The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity
Figures
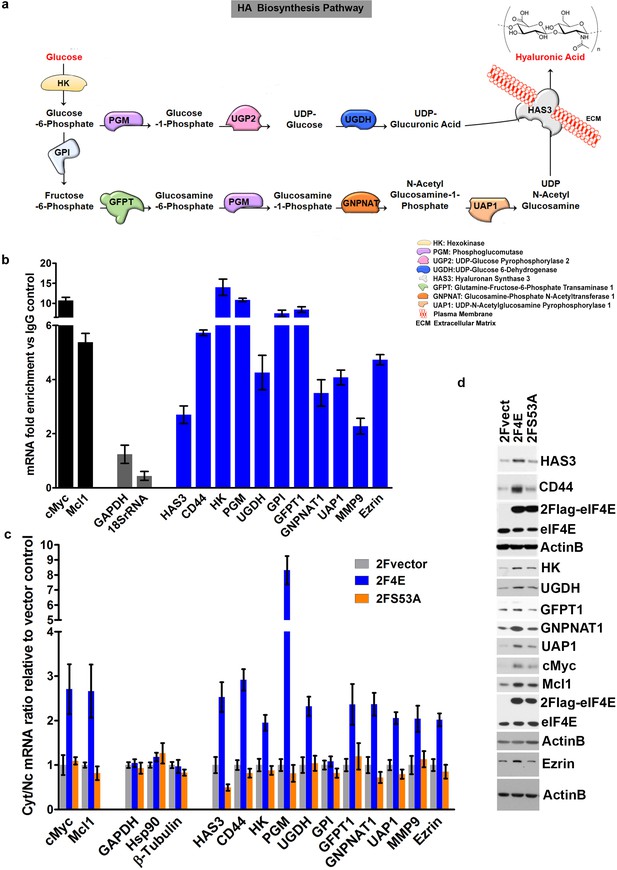
eIF4E regulates the expression of HA synthesizing enzymes and HA receptor CD44.
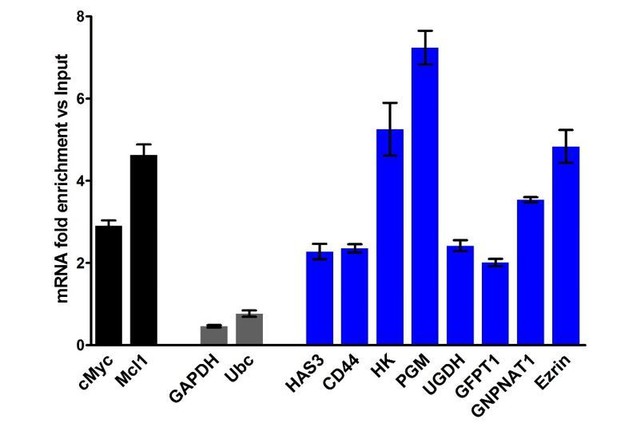
(A) HA biosynthesis pathway. (B) RT-qPCR of HA synthesizing enzymes and its receptor CD44 following RNA immunoprecipitation (RIP) using anti-eIF4E antibody from nuclei of U2Os 2Flag-eIF4E cells. Data are normalized to IgG control and presented as fold change. c-Myc and Mcl-1 are positive targets of eIF4E and thus serve as positive controls, whereas GAPDH and 18S rRNA served as negative controls. We used standard deviation to denote statistical significance. One representative experiment is shown, which was carried out in triplicate. (C) RT-qPCR of HA synthesizing enzymes in cytoplasmic versus nuclear fractions of U2Os cells overexpressing 2Flag-eIF4E (2F4E), S53A mutant (2FS53A) or vector control (2Fvect). Data are normalized to vector control. c-Myc and Mcl-1 served as known eIF4E targets, whereas GAPDH, Hsp90 and β-Tubulin served as negative controls. One representative experiment is shown, which was carried out in triplicate. (D) Western blot of HA synthesizing enzymes and CD44 as a function of eIF4E or S53A mutant overexpression. Mcl-1 served as positive eIF4E target control. Actin was used as a loading control. Each Actin blot corresponds to the western blots above it. Both 2Flag-eIF4E and endogenous eIF4E are shown. HK: Hexokinase; HAS3: Hyaluronan Synthase 3; PGM5: Phosphoglucomutase 5; UGP2: UDP glucose pyrophosphorylase; UGDH: UDP glucose dehydrogenase; GFPT1: Glutamine fructose 6 phospho transaminase; GNPNAT1: Glucosamine phosphate N-acetyltransferase; UAP1: UDP N-acetyl pyrophosphorylase; GPI: Glucose-6-phosphate isomerase; CD44: HA receptor; MMP9: Matrix Metalloproteinase 9. For bar graphs, the mean ± standard deviation are shown. Experiments were carried out in triplicate, at least three independent times.
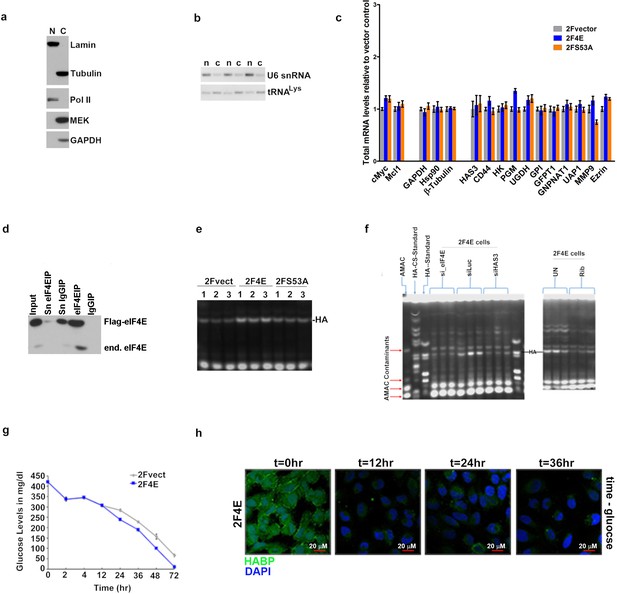
eIF4E regulates HA synthesis.
(A) Control Western blot for cellular fractionation corresponding to eIF4E RIP shown in Figure 1B. Lamin and Pol II are used as nuclear markers while GAPDH, β-Tubulin and MEK are used as cytoplasmic markers. (B) Semi-qPCR for tRNAlys and U6 snRNA as control for the cytoplasmic and nuclear fractions, respectively, corresponding to the export assay shown in Figure 1C. (C) RT-qPCR of total mRNA levels corresponding to mRNA export assay shown in Figure 1C. One representative experiment is shown that was carried out in triplicate. (D) Control western blot of eIF4E recovered in immune complexes with eIF4E antibodies and IgG relative to input amount of eIF4E is shown for the RIP data presented in Figure 1b. (E–F) Fluorophore-assisted carbohydrate electrophoresis (FACE) gels corresponding to bar graph presented in Figure 2c. (G) Glucose levels were measured in mg/dl using Clarity Plus Blood Glucose Monitoring Kit in Vector or eIF4E U2Os cells as a function of time (in hours). Experiments were carried out in triplicate, at least three independent times. Representative experiment is shown. (H) Fluorescence staining of HA (in green) of U2Os cells overexpressing eIF4E collected at different time points following media change (to DMEM containing 2 g/l glucose). DAPI is in blue. A × 40 objective with no digital zoom used. Experiments were carried out in triplicate, at least three independent times. For bar graphs, the mean ±standard deviation are shown.
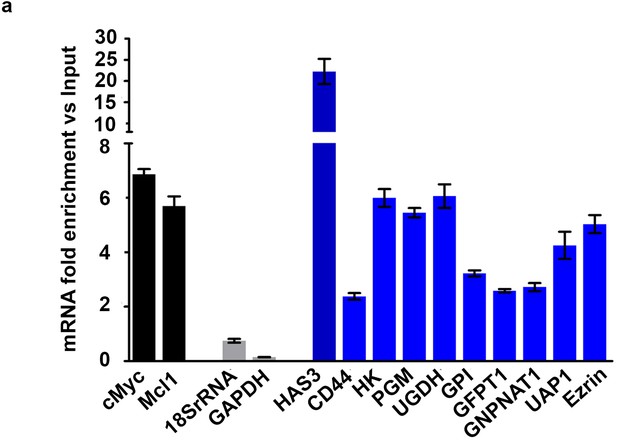
eIF4E regulates HA synthesis.
(A) RT-qPCR of HA synthesizing enzymes and its receptor CD44 following RNA immunoprecipitation of nuclear eIF4E in U2Os 2Flag-eIF4E cells using anti-eIF4E antibody. Data are normalized to nuclear input control and presented as fold change. c-Myc and Mcl-1 are targets of eIF4E and thus serve as positive controls while GAPDH and 18S rRNA serve as negative controls. Experiments were carried out in triplicate, at least three independent times. The mean ±standard deviation is shown.
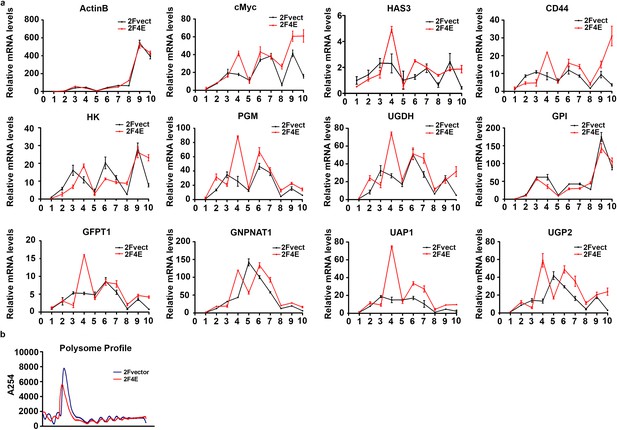
eIF4E regulates HA synthesis.
(A) Polysomal profiling of transcripts of HA synthesizing enzymes and its CD44 in U2Os 2Flag-eIF4E (red line) vs 2Flag-vector (black line) cells. Fractions were normalized to corresponding input RNAs, and data are expressed as fold of mRNA to monosomes. (B) Total polysomal profiling of U2Os 2Flag-vector and 2Flag-eIF4E cells. Experiments were carried out two independent times.
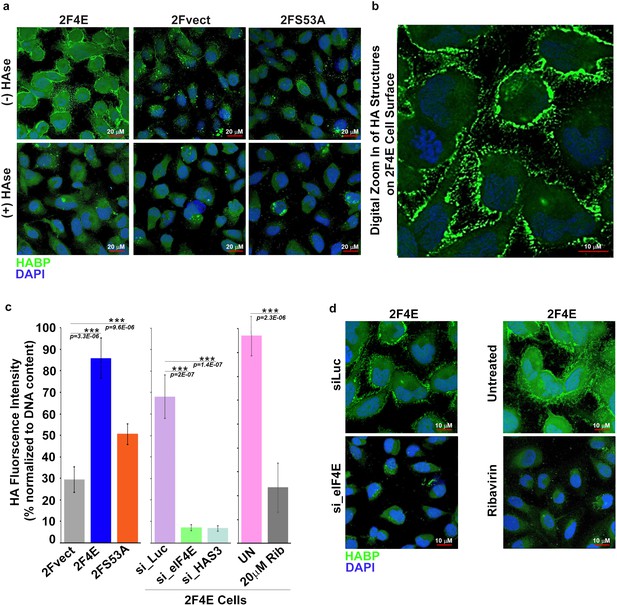
eIF4E overexpression correlates with increased HA synthesis.
(A) Fluorescence staining of HA (in green) using biotinylated HA-binding protein with streptavidin-FITC in U2Os cells overexpressing eIF4E, S53A mutant or vector control in the presence or absence of Streptomyces Hyaluronidase treatment. DAPI is in blue. Note cell surface expression of HA in response to eIF4E overexpression. All confocal settings are identical between specimens and thus lower signal is indicative of less HA. A × 40 objective with no digital zoom was used. (B) 2x digital zoom in confocal images of HA from part (A). (C) Quantification of fluorophore-assisted carbohydrate electrophoresis (FACE) gels (Sup Figure 1e&f) for HA levels in U2Os cells expressing eIF4E, S53A mutant or vector control, and U2Os cells overexpressing eIF4E following HAS3/eIF4E knockdown or pharmacological inhibition with ribavirin (Rib). (D) Fluorescence staining of HA (in green) following siRNA to eIF4E or ribavirin treatment in U2Os cells overexpressing eIF4E. DAPI is in blue. A × 63 objective with no digital zoom used. For bar graphs, the mean ± SD are shown. Experiments were carried out in triplicate, at least three independent times. **p < 0.01, ***p < 0.001 (Student’s t-test).
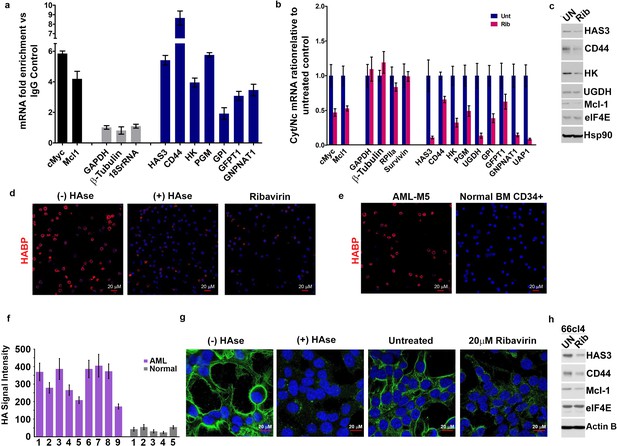
eIF4E elevates HA in cancer cell lines and primary specimens.
(A) RT-qPCR of HA synthesizing enzymes and its receptor CD44 following RNA immunoprecipitation (RIP) from nuclei of MM-6 cells using anti-eIF4E antibody. Data are presented as fold change relative to IgG controls. c-Myc and Mcl-1 served as endogenous eIF4E targets, while GAPDH, β-Tubulin and 18SrRNA served as a negative control. (B) RT-qPCR of HA synthesizing enzymes in cytoplasmic versus nuclear fractions of MM-6 cells treated with Ribavirin (Rib). Data are normalized to untreated control. Error bars are means ± S.D. c-Myc and Mcl-1 served as positive controls since they are known eIF4E targets, while GAPDH, β-Tubulin, RPIIa and Survivin served as negative controls. (C) Western blot of HA synthesizing enzymes and CD44 as a function of Ribavirin treatment in MM-6 cell line. Mcl-1 served as endogenous eIF4E target, while Hsp90 served as a loading control. (D) Fluorescence staining of HA (in red) in MM-6 cell lines treated with Ribavirin (Rib) versus untreated (UN) in the presence or absence of Hyaluronidase treatment. DAPI is in blue. A 63x objective with no digital zoom was used. (E) Fluorescence staining of HA (in red) in human CD34+ specimens from healthy volunteer compared with leukemic cells from M5 AML Patient. (F) Quantification of HA fluorescence staining in 9 M4/M5 AML patients and CD34+ specimens from five healthy volunteers using ZEN software. HA signal intensity is presented as the geometric means of the HA signal. The mean ± standard deviations are shown. (G) Fluorescence staining of HA (in green) in 66cl4 cells in the presence or absence of Hyaluronidase or Ribavirin treatment. DAPI is in blue. A × 63 objective with no digital zoom used. (H) Western blot control of HAS3 and CD44 as a function of ribavirin treatment in 66cl4 cell line. Mcl-1 served as endogenous eIF4E target, while ActinB served as loading control. Experiments were carried out in triplicate, at least three independent times. For bar graphs, the mean ±standard deviation are shown.
eIF4E elevates HA in cancer cell lines and primary specimens.
(A) Control western blot for cellular fractionation corresponding to eIF4E RIP. Lamin and Pol II are used as nuclear markers while Raf1 is used as cytoplasmic marker. (B) Control western blot of eIF4E recovered in immune complexes with eIF4E antibodies and IgG relative to input amount of eIF4E corresponding for RIP in MM6 cells in Figure 3. (C) RT-qPCR of total mRNA levels corresponding to mRNA export assay in untreated cells (UN) or Ribavirin treated (Rib). (D) Semi-qPCR for tRNAlys and U6 snRNA served as controls for the cytoplasmic and nuclear fractions, respectively, for Figure 3.
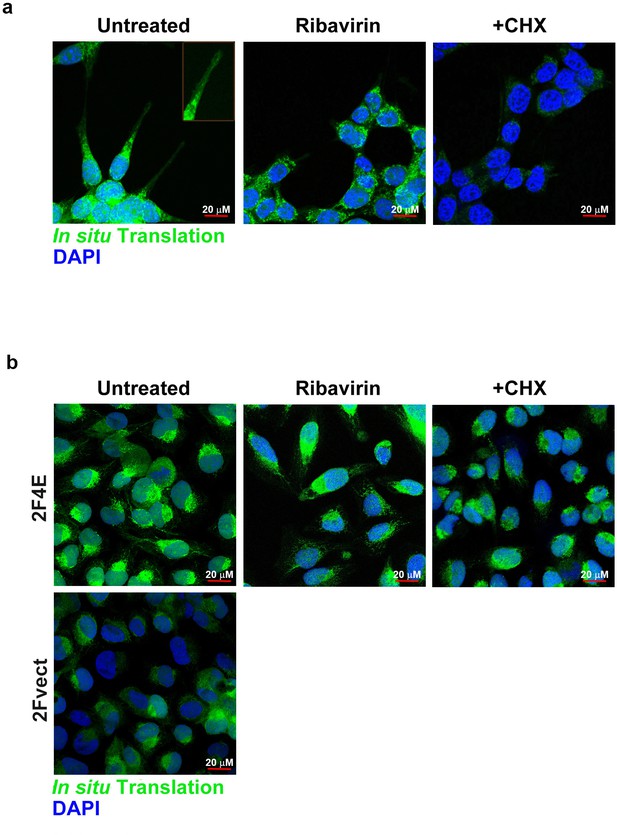
eIF4E concentrates in HA rich protrusions and correlates with sites of active translation.
(A–B) Fluorescence staining of Click_iT HPG Alexa Flour 488 in 66cl4 or U2Os cells overexpressing eIF4E cells (2F4E), respectively, following treatment with 20 µM Ribavirin versus untreated control. Cyclohexamide treatment (+CHX) is used as a negative control. A x40 or 63 objectives is used. Experiments were carried out in triplicate, at least three independent times.
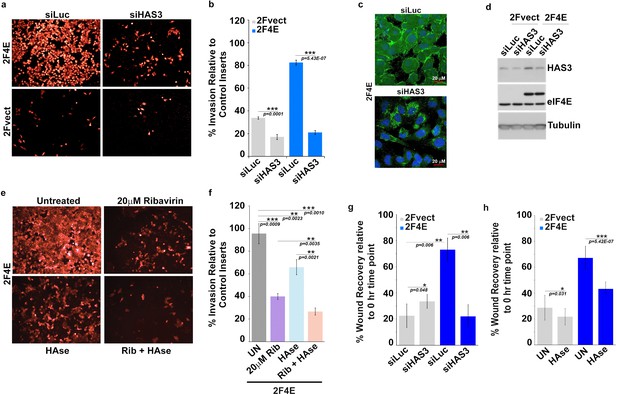
Surface HA is required for eIF4E-mediated invasion and migration of cancer cells.
(A–B) Cell invasion through matrigel assessed following siRNA-mediated knockdown of HAS3 or scrambled control (Luciferase (siLuc)) in U2Os cells overexpressing eIF4E (2F4E) or vector (2FVect). Invasion is measured as the percentage of fluorescence staining intensity in matrigel coated inserts versus that of the control inserts. (C) Fluorescence staining of HA (in green) following siRNA to HAS3. DAPI is in blue. A × 40 objective with no digital zoom used. (D) Western blot to demonstrate knockdown efficiency of HAS3. Tubulin served as loading control. (E–F) Cell invasion through matrigel assessed in U2Os cells overexpressing eIF4E following treatment with Ribavirin (Rib) and/or Hyaluronidase. Invasion is measured as the percentage of fluorescence staining intensity in matrigel-coated inserts versus that of the control inserts. (G–H) Cell migration across a scratch assessed in U2Os cells overexpression eIF4E or vector control following knockdown of HAS3 or treatment with Streptomyces Hyaluronidase. Migration is measured as the percentage of the area not filled with cells at t = 16 hr normalized to that of the t = 0 hr time point. For bar graphs, the mean ±standard deviation are shown. Experiments were carried out in triplicate, at least three independent times. **p < 0.01, ***p < 0.001 (Student’s t-test).
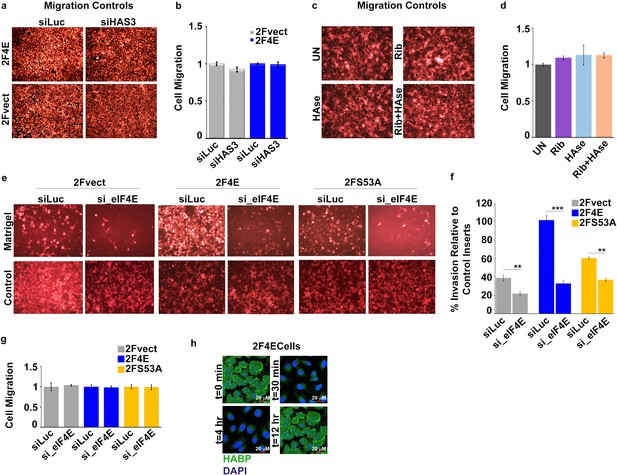
Surface HA is required for eIF4E-mediated invasiveness of cancer cells.
(A–D) Control inserts for Matrigel invasion assays shown in Figure 4. (E–F). In vitro Matrigel invasion assay following siRNA-mediated knockdown of eIF4E in U2Os cells overexpressing eIF4E, S53A mutant or vector control. (G) Control inserts for invasion assays shown in parts E and F at 48 hr. (H) Fluorescence staining of HA (in green) in U2Os cells overexpressing eIF4E collected at different time points following HAse treatment. DAPI is in blue. A × 40 objective with no digital zoom used. Experiments were carried out in triplicate, at least three independent times. For bar graphs, the mean ±standard deviation are shown. **p < 0.01, ***p < 0.001 (Student’s t-test).
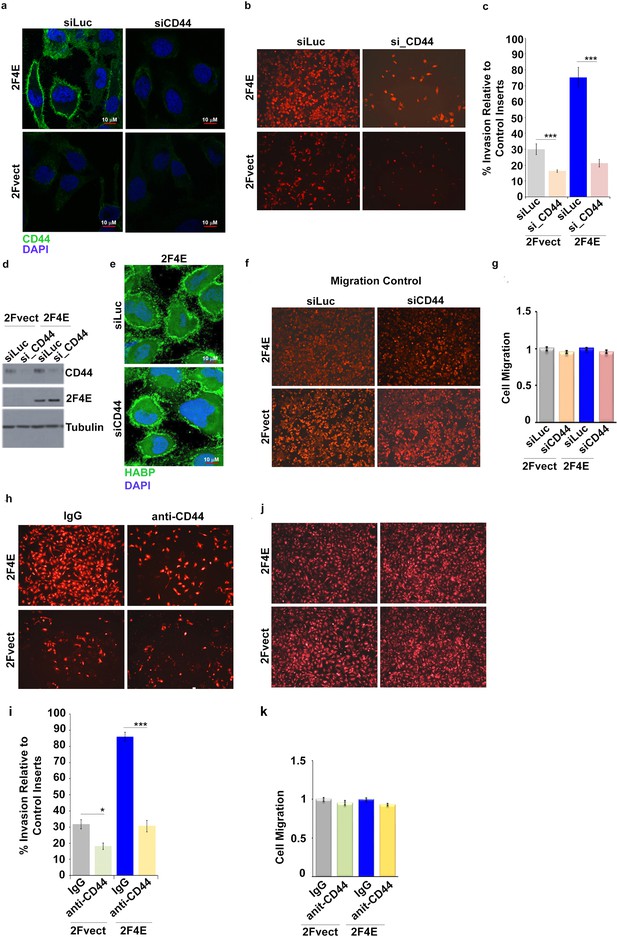
CD44 is required for the invasion of eIF4E cells.
(A) Immunofluorescence staining of CD44 (in green) following siRNA-mediated knockdown of CD44 in U2Os cells overexpressing eIF4E or vector control. DAPI is in blue. A × 40 objective with no digital zoom used. (B–C) Cell invasion through matrigel assessed 48 hr following siRNA-mediated knockdown of CD44 in U2Os cells overexpressing eIF4E (2F4E) or vector control (2FVect). siRNA against Luciferase was used as a control. Invasion was measured as a percentage of fluorescence staining intensity of matrigel-coated inserts normalized to control inserts. (D) Western blot control to assess knockdown efficiency of CD44. (E) Fluorescence staining of HA (in green) following siRNA to CD44 versus scrambled control in U2Os cells overexpressing eIF4E. DAPI is in blue. A × 63 objective with no digital zoom used. (F–G) Control inserts for Matrigel invasion assays shown in part (B). (H–I) Cell invasion through matrigel assessed following treatment with anti-CD44 blocking antibody (A3D8) known to bind the HA-binding domain of CD44. Invasion is measured as percentage of fluorescence staining intensity of matrigel-coated inserts normalized to control inserts. (J–K) Control inserts for Matrigel invasion assays shown in part (H–I). For bar graphs, the mean ±standard deviation are shown. Experiments were carried out in triplicate, at least three independent times. **p < 0.01, ***p < 0.001 (Student’s t-test).
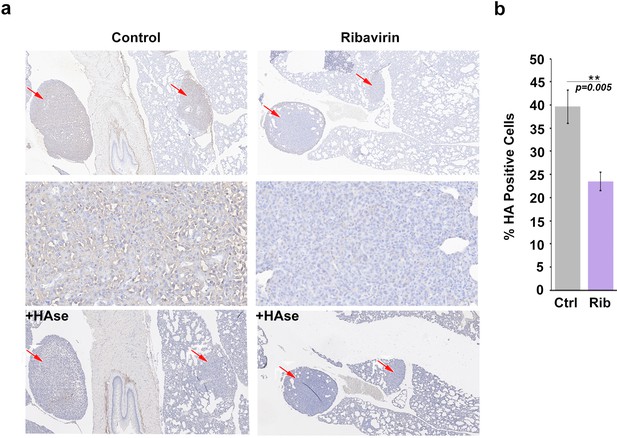
HA biosynthesis is required for eIF4E-mediated lung metastasis in mice.
(A) Histochemical staining of HA using biotinylated HABP in metastatic mouse tumors. +HAse indicates sections were treated with HAse prior to biotinylated HABP to ensure HA staining was specific. Red arrows indicate tumors. 3X (first and third row) and 50X (second row) magnification are presented. (B) Quantification from Visiomorph for all 19 mice over multiple sections per animal. For bar graphs, the mean ± standard deviation are shown. *p < 0.05 (Student’s t-test).

Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29830.014





