Key steps in unconventional secretion of fibroblast growth factor 2 reconstituted with purified components
Figures
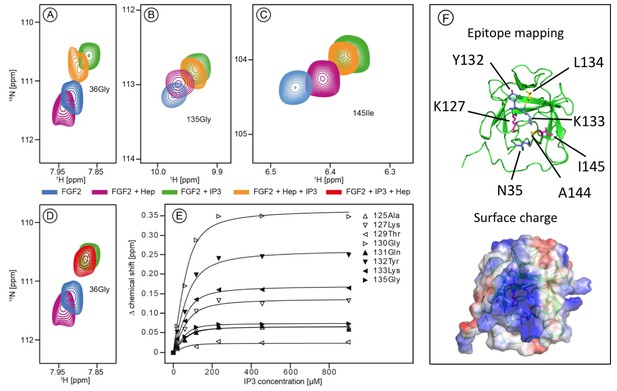
Structural analysis of the FGF2 binding epitopes for IP3 [head group of PI(4,5)P2] and heparin employing NMR spectroscopy.
Enlarged regions of exemplary resonance peaks from a two-dimensional 15N-1H correlation spectrum that are shifted upon addition of IP3 and Heparin or both are shown on panels (A to –D). In panel (E), titration curves for individual 15N-1H resonances are shown. Binding curves were fitted according to a simple two-state binding model and a KD was derived from the mean of the individual titration curves. HSQC for all NMR titration experiments are given in Figure 1—source data 1. In panel F (top), a cartoon of the FGF2 NMR structure is shown (PDB: 1BLD) with the side-chain of residues most significantly shifted upon IP3 binding (blue), heparin binding (yellow) or affected by both binding partners (magenta). In addition, surface mapping of residues shown in panel E using the known structure (PDB: 1BLD) of FGF2 is illustrated in the bottom part of panel (F). Both IP3 and heparin binding epitopes map to the same positively charged region of FGF2 highlighted in blue.
-
Figure 1—source data 1
Data for Figure 1, panels A-E.
This source file provides HSQC for all NMR titration experiments, of which examples are shown in Figure 1.
- https://doi.org/10.7554/eLife.28985.003
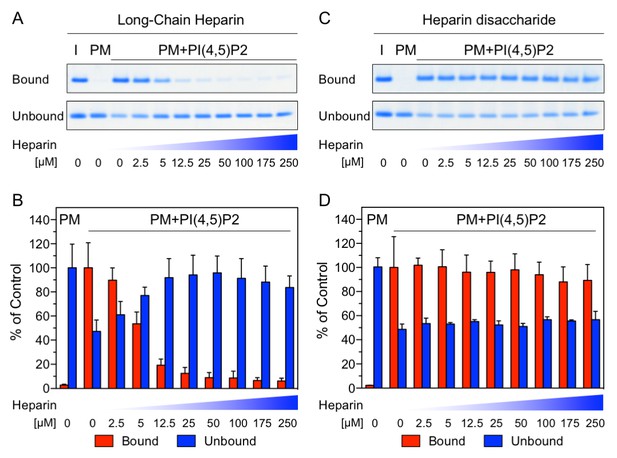
Binding of FGF2 to PI(4,5)P2 and heparin is mutually exclusive.
Biochemical analysis employing plasma-membrane-like liposomes either lacking (PM) or containing 2 mol% PI(4,5)P2 (PM +PIP2). Large unilamellar vesicles (LUVs) with bound His-FGF2-Y81pCMF-WT were incubated with increasing concentrations of either long-chain heparins or a defined heparin disaccharide as indicated. After 1 hr of incubation liposome-associated material (bound) was separated from supernatants (unbound). 50% of bound, 13.5% unbound and 14.8% of input material (I) were analyzed by SDS-PAGE. Coomassie-derived signals were quantified and normalized to controls. The fraction of FGF2-Y81pCMF-WT bound to PM-like liposomes containing PI(4,5)P2 in the absence of heparin was set to 100% (red bars in Figure 2B and D). The unbound fraction of FGF2-Y81pCMF-WT was normalized using PM-like liposomes lacking PI(4,5)P2 (blue bars in Figure 2B and D). Mean values with standard deviations are shown (n = 3). Raw and normalized data of individual experiments as well as calculations of mean values with standard deviations are shown in Figure 2—source data 1.
-
Figure 2—source data 1
Data for Figure 2, panels B and D.
This source file provides raw and normalized data, and calculations of mean values with standard deviations that are plotted in Figure 2, panels B and D.
- https://doi.org/10.7554/eLife.28985.005
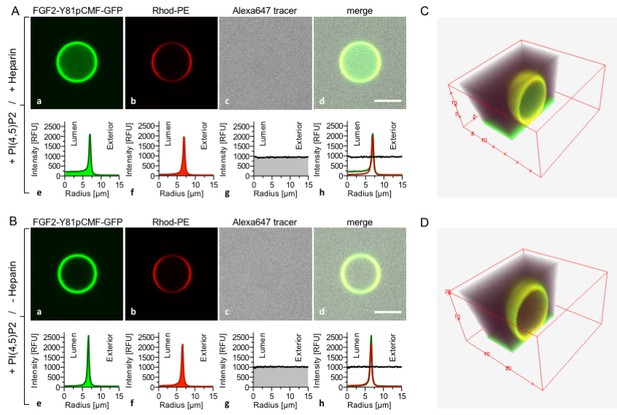
Reconstitution of FGF2 membrane translocation with purified components.
Giant unilamellar vesicles with a plasma membrane-like lipid composition containing PI(4,5)P2 were prepared in the presence (panel A and C) or absence (panel B and D) of long-chain heparins. Rhodamine-PE was incorporated into the lipid bilayer during GUV preparation as membrane marker. After removal of excess heparin by low speed centrifugation, GUVs were incubated with FGF2-Y81pCMF-GFP (200 nM) and a small fluorescent tracer (Alexa647). Following 180 min of incubation luminal penetration of GUVs by FGF2-Y81pCMF-GFP and small tracer molecules was analyzed by confocal microscopy (scale bar = 10 µm). GUVs were analyzed in all three channels using the plugin ‘Radial profile’ of the ImageJ software as explained under ‘Materials and methods’. Profile plots of normalized integrated intensities around concentric circles as a function of distance from the center of the GUV are given in relative fluorescence units (RFU). FGF2 membrane translocation is indicated by increased GFP fluorescence intensity in the lumen of GUVs compared to the exterior as exemplified in sub-panel e of panel A. The dependence of FGF2-Y81pCMF-GFP membrane translocation on luminal heparin is further documented by 3D reconstruction images (panels C and D) providing a spatial view into the GUV interior.
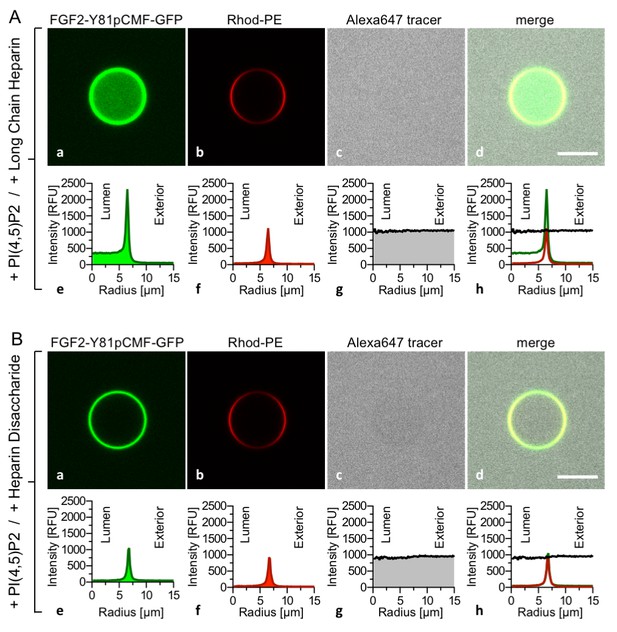
FGF2 membrane translocation depends on luminal long-chain heparins that cannot be substituted by low affinity heparin disaccharides.
Giant unilamellar vesicles with a plasma membrane-like lipid composition containing PI(4,5)P2 were prepared that contain either long-chain heparins (panel A) or a defined heparin disaccharide (panel B). Incubation with FGF2-Y81pCMF-GFP and data analysis were conducted as described in the legend to Figure 3 and under ‘Materials and methods’.
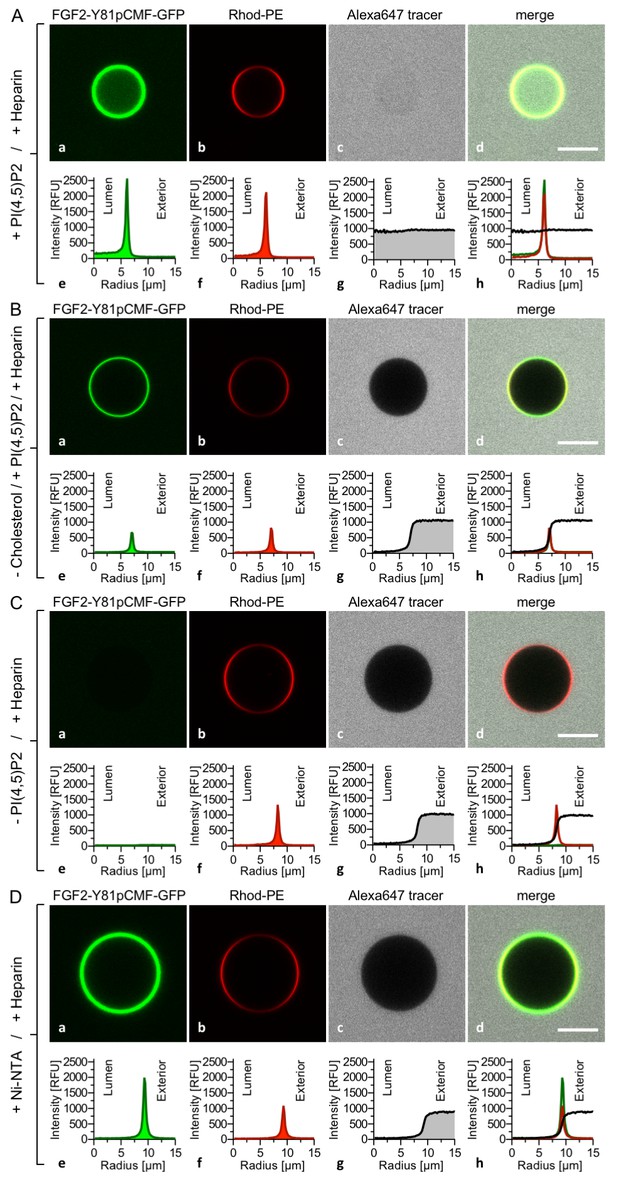
PI(4,5)P2 is required for both membrane pore formation and FGF2 membrane translocation.
Giant unilamellar vesicles were prepared with a plasma membrane-like lipid composition either containing PI(4,5)P2 (panel A), containing PI(4,5)P2 but lacking cholesterol (panel B), lacking PI(4,5)P2 (panel C) or containing a Ni-NTA lipid substituting PI(4,5)P2 (panel D). All four types of GUVs contained luminal long-chain heparins. Incubation with FGF2-Y81pCMF-GFP and data analysis were conducted as described in the legend to Figure 3 and under ‘Materials and methods’. Note increased GFP fluorescence in the lumen of GUVs as exemplified in sub-panel e of panel A indicating FGF2-Y81pCMF-GFP membrane translocation.
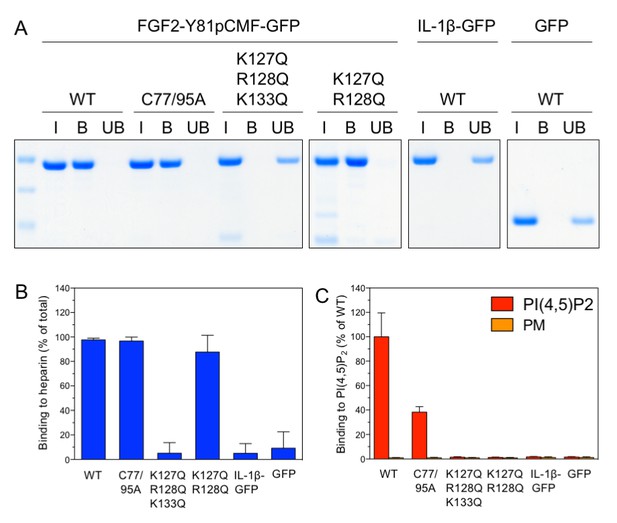
Analysis of FGF2 variant forms with differential defects in binding to PI(4,5)P2 and heparin.
The variant forms of FGF2-Y81pCMF-GFP indicated were tested for binding to heparin beads (panels A and B) and PI(4,5)P2 (panel C). Heparin sepharose beads were incubated with the FGF2 fusion proteins indicated. Bound and unbound material was separated by centrifugation. Bound proteins were eluted with SDS sample buffer and analysed by SDS-PAGE and Coomassie staining [5% input (I), 20% bound (B) and 5% unbound (UB)]. Signals were quantified using a Li-COR Odyssey infrared imaging system. Mean values with standard deviations of three independent experiments are shown (panel B). Raw and normalized data of individual experiments as well as calculations of mean values with standard deviations are shown in Figure 6—source data 1. Binding of the FGF2 fusion proteins to PI(4,5)P2 contained in plasma membrane-like liposomes was assessed using a flow-cytometry assay (Temmerman et al., 2008; Temmerman and Nickel, 2009) (panel C). Data were normalized by defining binding of FGF2-Y81pCMF-GFP to PI(4,5)P2 as 100% binding efficiency. Mean values with standard deviations are shown (n = 4). Consider Figure 6—source data 1 for more details.
-
Figure 6—source data 1
Data for Figure 6, panels B and C.
This source file provides raw and normalized data of SDS-PAGE and Coomassie analysis (Li-COR Odyssey infrared imaging system) with calculations of mean values with standard deviations that are plotted in Figure 6B. Furthermore, corrected fluorescent units of individual experiments (each experiment represents the mean of a duplicate) plus calculations of mean values with standard deviations of FACS-based experiments that are shown in Figure 6C are given.
- https://doi.org/10.7554/eLife.28985.010
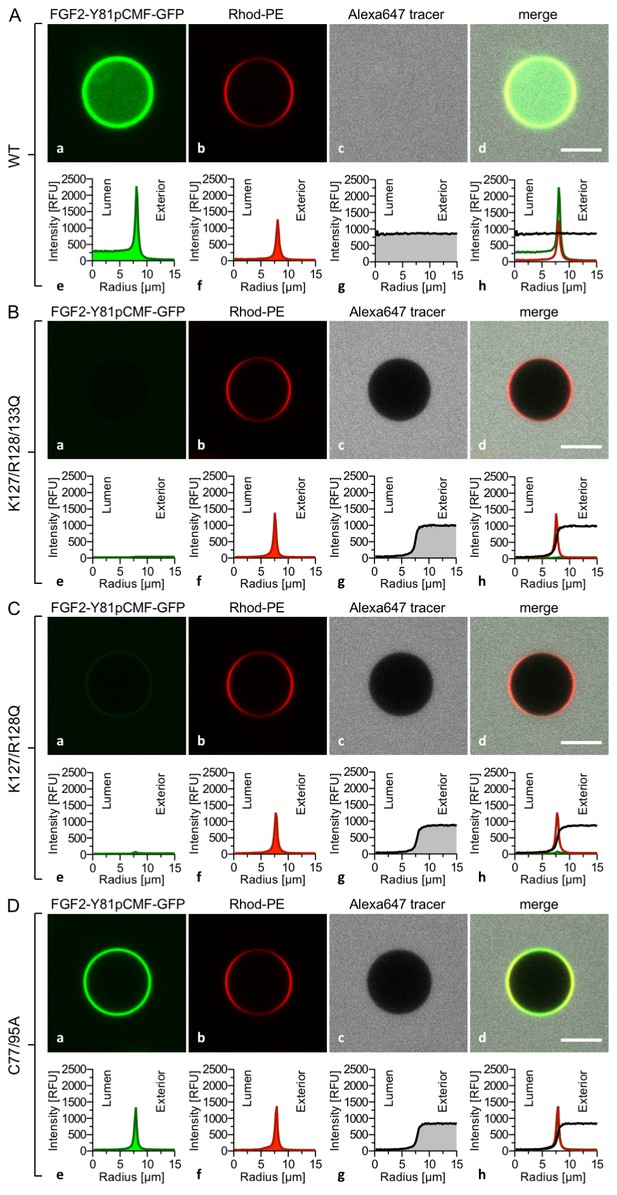
FGF2 membrane translocation depends on cis-elements mediating binding to PI(4,5)P2 and heparin as well as driving FGF2 oligomerization and membrane pore formation.
Giant unilamellar vesicles with a plasma membrane-like lipid composition containing both PI(4,5)P2 and luminal long-chain heparins were prepared as described in the legend to Figure 3 and under ‘Materials and methods’. GUVs were incubated with variant forms of FGF2-Y81pCMF-GFP as indicated. These included the wild-type form (panel A), the K127Q/R128Q/K133Q form deficient in binding to PI(4,5)P2 and heparin (panel B), the K127Q/R128Q form deficient in binding to PI(4,5)P2 (panel C) and the C77A/C95A form deficient in oligomerization and membrane pore formation (panel D). Incubation conditions and data analysis were conducted as described in the legend to Figure 3 and under ‘Materials and methods’. Note increased GFP fluorescence in the lumen of GUVs as exemplified in sub-panel e of panel A indicating membrane translocation of the wild-type form of FGF2-Y81pCMF-GFP.
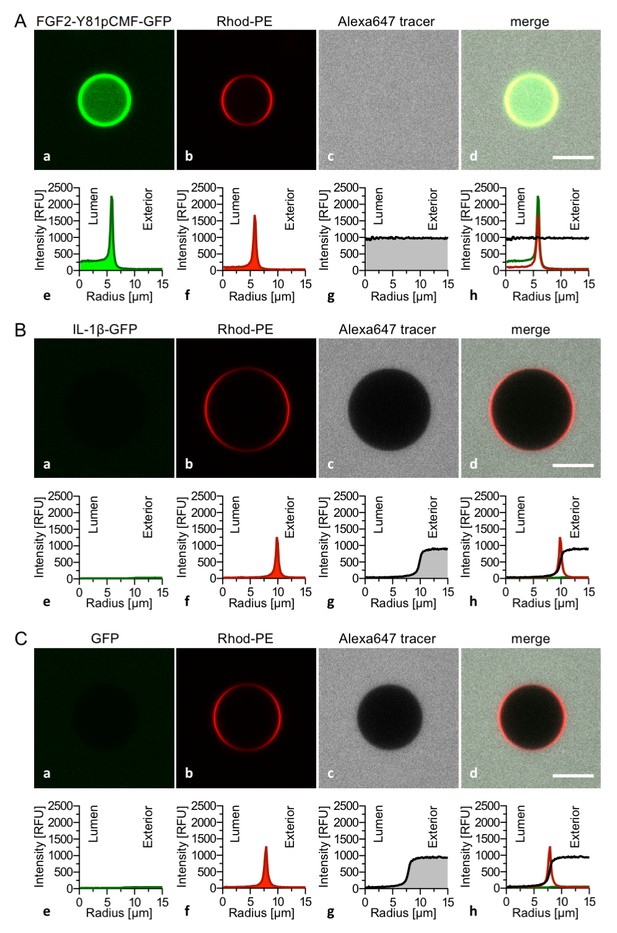
Interleukin 1β, a structural homologue of FGF2, is incapable of binding to PI(4,5)P2, membrane pore formation and membrane translocation.
Giant unilamellar vesicles with a plasma membrane-like lipid composition containing both PI(4,5)P2 and luminal long-chain heparins were prepared as described in the legend to Figure 3 and under ‘Materials and methods’. Incubation conditions using the wild-type form of FGF2-Y81pCMF-GFP (panel A), the mature form of Interleukin 1β-GFP (panel B) and GFP as control protein (panel C) as well as data analysis were conducted as described in the legend to Figure 3 and under ‘Materials and methods’. Note increased GFP fluorescence in the lumen of GUVs as exemplified in sub-panel e of panel A indicating membrane translocation of the wild-type form of FGF2-Y81pCMF-GFP.
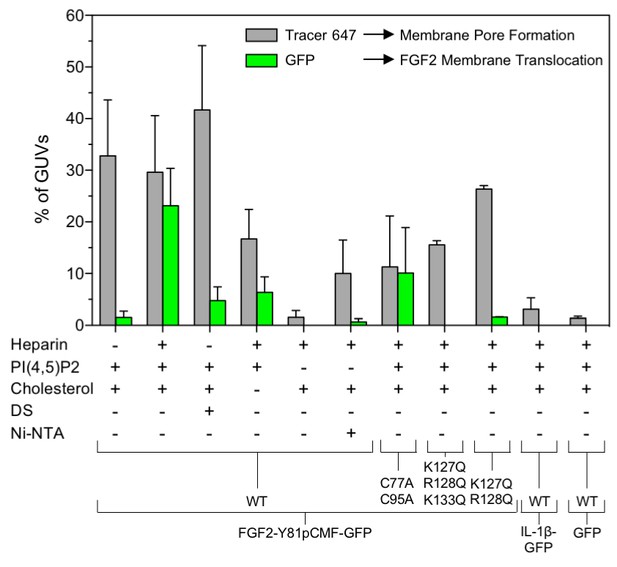
Quantification and statistical analysis of FGF2-Y81pCMF membrane translocation and its dependence on both cis-elements and trans-acting factors known to be required for FGF2 secretion from cells.
A quantitative analysis of membrane translocation and pore formation by the various proteins indicated was conducted based upon the experiments shown in Figures 3, 4, 5, 7 and 8. Various types of GUVs with a plasma membrane-like composition were used that differed with regard to the presence of the components indicated. For all conditions, data were derived from at least three independent experiments each of which involved the analysis of 20–120 GUVs per experimental condition. Gray bars indicate the percentage of GUVs with membrane pores with a ratio of Alexa647 tracer fluorescence in the lumen versus the exterior of ≥0.6. Green bars indicate the percentage of GUVs where membrane translocation of GFP-tagged proteins had occurred with a ratio of GFP fluorescence in the lumen versus the exterior of ≥1.6 being used as a threshold value. Standard deviations are shown (n ≥ 15 for experiments shown in Figure 3 and n ≥ 3 for all other conditions shown in Figures 4, 5, 7 and 8). Detailed information on each individual experiment is provided in Figure 9—source data 1.
-
Figure 9—source data 1
Data for Figure 9.
This source file provides additional information on each sample including number of experiments performed, number of GUVs analyzed in each single experiment, original counts and percentage of GUVs positive for the small tracer or FGF2 translocation, respectively, as well as calculations of mean values with standard deviations plotted in Figure 9.
- https://doi.org/10.7554/eLife.28985.014
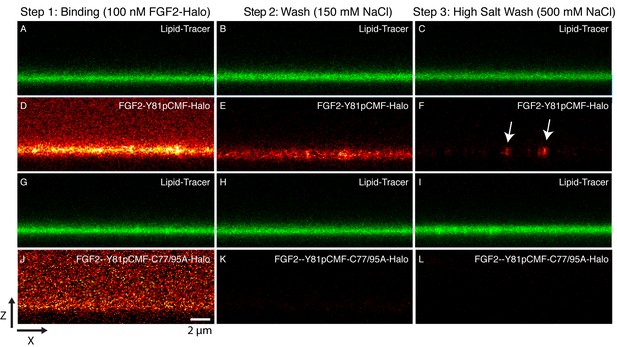
Binding and membrane insertion of FGF2-Halo-StarRed fusion proteins into supported lipid bilayers containing PI(4,5)P2.
FGF2-Y81pCMF-Halo-StarRed (Panels A–F) and FGF2-Y81pCMF-C77/95A-Halo-StarRed (panels G–L) were added at a final concentration of 100 nM to supported lipid bilayers (SLBs) containing 68 mol% POPC, 30 mol% cholesterol and 2 mol% PI(4,5)P2 plus trace amounts of DPPE-OregonGreen to image the bilayer. FGF2-Y81pCMF-Halo-StarRed and FGF2-Y81pCMF-C77/95A-Halo-StarRed were bound to SLBs (panels A, D, G and J) followed by a 150 mM NaCl washing procedure (panels B, E, H and K). In a final step, a 500 mM salt wash was applied to remove FGF2 monomers (panels C, F, I and L). FGF2-Y81pCMF-Halo-StarRed and FGF2-Y81pCMF-C77/95A-Halo-StarRed bound to SLBs were imaged as explained in ‘Materials and methods’.
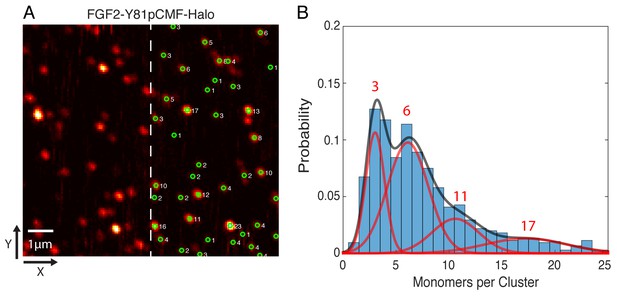
Single molecule imaging and brightness analysis of FGF2-Y81pCMF-Halo-StarRed to determine the oligomeric state of membrane inserted FGF2 clusters.
(A) Immobile FGF2-Y81pCMF-Halo-StarRed clusters associated with SLBs following a high salt wash (Figure 10F) were imaged by confocal microscopy. The brightness of individual clusters was determined by fitting a 2D Gaussian to each diffraction limited spot using a single molecule tracking software in MATLAB. The number of monomers in each cluster (right part of image) was estimated by normalizing the brightness of each cluster to the brightness of monomeric HALO-StarRed). Original peak intensities and cluster analyses are available in Figure 11—source data 1. (B) Gaussian mixture analysis of the oligomeric state of membrane inserted FGF2-Y81pCMF-Halo-StarRed (>1000 clusters from six independent experiments). The distribution of monomers per cluster was complex. A Gaussian mixture analysis found 4 components with 3, 6, 11 and 17 monomers per cluster.
-
Figure 11—source data 1
Data for Figure 11.
This source file provides peak intensities that were used to determine the number of monomers per cluster and the brightness of the StarRed monomer control (see ‘Material and methods’ for detail). In addition, original data of FRAP experiments mentioned in the text to Figure 11 are shown.
- https://doi.org/10.7554/eLife.28985.017
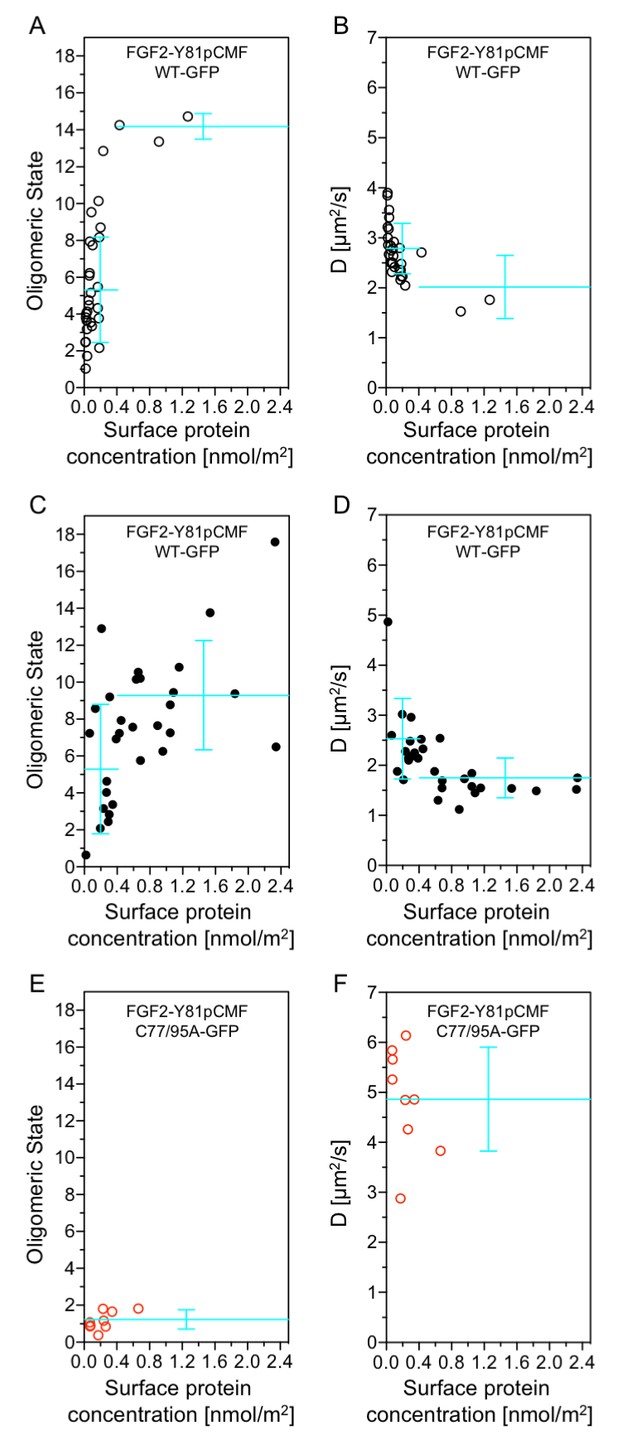
Functional correlation of the oligomeric state of membrane associated FGF2 translocation intermediates and membrane pore formation.
Giant unilamellar vesicles with a plasma membrane-like lipid composition containing PI(4,5)P2 and the membrane tracer DOPE-Atto633 were prepared as described in ‘Materials and methods’. After pre-incubation with either the wild-type form of FGF2-Y81pCMF-GFP (panels A-D, black circles) or FGF2-Y81pCMF-C77/95A-GFP (panels E and F; red circles) for at least 30 min, z-scan FCS measurements using 515/50 nm (FGF2-GFP) and 697/58 nm (DOPE-Atto633) emission channels were conducted on single GUVs. The small free tracer AlexaFlour532 was added to the buffer in order to visualize FGF2 membrane pore formation. Accordingly, GUVs were classified into two groups with (panels C and D, filled circles) and without membrane pores (panels A, B, E, and F; empty circles). Z-scan measurements and analyses are described in detail under ‘Materials and methods’. Average oligomeric state values (panels A, C, E) and diffusion constants (panels B, D, F) were plotted as a function of protein surface concentration. A total of 60 individual GUVs incubated with FGF2-Y81pCMF-WT-GFP (panels A-–D) and 9 GUVs incubated with FGF2-Y81pCMF-C77/95A (panels E and F) were analyzed. Additional data of Z-scan FCS for each individual GUV, monomer control reference measurements as well as calculations of mean values with standard deviations are provided in Figure 12—source data 1.
-
Figure 12—source data 1
Data for Figure 12.
This source file provides extensive information of each individual GUV analyzed by Z-scan FCS including pore formation state, particle number, brightness of a cluster, oligomeric state, two independent determinations of surface protein concentration, diffusion times, and diffusion coefficients. Furthermore, reference measurements of monomer units (see ‘Material and methods’ for details) as well as calculations of mean values with standard deviations that are plotted in Figure 12 are given.
- https://doi.org/10.7554/eLife.28985.019
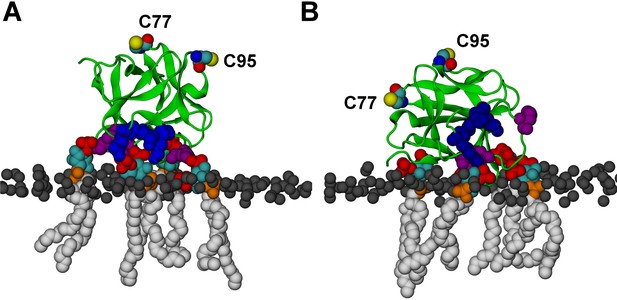
FGF2 orientation on the membrane surface.
(A) High-affinity orientation of FGF2, showing all the known PI(4,5)P2-binding site residues (K127, R128, K133) as well as the previously undetermined binding site residues (K34, K137, K143). (B) Low-affinity orientation of FGF2 in which the binding site residues lose contact with PI(4,5)P2 and point away from the PI(4,5)P2 head groups. FGF2 is rendered as green cartoon, and its C95 and C77 residues are shown as van der Waals (vdW) spheres and highlighted by text in the figure. The key binding pocket residues (K127, R128, K133) are shown as blue vdW spheres, and the additional binding site residues (K34, K137, K143) are shown as purple vdW spheres. Lipids are colored as gray vdW spheres (POPC phosphate atoms), red vdW spheres [PI(4,5)P2 bisphosphates], cyan vdW spheres [inositol ring in PI(4,5)P2], orange vdW spheres [phosphate linking the fatty acid chains and the inositol ring in PI(4,5)P2], and white vdW spheres [fatty acid chains in PI(4,5)P2]. Water molecules and ions are not shown for clarity.
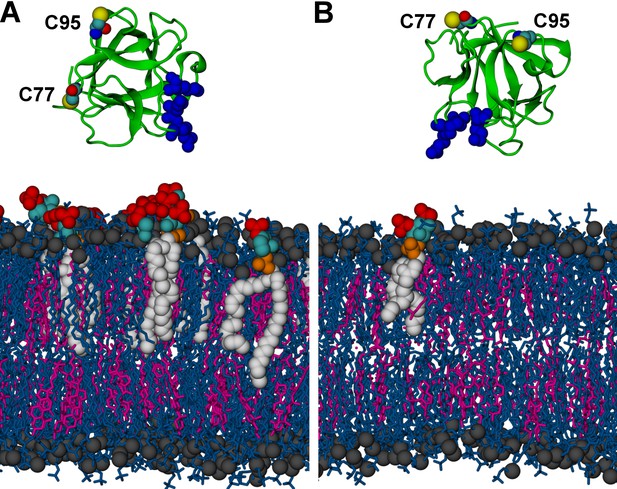
Initial structures in systems (A) M1 (POPC/cholesterol/PI(4,5)P2 (65/29.5/5.5 composition on the cytosolic side interacting with FGF2); see text and Table 1) and (B) M2 (a single PI(4,5)P2 molecule allowed to interact with FGF2).
FGF2 is rendered as green cartoon, C95 and C77 residues are shown as van der Waals (vdW) spheres, and binding pocket residues (K127, R128, K133) are shown as blue van der Waals spheres. Lipids are colored as gray vdW spheres (POPC phosphate atoms), blue licorice (rest of POPC), magenta (cholesterol), red vdW spheres [PI(4,5)P2 bisphosphates], cyan vdW spheres [inositol ring in PI(4,5)P2], orange vdW spheres [phosphate linking the fatty acid chains and the inositol ring in PI(4,5)P2], and white vdW spheres [fatty acid chains in PI(4,5)P2]. Water molecules and ions are not shown for clarity.
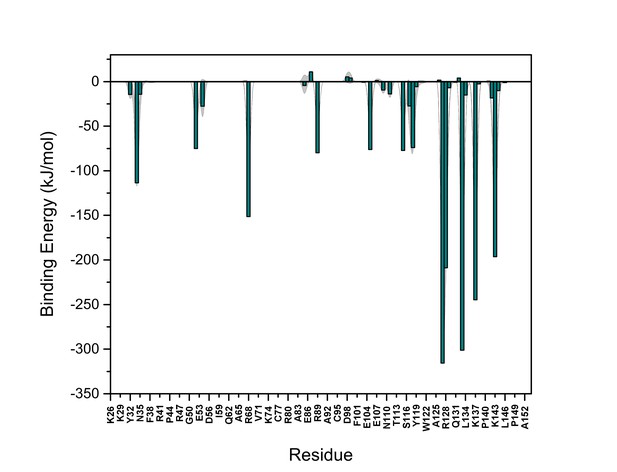
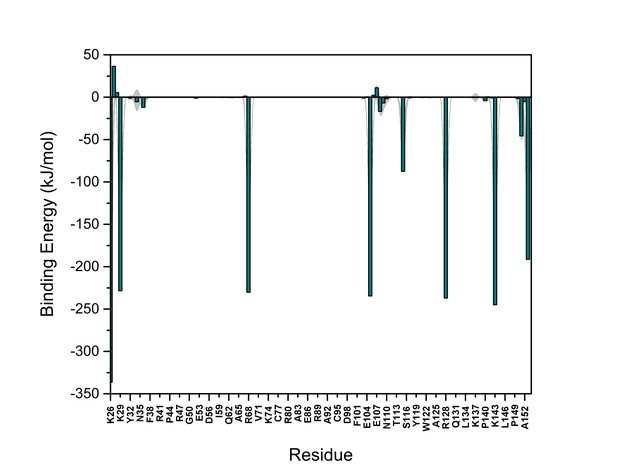
PI(4,5)P2 binding energy based on electrostatics and van der Waals interactions.
The average interaction energy (electrostatic and van der Waals contributions) of PI(4,5)P2 in system M1 over the residues of FGF2 in the high-affinity orientation binding to PI(4,5)P2. The standard errors are shown as shadows.

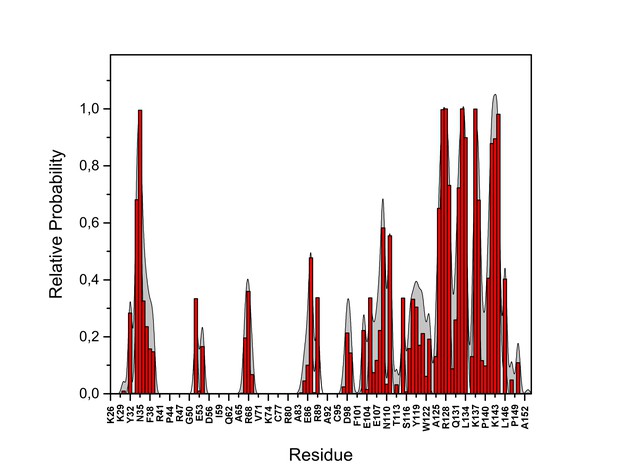
PI(4,5)P2 interaction based on hydrogen bond analysis.
Relative probability of hydrogen bonds formed between the residues of FGF2 in the high-affinity orientation and PI(4,5)P2 (system M1). The standard errors are shown as shadows.
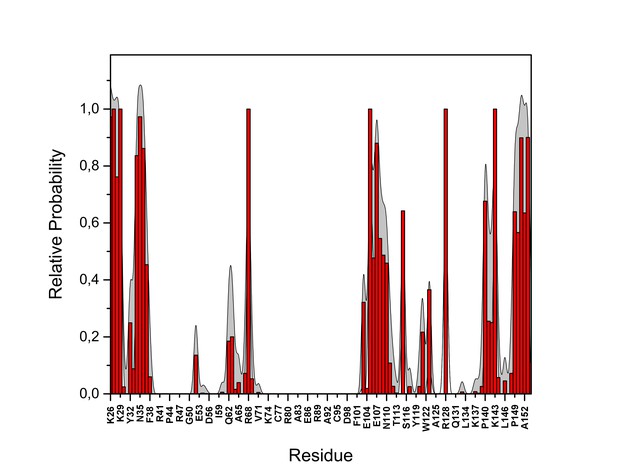
PI(4,5)P2 contacts with FGF2.
Relative probability of contacts (<0.6 nm) between the FGF2 residues and PI(4,5)P2 in the high-affinity orientation (system M1). The standard errors are shown as shadows.
PI(4,5)P2 binding energy based on electrostatics and van der Waals interactions.
The average interaction energy (electrostatic and van der Waals contributions) of PI(4,5)P2 in system M1 over the residues of FGF2 in the low-affinity orientation binding to PI(4,5)P2. The standard errors are shown as shadows.

PI(4,5)P2 interaction based on hydrogen bond analysis.
Relative probability of hydrogen bonds formed between the residues of FGF2 in the low-affinity orientation and PI(4,5)P2 (system M1). The standard errors are shown as shadows.
PI(4,5)P2 contacts with FGF2.
Relative probability of contacts (<0.6 nm) between the FGF2 residues and PI(4,5)P2 in the low-affinity orientation (system M1). The standard errors are shown as shadows.
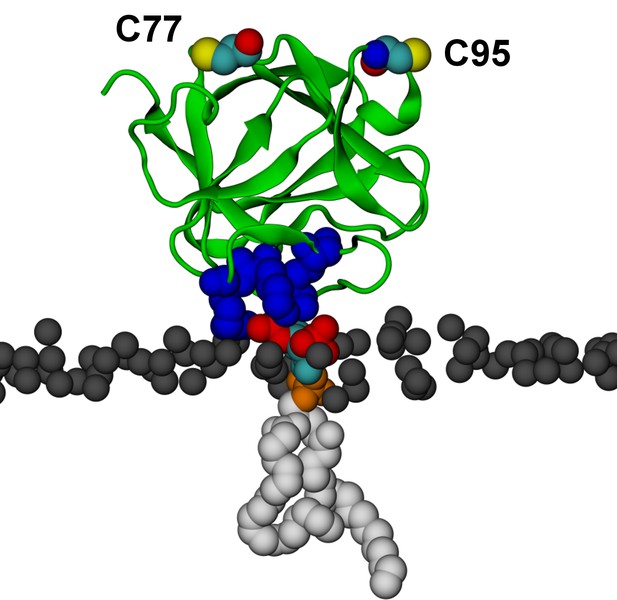
Binding of FGF2 to a single PI(4,5)P2.
Binding of FGF2 to a single PI(4,5)P2 molecule (system M2) with its known binding site residues (K127, R128, K133) colored as blue van der Waals spheres. During the simulation the orientation of FGF2 in respect to the membrane fluctuated between the high-affinity (shown here) and low-affinity orientations. Color coding as in Figure 13.
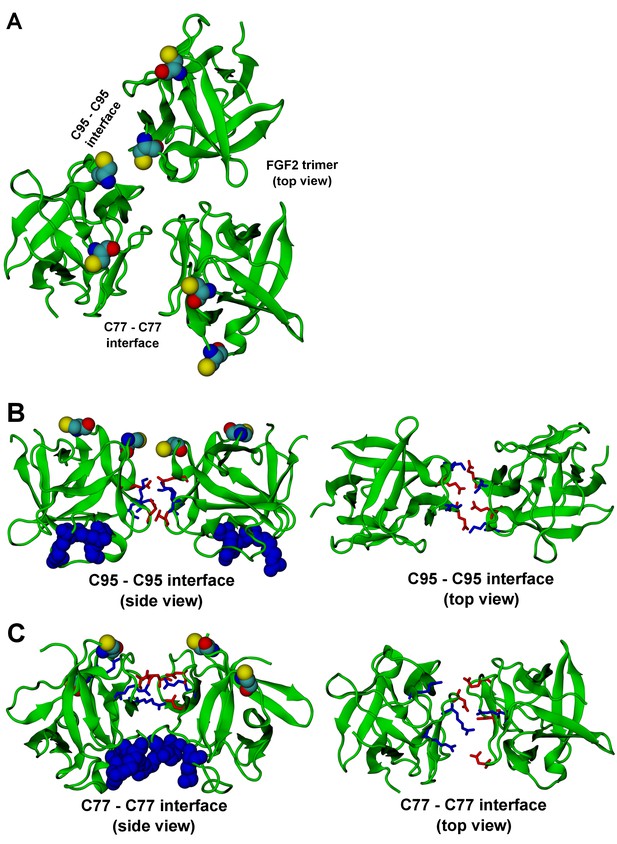
FGF2 trimer configurations.
Snapshots representing the most populated structures in FGF2 trimer simulations (system T). (Panel A) depicts the top view of the FGF2 trimer aggregate with C95 – C95 and C77 – C77 interfaces labeled, where C95 and C77 are colored as van der Waals spheres. The trimer is split into two dimer interfaces shown in (panels B and C). (B) The interface residues involved in C95 – C95 disulfide-linked dimers. (C) The interface residues involved in C77 – C77 disulfide-linked dimers. The interface residues are depicted in stick representation, where negatively charged residues (D, E) are colored as red and positively charged residues (K, R) as blue. The PI(4,5)P2 binding pocket residues (K127, R128, K133) are rendered as blue van der Waals spheres. For clarity, POPC, PI(4,5)P2, cholesterol, water, and ions are not shown.
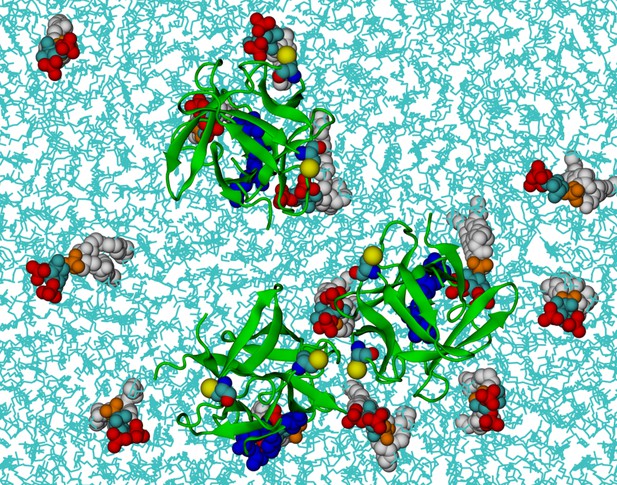
FGF2 trimer in the beginning of the simulations.
Top view of the initial positions of three FGF2 monomers on the membrane surface in a simulation of FGF2 trimers (system T). POPC and cholesterol molecules are colored as cyan lines. The color coding of the proteins and PI(4,5)P2 is as in Figure 13.
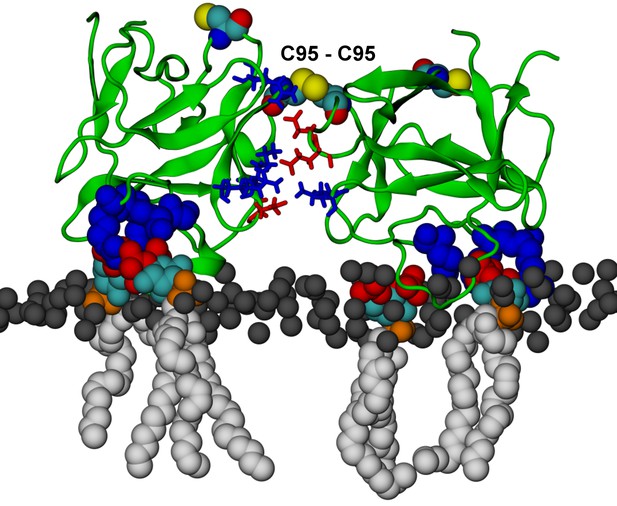
FGF2 dimer simulation.
FGF2 dimer with a covalently linked C95 – C95 bridge (system D). C95 and C77 are shown as van der Waals spheres. The interface residues involved in ion pairs are shown in stick representation, where negatively charged residues (D, E) are colored as red and positively charged residues (K, R) as blue. The PI(4,5)P2 binding pocket residues (K127, R128, K133) are depicted as blue van der Waals spheres. The rest of the protein and membrane is colored as in Figure 13.
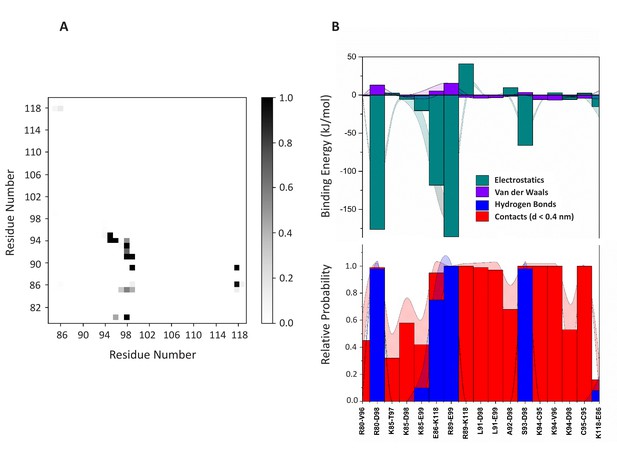
Dimerization interface.
(A) Contact map analysis for the dimer (system D). (B) Based on the contact map, 17 residue pairs (distance <0.4 nm) were identified for which electrostatic and van der Waals interactions (panel B (top)) were computed. The hydrogen bonds and contacts are shown in panel B (bottom). The standard error is shown as shadows.
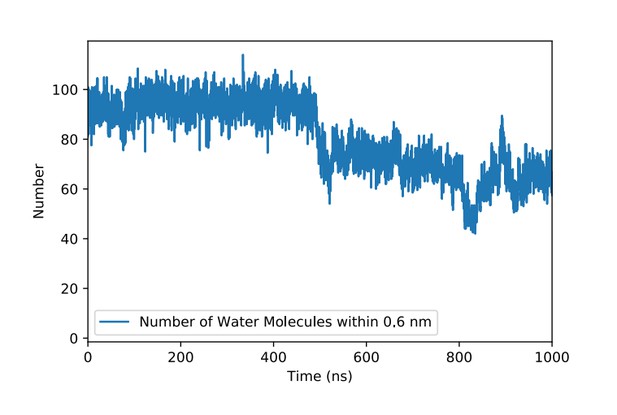
Number of water molecules in the vicinity of the PI(4,5)P2 binding pocket in FGF2 in the course of time.
The PI(4,5)P2 head group binds to the binding pocket at about 500 ns. Here, a water molecule is considered to be in the vicinity of the binding pocket if its distance from the main residues (K127, R128, K133) is 0.6 nm or less.
Videos
FGF2 binding simultaneously to several PI(4,5)P2 molecules (system M1).
FGF2 binds to the membrane surface in the high-affinity orientation and interacts simultaneously with multiple PI(4,5)P2, including the key residues in the main binding pocket (K127, R128, K133) and the residues in its vicinity (K34, K137, K143). The final frame of the video at 913 ns matches the snapshot in Figure 13A taken though from a slightly different perspective. The color coding used for the protein and PI(4,5)P2 is consistent with Figure 13. In the membrane, cholesterols are shown in light purple, the phosphorous atoms of POPC molecules are depicted as a transparent surface, and for clarity’s sake water and ions are not shown.
FGF2 binding to a single PI(4,5)P2 (system M2).
FGF2 adhering to the membrane surface through binding to a single PI(4,5)P2. In the binding process, FGF2 first interacts with the residues close to the main binding site (K34, K137, K143, shown in purple van der Waals spheres), and then, at about 500 ns, rotates itself and interacts with the residues in the binding pocket (K127, R128, K133, shown in blue van der Waals spheres). The color coding used for the protein and PI(4,5)P2 is consistent with Figure 13. In the membrane, cholesterols are shown in light purple, the phosphorous atoms of POPC molecules are depicted as a transparent surface, and for clarity’s sake water and ions are not shown.
Legends for source data
Tables
Descriptions of model systems and their simulation details.
| System | No. of FGF2 molecules | No. of POPC molecules | No. of CHOL molecules | No. of PI(4,5)P2 molecules | No. of water molecules | Distance from membrane surface (nm) | No. of repeats | Simulation time (ns) |
|---|---|---|---|---|---|---|---|---|
| M1 | 1 | 348 | 152 | 14 | 34783 | 1.5 | 5 | 1000 × 5 |
| M2 | 1 | 177 | 76 | 1 | 15684 | 1.5 | 1 | 1000 |
| T | 3 | 348 | 152 | 14 | 31575 | 0.5 | 2 | 1000 × 2 |
| D | 2 | 348 | 152 | 14 | 29770 | * | 1 | 130 |
-
*The initial FGF2 dimer orientation and position on the membrane surface were the same as those in the final frame of the system T (repeat 2).





