The Calcineurin-FoxO-MuRF1 signaling pathway regulates myofibril integrity in cardiomyocytes
Figures
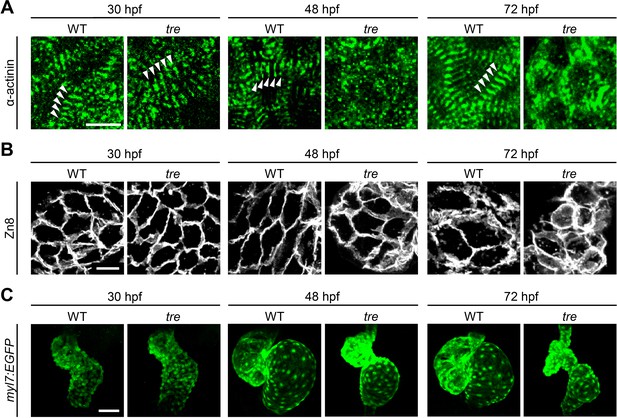
Disorganized myofibril structure in tre/ncx1h cardiomyocytes.
Wild type (WT) and tre/ncx1h (tre) mutant hearts at 30, 48 and 72 hpf. (A) Zebrafish hearts stained for α-actinin to visualize Z-lines. At 30 hpf, periodic α-actitin staining was observed in wild type and tre hearts (arrowheads). By 48 hpf, sarcomeres are disassembled in tre hearts. Scale bar, 10 μm. (B) The cell shape of cardiomyocytes was visualized by Zn8 staining. Scale bar, 10 μm. (C) Embryonic fish hearts were visualized by GFP expression in the myl7:EGFP transgenic background. Note that tre hearts become dysmorphic after two days of development. Scale bar, 50 μm.
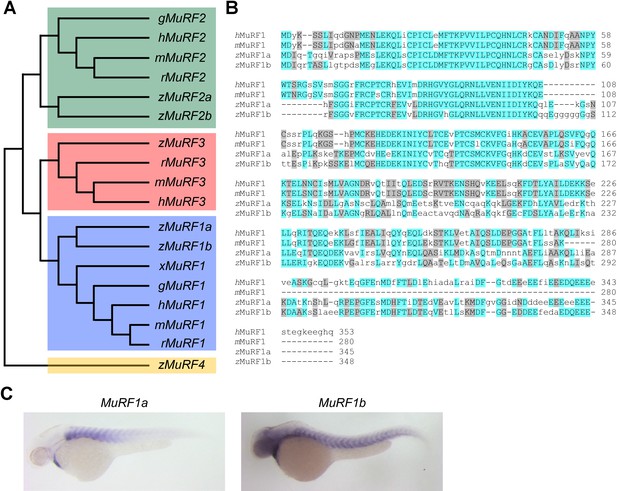
Zebrafish murf1 genes.
(A) Phylogenetic tree of vertebrate murf1, 2, 3 and 4 (also known as trim63, 55, 54 and 101, respectively). The tree was constructed using ClustalX with the neighbor-joining method. Zebrafish (z), Human (h), mouse (m), rat (r), chick (g), frog (x). (B) Alignment of murf1 genes from human, mouse and zebrafish. Blue boxes highlight identical amino acids and grey boxes indicate similar residues. (C) Whole-mount in situ hybridization demonstrating the expression patterns of murf1a and murf1b in the zebrafish embryo.
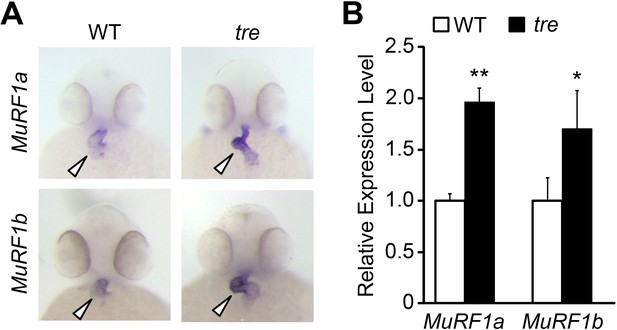
Upregulation of MuRF1 in tre/ncx1h deficient hearts.
(A) In situ hybridization analysis shows a significant increase of murf1a and murf1b expression in tre hearts. Arrowheads point to the heart. (B) Quantitative RT-PCR analysis shows an upregulation of murf1a and murf1b in tre hearts. * p<0.05; ** p<0.01.
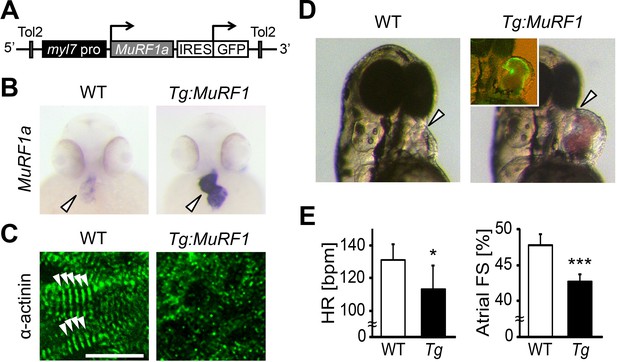
Upregulation of MuRF1a leads to myofibril disarray.
(A) Schematic representation of the construct that drives cardiac-specific MuRF1a expression. (B) Murf1a expression is upregulated in the 2-day-old myl7:MuRF1a-IRES-GFP heart (right panel) compared to the wild type heart (left panel). (C) α-actinin staining in 3-day-old wild type (left panel) and transgenic (right panel) cardiomyocytes. Note that sarcomeres are disassembled in myl7:MuRF1a-IRES-GFP cardiomyocytes. (D) Live images of wild type and Tg(myl7:MuRF1a-IRES-GFP) fish at 72 hpf (left panels). Transgenic hearts are GFP positive and become dilated (inset). (E) Heart rate (HR) and atrial fractional shortening (FS) in wild type and myl7:MuRF1a-IRES-GFP embryos at 72 hpf. *p<0.05; ***p<0.001.
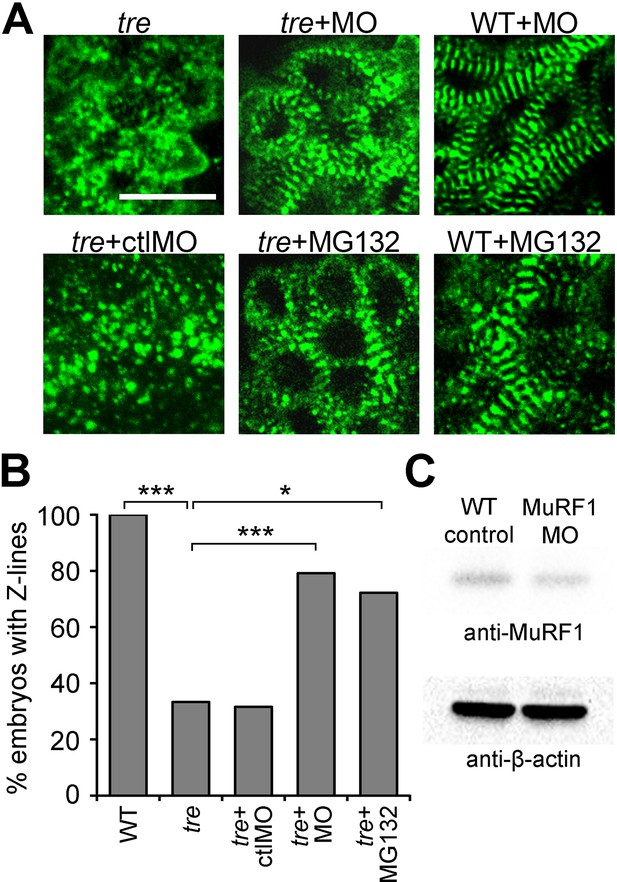
Blocking MuRF1-mediated proteasome degradation preserves myofibril integrity in tre/ncx1 deficient hearts.
(A) Z-lines were visualized by α-actinin staining. By 72 hpf, sarcomeres are disassembled in hearts of uninjected (tre) and control morpholino-injected (tre +ctlMO) tremblor embryos. Murf1a/murf1 b knockdown does not affect sarcomere integrity in wild type embryos (WT +MO), but prevents sarcomere degradation in tre (tre +MO). Similarly, treatment with a proteasome inhibitor, MG132, preserves myofibril integrity in tre cardiomyocytes (tre +MG132). Scale bar, 10 μm. (B) Graph shows % of embryos with periodic α-actinin staining at 72 hpf. (C) Western blot detecting MuRF1 and β-actin proteins in uninjected control (WT control) and murf1a/murf1 b knockdown (MuRF1 MO) embryos. Chi-squared test, *p<0.05; **p<0.01; ***p<0.001.
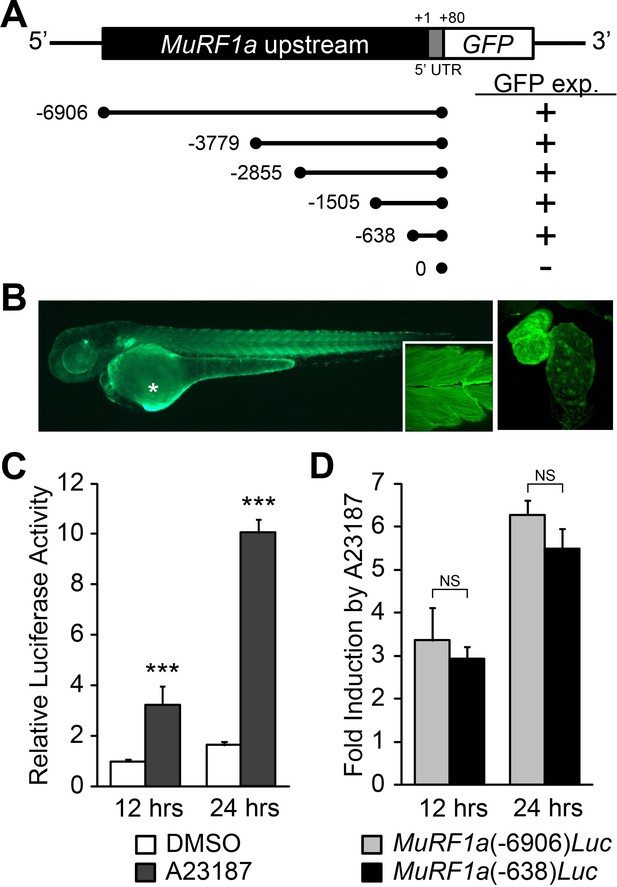
Identification of a MuRF1a regulatory element.
(A) Schematic representation of murf1a reporter constructs. + denotes the presence of GFP expression in the heart and somites. (B) A MuRF1a (−6906)-GFP transgenic embryo exhibits GFP expression in the heart and the somites (inset shows higher magnification image of the somites). The right panel shows a higher magnification image of the heart. The asterisk denotes auto-fluorescence from the yolk. (C) A23187 treatment induces luciferase activity driven by the MuRF1a (−6906) promoter. Values on the y-axis represent the luciferase activity relative to cells treated with DMSO for 12 hr. (D) Comparison of Ca2+ responsiveness between the MuRF1a (−638) and MuRF1a (−6906) promoters. Values on the y-axis represent the fold increase in luciferase activity in response to A23187 treatment compared to DMSO-treated cells at each time point. ***p<0.001; NS, not significant.
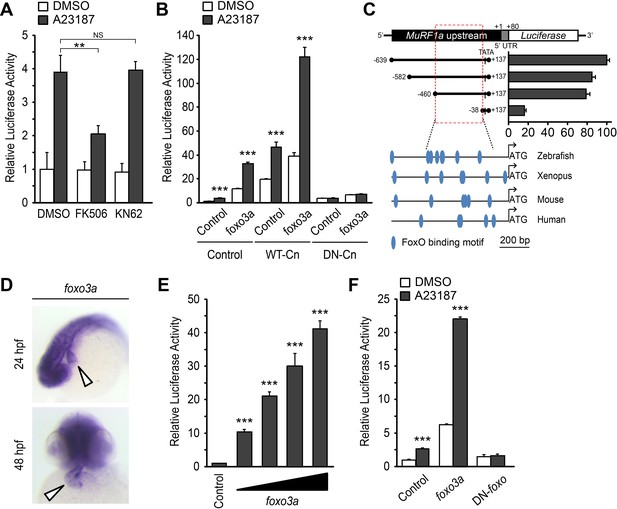
A Ca2+-Cn-FoxO signaling pathway regulates MuRF1 expression.
(A) HEK293T cells were transiently transfected with the MuRF1a (−638) luciferase reporter construct. Cells were incubated with FK506 or KN62 before DMSO or A23187 treatment. (B) Luciferase assay of the MuRF1a (−638) reporter cotransfected with foxo3a, wild type calcineurin (WT-Cn) or dominant-negative calcineurin (DN-Cn). (C) Diagram of the MuRF1a (−638) reporter serial deletion constructs generated for this study. The bar graph (right) shows the luciferase activity of each reporter construct relative to that of the empty expression plasmid. The red dotted box indicates the minimal cis-regulatory element of MuRF1a. The lower diagrams represent an alignment of the zebrafish, Xenopus, mouse and human MuRF1 promoters. Blue circles indicate putative FoxO binding sites (D) Whole-mount in situ hybridization detects foxo3a expression in the zebrafish heart. White arrowheads point to the heart. (E) HEK293T cells were transfected with the MuRF1a (−638) luciferase reporter and foxo3a expression plasmid. (F) HEK293T cells were transfected with the MuRF1a (−638) reporter plasmid and either a wild type or dominant negative foxo3a expression plasmid. Values on the y-axis are expressed relative to the luciferase activity of DMSO treated cells. **p<0.01; ***p<0.001; NS, not significant.
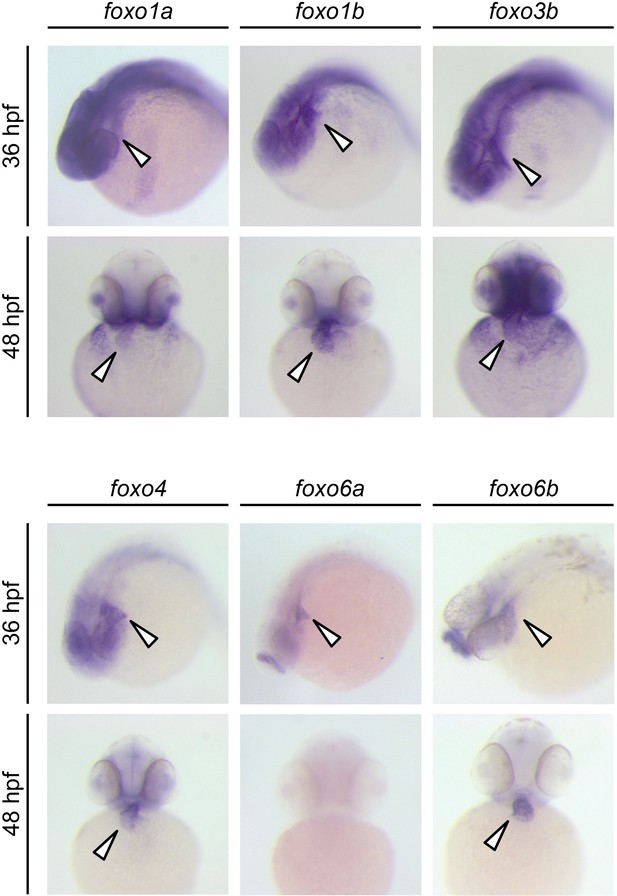
Expression patterns of zebrafish foxo genes.
Whole-mount in situ hybridization analysis showing foxo expression in zebrafish embryos. While foxo6a expression is diminished by 48 hpf, all the other foxo genes examined (foxo1a, 1b, 3b, 4, 6a and 6b) are persistently expressed in the heart. Arrowheads point to the heart.
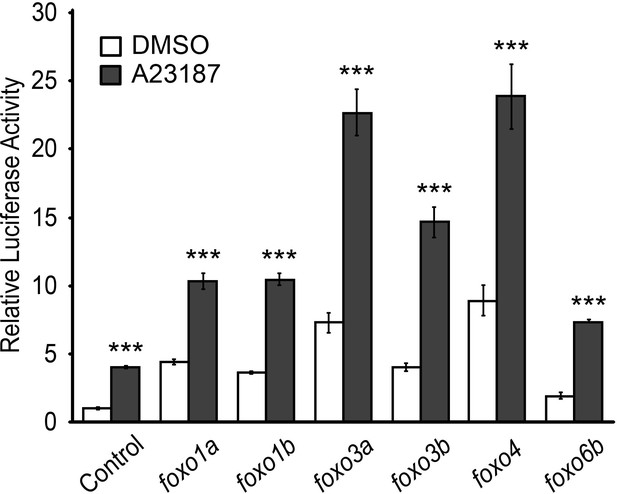
FoxO induces MuRF1 expression.
Luciferase activity of the MuRF1a (−638) reporter cotransfected with different foxo genes. Values on the y-axis are expressed relative to the luciferase activity of DMSO treated cells. ***p<0.001.
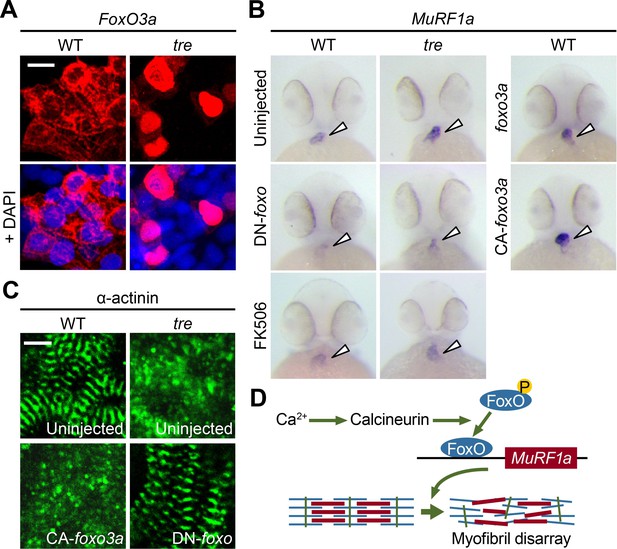
FoxO3a regulates MuRF1 expression in the heart.
(A) FoxO3a is predominantly localized in the cytoplasm of 2-day-old wild type cardiomyocytes, but is concentrated in the nuclei of tre cardiomyocytes. FoxO3a is pseudo colored in red and nuclei are labeled by DAPI in blue. Scale bar: 10 μm. (B) In situ hybridization showing stronger MuRF1 signals in tre mutant and CA-foxo3a injected two dpf hearts compared to wild type siblings. Murf1a expression in tre hearts is suppressed by DN-foxo overexpression or FK506 treatment. (C) Immunostaining of α-actinin in three dpf hearts. Intact sarcomeres were detected in control (uninjected) and DN-foxo3a injected tre hearts whereas disassembled sarcomeres were observed in tre and CA-foxo3a injected hearts. Scale bar: 5 μm. (D) Model for Ca2+ overload-induced myofibril disarray. Calcineurin dephosphorylates FoxO leading to FoxO nuclear translocation, MuRF1 expression and sarcomere disassembly.
Videos
Three-day-old myl7:EGFP transgenic heart.
https://doi.org/10.7554/eLife.27955.006Three-day-old myl7:MuRF1a-IRES-GFP transgenic heart.
https://doi.org/10.7554/eLife.27955.007Two-day-old ncx1h/murf1a/murf1b triple deficient heart.
https://doi.org/10.7554/eLife.27955.009Tables
Primers and morpholinos used in this manuscript
https://doi.org/10.7554/eLife.27955.015| Experiment | Target gene | Sequence |
|---|---|---|
| Quantitative RT-PCR | murf1a | F: 5'- GGAAGAAAACTGCCAGGCACAG −3' R: 5'- CTGGGTGATCTGCTCCAGAAGATG −3' |
| murf1b | F: 5'- CAGGACAATGCTCAACGTGCC −3' R: 5'- CTTGCTCTTTGCCAATACGCTCTAAGAG −3' | |
| Molecular cloning* | murf1a | F: 5'- CTGAGGTACCAAGCAGTGAAGGTTA −3' R: 5'- GCTAGGTACCAGTCTCTCATTGCTT −3' |
| murf1b | F: 5'- CTATGGATCCCTGCAGGGAATCATTTAC −3' R: 5'- GTTACTCGAGCATTTGTCAATGACCTTG −3' | |
| foxo1a | F: 5'- GTCTGAATTCCAGTATTGCTGGTACCATG −3' R: 5'- CATTGCTAGCACTACCCAGACACCCAG −3' | |
| foxo1b | F: 5'- GTATGGATCCTTGGTGATGGCAGAACC −3' R: 5'- GTATCTCGAGCAGCAGATGACATGTCTATC −3' | |
| foxo3a | F: 5'- GTATGGATCCGGAGTCGAGGAAATATGG −3' R: 5'- GTATCTCGAGCAGTTGCTTTACAGTGGAC −3' | |
| foxo3b | F: 5'- GTATGGATCCCGACCAAGACAGTAAAGAG −3' R: 5'- GTATCTCGAGCTGAGCAATTCCCATCAG −3' | |
| foxo4 | F: 5'- GTCTGAATTCCATCGCACAATGGAGG −3' R: 5'- CATTGCTAGCCAACAGTGGAGTTAGCT −3' | |
| foxo6a | F: 5'- ATGAGGATCCAACTCCATTAGACACAACC −3' R: 5'- GCTAGAATTCGTGTGATTGTTGAGGTCC −3' | |
| foxo6b | F: 5’- ATGAGGATCCCGGTTTCTTAAGCACAGAAG- 3’ R: 5’- ATGAGAATTCGACATTTATCCAGGCACC- 3’ | |
| MuRF1a(−6906) MuRF1a(−639) MuRF1a(−582) MuRF1a(−460) MuRF1a(−38) Common Reverse primer for MuRF1 | F: 5'- GTTAGCTAGCCGACTTACTCACTCC −3' F: 5'- GTACTTGGAGCGGCCGCAATAA −3' F: 5'- GTCAGCTAGCCCAACCCAGACAATATATTACT −3' F: 5'- GTCGGCTAGCGGGAAATAATAATATTGTGATTG −3' F: 5'- GACTGCTAGCCGGCTGGTATATAAGAC −3' R: 5'- GAATCTCGAGTGCTGAGGTAGAGTC −3' | |
| Morpholino | Control murf1a murf1b | 5’-CCTCTTACCTCAGTTACAATTTATA-3’ 5’-TTTGACCCGTTTGGATGTCCATTGC-3 5’-AAGAGGCAGTTCGCTGAATGTCCAT-3’ |
-
*Restriction enzyme sites are underlined.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.27955.016





