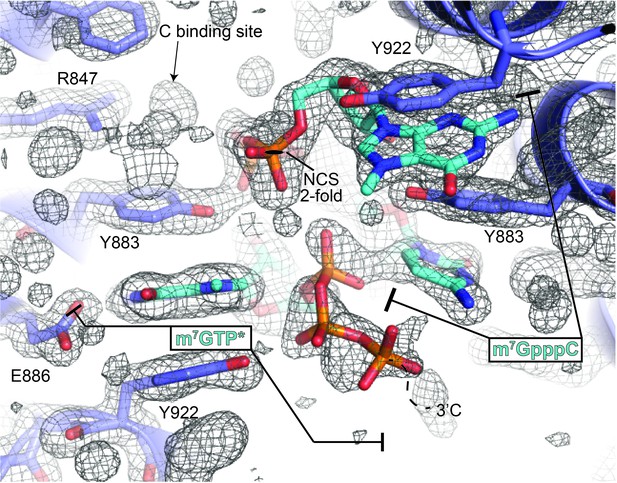
The m7GpppC-DM15 co-crystal non-crystallographic symmetry reveals one ordered dinucleotide and one partially ordered dinucleotide.
The composite omit map shown is contoured at 1σ and reveals an ordered m7GpppC dinucleotide bound to the DM15 NCS-mate on the right. The DM15 NCS-mate on the left binds the m7G in the G-binding pocket, while the triphosphate moiety is ordered and facing away from the C-binding pocket (m7GTP*). Presumably, the density for the cytidine is mostly disordered: based on the path of the m7G triphosphate moiety (dotted line and 3'C label), the cytidine is most likely in the solvent channel; density for the Watson-Crick face of a cytidine is visible in the C-binding site in this copy of DM15, but is presumably of very low occupancy. Based on this data and the data shown in Figure 1—figure supplement 2B, only one inverted triphosphate per asymmetric unit can be accommodated in the anion binding site that overlaps with the non-crystallographic 2-fold axis.
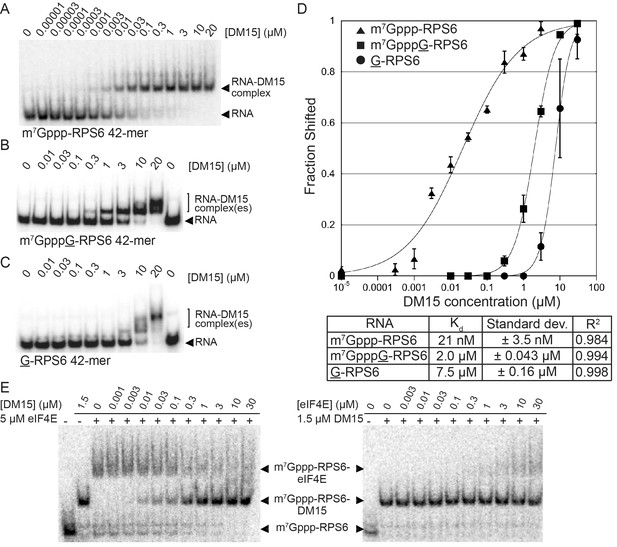
The LARP1 DM15 region recognizes capped TOP sequences and outcompetes eIF4E for their binding.
(A–C) Electrophoretic mobility shift assays using the indicated RNA. (D) Quantitation of 3 replicate EMSAs; error bars represent standard deviation. (E) Competition assays analyzed by native gel electrophoresis using labeled m7Gppp-RPS6 as substrate with the indicated protein concentrations.
-
Figure 2—source data 1
Data for graphed EMSAs.
- https://doi.org/10.7554/eLife.24146.010
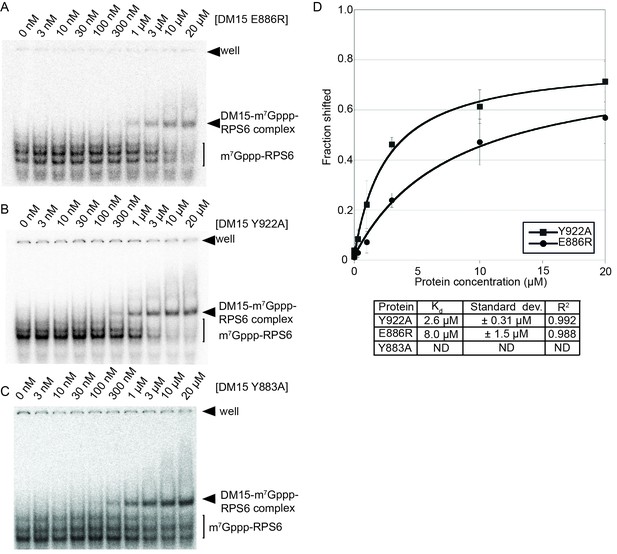
Mutation of amino acids that stabilize the cap and the RNA decreases the affinity of the DM15 region for capped TOP RNA.
(A–C) EMSA of capped RPS6 42-mer 5’UTR with each indicated point mutant. (D) Quantification of three replicates of each indicated EMSA. ND, not determined.
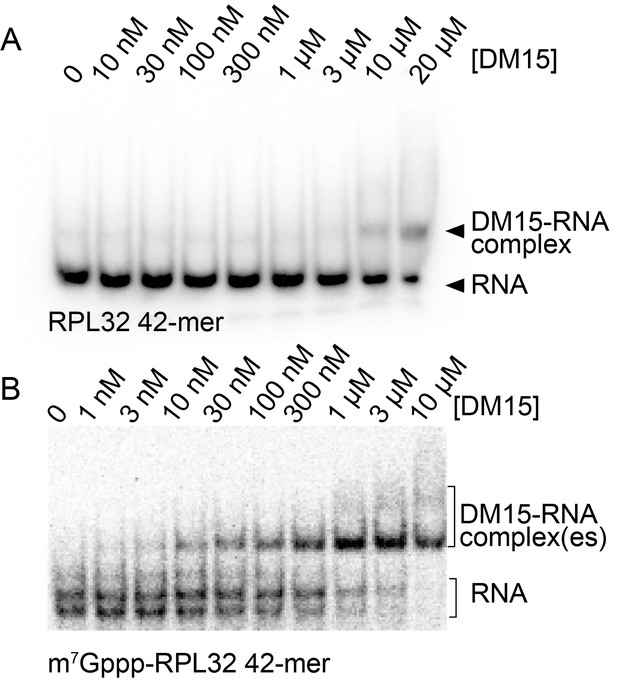
Capping another TOP sequence enhances the affinity of DM15 for the RNA.
(A) EMSA of DM15 with an uncapped RNA representing the first 42 nucleotides of the 5’UTR of RPL32. (B) EMSA of DM15 with the capped version of the RNA from panel (A).
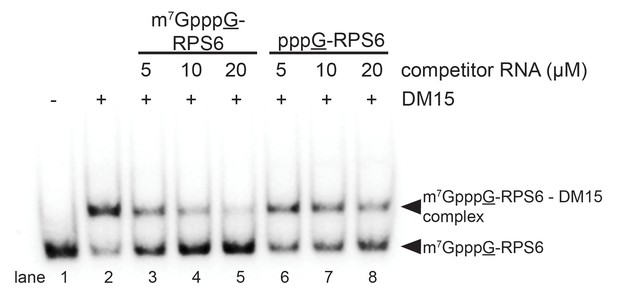
The interaction between the DM15 region of LARP1 and the cap moiety is specific.
Competition assay challenging DM15 bound to labeled capped non-TOP RNA with capped (lanes 3–5) or uncapped (lanes 6–8) cold competitor RNA.
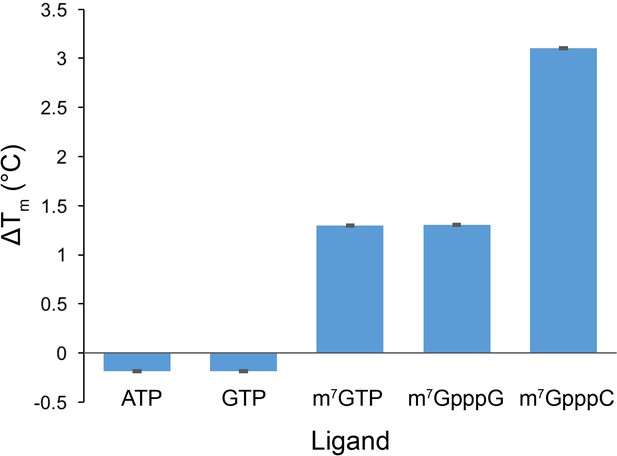
Cap analog, m7GTP, and m7GpppC stabilize the protein fold of the DM15 region of LARP1.
Quantification of thermal shift assays conducted in the presence of GTP, m7GTP, ATP, m7GpppG, or m7GpppC; n = 3 for all assays; error bars show 1 standard deviation.
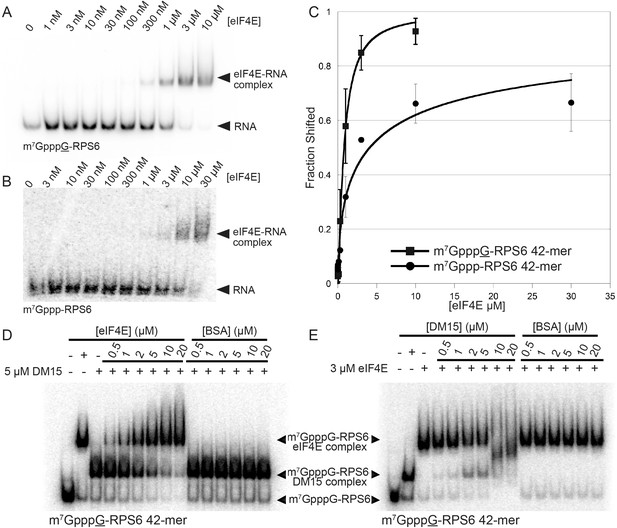
eIF4E binds non-TOP mRNAs and outcompetes the DM15 region of LARP1 for their binding.
(A–B) EMSAs with the indicated substrates and increasing concentrations of recombinant human eIF4E. (C) Quantitation of EMSAs, n = 3. Error bars indicate standard deviation. (D–E) Competition assays for the capped non-TOP substrate.
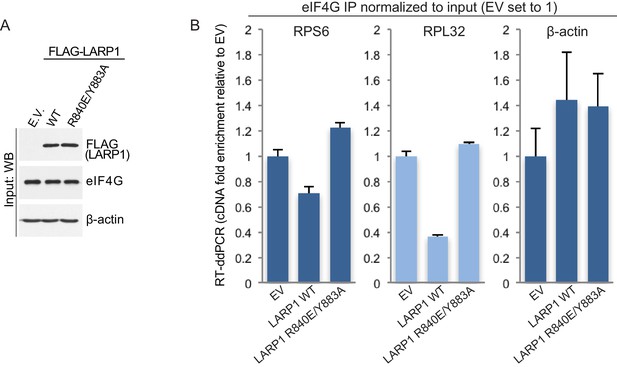
LARP1 prevents eIF4F assembly on 5’TOP mRNAs.
(A and B). Extracts of HEK293T cells that were transfected with empty vector (EV), FLAG-tagged wild type LARP1 (WT) or FLAG-LARP1 double-mutant (R840E/Y883A), were immunoprecipitated with anti-eIF4G antibody. Inputs were analyzed by Western blot (A) and eIF4G-IPs were analyzed for TOP mRNA abundance by RT-ddPCR (B). Data were normalized to input mRNA levels. Three biological replicates were performed and error bars denote propagated standard deviation.
-
Figure 3—source data 1
Data analysis for eIF4G IPs.
- https://doi.org/10.7554/eLife.24146.017
Tables
X-ray data collection and refinement statistics.
| Data collection | m7GTP | RNA-bound | m7GpppC |
|---|---|---|---|
| Space group | P21 | I4 | P21 |
| Unit cell dimensions | |||
| a, b, c (Å) | 48.177, 60.163, 60.609 | 107.095, 107.095, 29.113 | 47.959, 59.962, 60.321 |
| β | 101.283 | 90 | 100.653 |
| Resolution (Å) | 33.90–1.77 | 37.86–2.59 | 30.00–1.69 |
| Rmerge (%) | 3.5 (27.7) | 15.6 (37.1) | 3.6 (12.5) |
| I/σ(I) | 16.30 (2.1) | 8.79 (1.36) | 19.85 (8.8) |
| Completeness (%) | 82 (73) | 98.5 (87.1) | 92.2 (93.2) |
| Redundancy | 2.9 (1.5) | 4.3 (2.1) | 3.4 (2.8) |
| Refinement | m7GTP | RNA-bound | m7GpppC |
|---|---|---|---|
| Resolution (Å) | 33.90–1.77 | 37.86–2.59 | 30.00–1.69 |
| No. reflections | 28525 | 5272 | 34328 |
| Completeness (%) | 82 (73) | 98.5 (87.1) | 92.2 (93.2) |
| Rwork/Rfree | 17.50/20.80 | 20.69/23.51 | 18.10/20.30 |
| RMSD bond angle (°) | 1.43 | 1.19 | 0.89 |
| RMSD bond length (Å) | 0.012 | 0.015 | 0.010 |
| Average B-factor | 42.0 | 36.4 | 33.9 |
| PDB ID | 5V4R | 5V7C | 5V87 |





