DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes
Figures
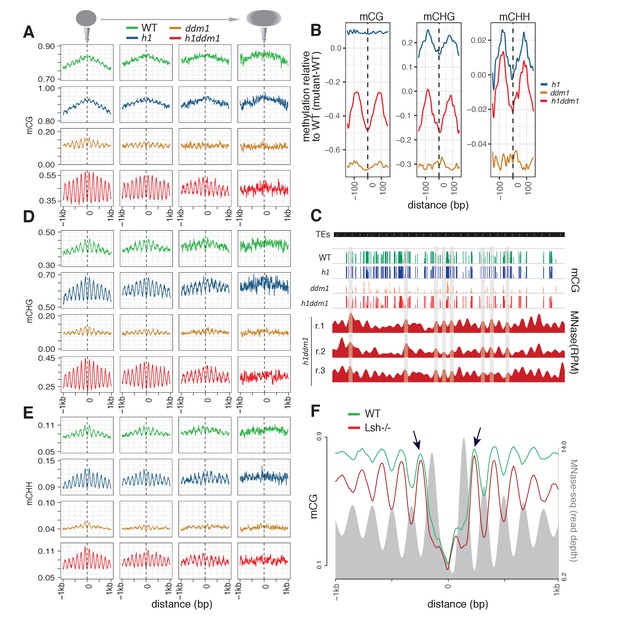
Arabidopsis and mouse nucleosomes are refractory to DNA methylation in the absence of DDM1/Lsh.
(A) Average CG methylation (mCG) plotted around heterochromatic nucleosomes grouped by positioning reliability, with better-positioned nucleosomes on the left. (B) Average methylation plotted around well-positioned heterochromatic nucleosomes (leftmost group in panel A) with WT methylation subtracted from that of the indicated mutant. (C) Genome browser view at AT1TE53390 of mCG in the indicated genotypes, with smoothed h1ddm1 MNase-seq shown in red (biological replicates shown as r.1–3). Vertical grey windows highlight well-positioned nucleosomes with core nucleosome depletion in ddm1 and h1ddm1. Methylation scale is 0 to 1, MNase-seq scale is 0 to 0.25 reads per million (RPM). (D–E) Average CHG methylation (D) or CHH methylation (E) is plotted around nucleosomes, as in (A). (F) Lsh null and WT mCG from mouse embryonic fibroblasts is plotted around CTCF binding sites. Nucleosome profiles from the same cell type are depicted in grey. Arrows point to methylated linkers closest to CTCF sites. Figure 1—figure supplement 1 presents the same analysis as Figure 1A,D,E but zoomed in around the anchoring nucleosome. Figure 1—figure supplement 2 shows genomic regions of Arabidopsis exhibiting specific loss of nucleosome core methylation in h1ddm1 plants. Figure 1—figure supplement 3 illustrates that well positioned nucleosomes shared by h1ddm1 and other genotypes exhibit the same methylation phenotype as shown in this figure.
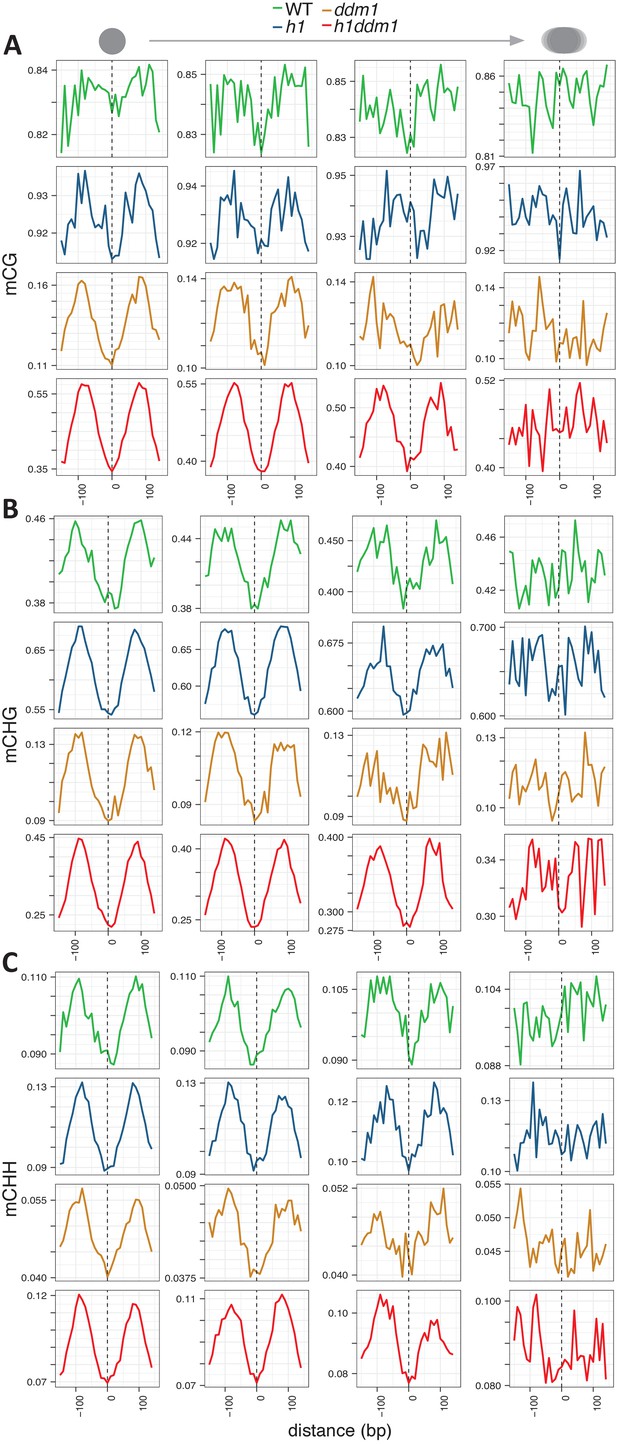
DNA methylation around nucleosome cores.
Methylation is plotted as described in Figure 1A,D and E for CG, CHG, and CHH contexts respectively. Note that Y-axis scales vary between genotypes.
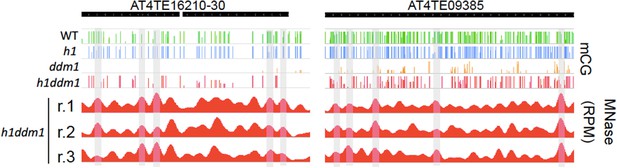
Linker-biased CG methylation in h1ddm1 plants.
Genome browser view of CG methylation (mCG) in the indicated genotypes, with smoothed h1ddm1 MNase-seq shown in red (biological replicates shown as r.1–3). Vertical grey windows highlight well-positioned nucleosomes with core nucleosome depletion in ddm1 and h1ddm1. Methylation scale is 0 to 1, MNase-seq scale is 0 to 0.25 reads per million (RPM).
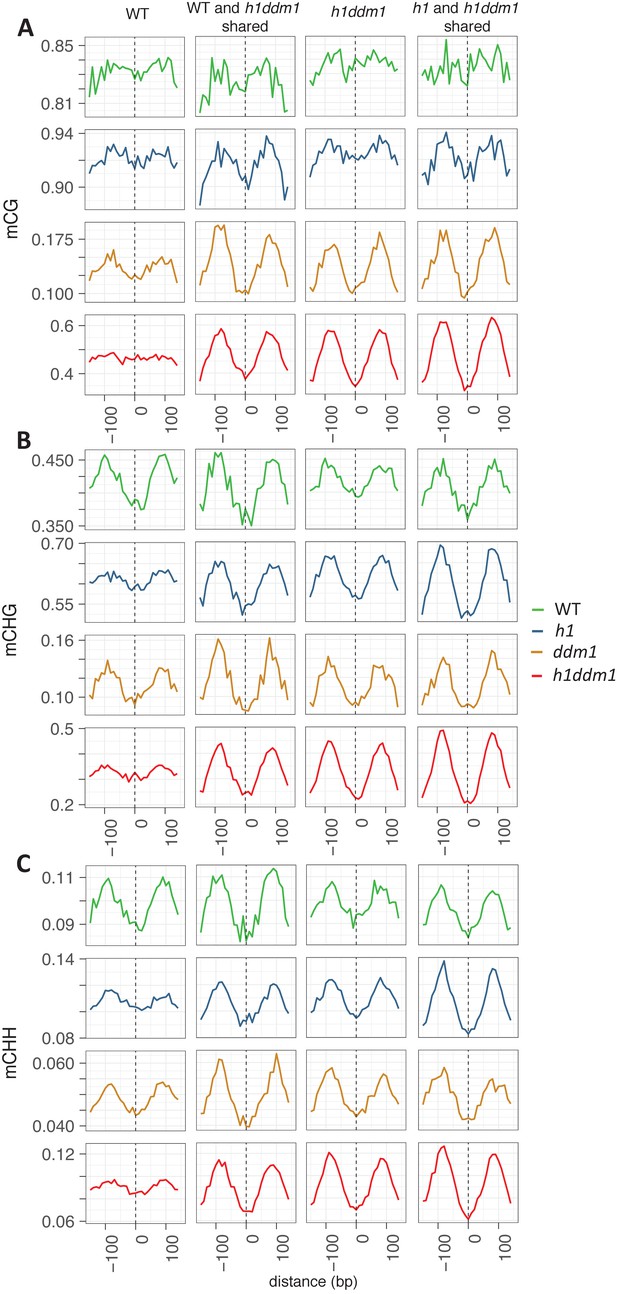
Well-positioned nucleosomes shared across genotypes exhibit nucleosome core depletion and linker enrichment.
All WT well-positioned nucleosomes (left column) were used to anchor methylation for mCG (A), mCHG (B), and mCHH (C). WT and h1ddm1 shared well-positioned nucleosomes, h1ddm1 only, and shared h1 and h1ddm1 well-positioned nucleosomes were also used for anchoring methylation in the given contexts. Note that use of all WT well-positioned nucleosomes largely prevents the detection of the nucleosome core depletion phenotype in the mCG context. Also note that Y-axis scales vary between genotypes. Nucleosome numbers: 19122 for WT, 4267 shared by WT and h1ddm1 (22.3% of WT), 22745 for h1ddm1, 6364 shared by h1 and h1ddm1 (28.0% of h1ddm1).
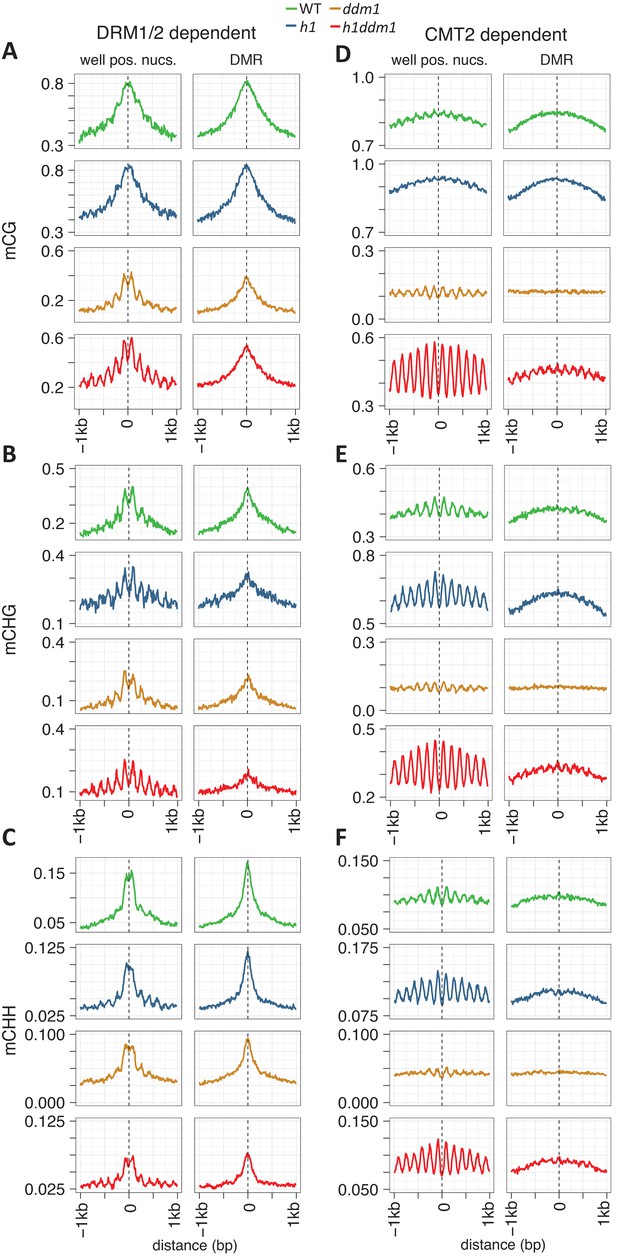
Nucleosomes within euchromatic TEs require DDM1 for DNA methylation.
(A–C) DNA methylation in DRM1/2-dependent differentially methylated regions (DMRs) for the given sequence context anchored at the dyads of the nucleosomes in the best-positioned group (defined as in Figure 1; left panels) or anchored at the centers of DMRs lacking positioned nucleosomes (right panels). (D–F) CMT2 DMRs; otherwise as in (A–C) (see Materials and methods for full description and definition of DMRs). Figure 2—figure supplement 1 shows the same data, but zoomed in on both axes.
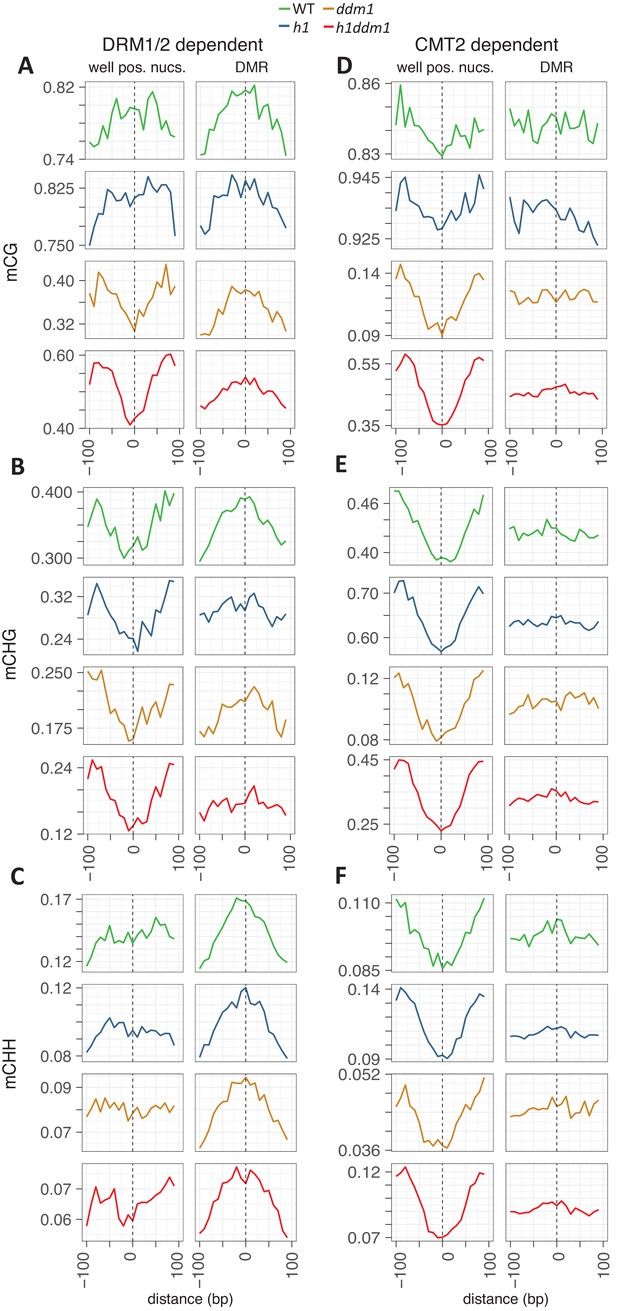
Zoomed-in view of DNA methylation around nucleosomes at DRM- or CMT2-dependent loci.
Methylation is plotted exactly as described for Figure 2, with DMRs of DMR1/2 on the left (A–C) and CMT2 on the right (D–F). Axes adjusted to enable visualization of nucleosome core depletion.
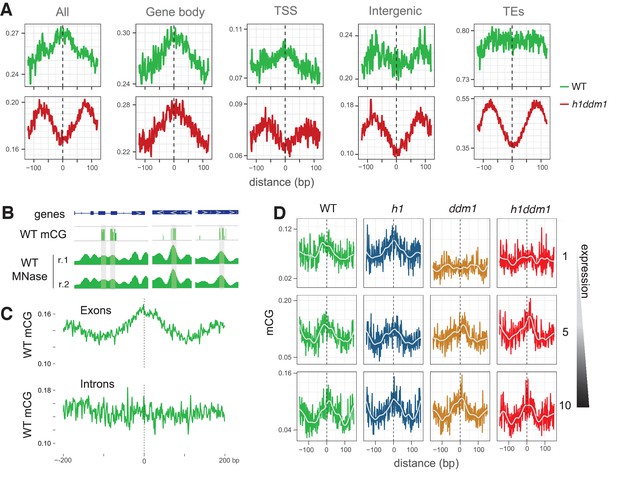
DDM1 facilitates the methylation of nucleosomes in genes.
(A) Average CG methylation at all nucleosomes, irrespective of positioning, for the indicated genomic context. TSS, transcription start site; TEs, transposable elements. (B) Representative genome browser snapshots showing CG methylation (mCG) at genic nucleosome cores in WT. Methylation scale is 0 to 1, MNase-seq scale is 0 to 0.2 reads per million (RPM). (C) All nucleosomes in genes (TSS and gene body combined), irrespective of positioning, were characterized as either exonic (any overlap with exon) or intronic (only 100% overlap with intron). Average CG methylation is plotted centered on the nucleosomes in the two groups. (D) Average CG methylation at well-positioned nucleosomes in genes; lowest expression decile (1), middle decile (5), and the highest expression decile (10) are shown. Figure 3—figure supplement 1 shows the enrichment of CG methylation on genic nucleosomes as a function of positioning reliability.
DNA methylation is enriched in genic nucleosomes.
Average CG methylation is plotted around genic nucleosomes grouped by positioning reliability, with better-positioned nucleosomes on top.
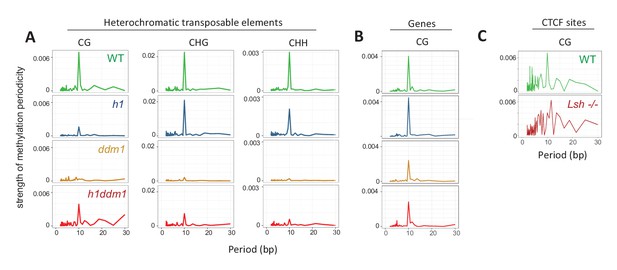
Methylation periodicity with respect to the histone octamer requires DDM1 and Lsh.
(A–B) Fast Fourier transform (FFT) periodogram of average DNA methylation across all 147 bp MNase-seq fragments in heterochromatic TEs (A) and genes (B) in Arabidopsis for the indicated genotype. (C) Same as (A–B), but with 147 bp MNase-seq fragments only from mouse CTCF sites, as in Figure 1F. Figure 4—figure supplement 1 shows the averaged Arabidopsis methylation data used for calculating periodicity by FFT while Figure 4—figure supplement 2 shows the averaged mouse methylation data.
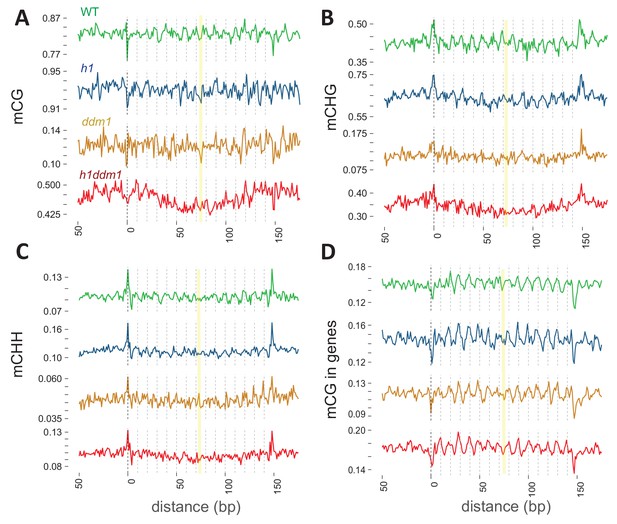
DDM1 is required for 10 bp methylation periodicity at heterochromatic nucleosomes.
Average DNA methylation in the indicated contexts across 147 bp MNase-seq fragments in heterochromatic TEs (A–C) and genes (D) for WT (green), h1 (blue), ddm1 (orange), and h1ddm1 (red). Yellow line indicates the approximate position of the presumptive dyad, where the DNA major groove faces nucleosomal histones.
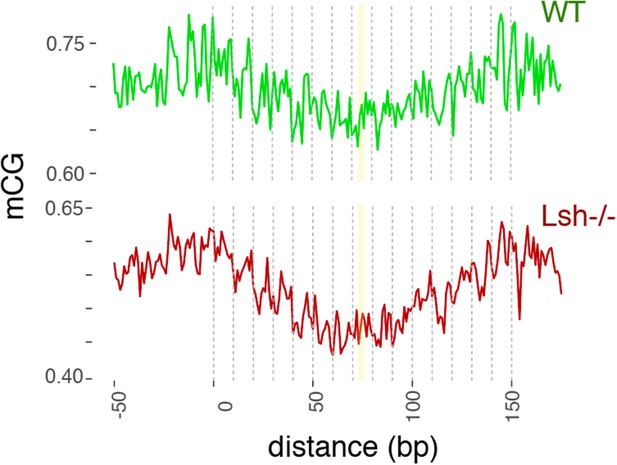
WT and Lsh-/- average CG methylation at WT 147 bp MNase-seq fragments overlapping CTCF sites.
Yellow line indicates the approximate position of the presumptive dyad, where the DNA major groove faces nucleosomal histones.
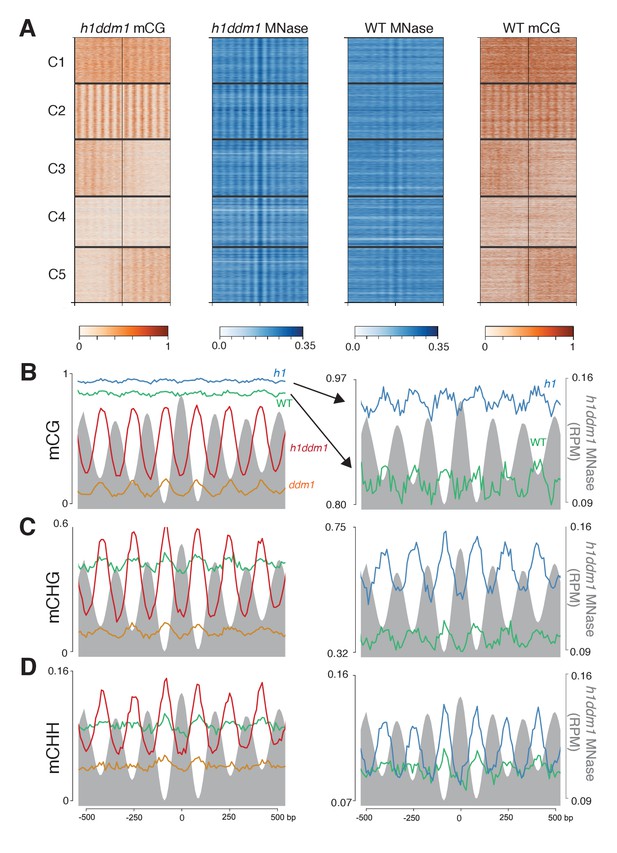
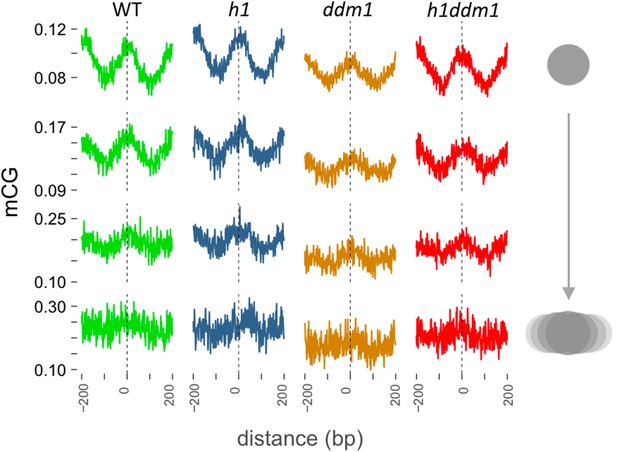
Loss of DDM1 and H1 produces linker-specific DNA methylation in Arabidopsis.
(A) h1ddm1 CG methylation (mCG) centered on heterochromatic well-positioned nucleosomes (n = 22,745) was grouped into five clusters by a self-organizing map algorithm. Nucleosomes (blue) and mCG (orange) are shown for h1ddm1 and WT; denser shades indicate increased values. (B–D) Average methylation plots for all genotypes in the indicated context at cluster C2, with h1ddm1 nucleosomes in grey fill. WT and h1 methylation plots are shown zoomed in on right panels, as indicated by arrows. Figure 5—figure supplement 1 contains non-CG methylation heatmaps. Figure 5—figure supplement 2 shows the phylogenetic distribution of species with naturally-occurring linker-specific DNA methylation. Figure 5—figure supplement 3 illustrates the absence of canonical linker histone in most species with linker-specific DNA methylation.
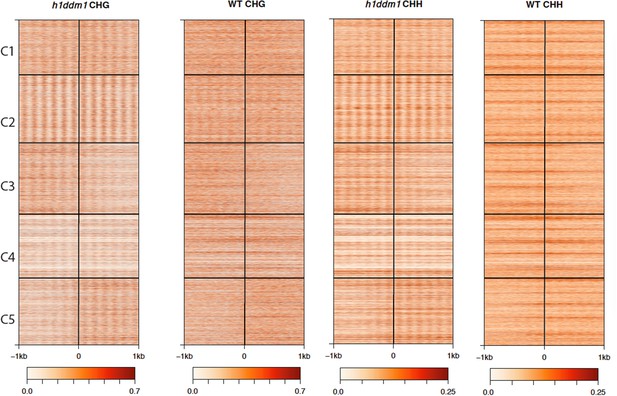
h1ddm1 and WT non-CG methylation at well-positioned arrays of nucleosomes in h1ddm1 plants.
CHG and CHH methylation exhibit similarly dramatic core depletion of methylation in h1ddm1 for C2, as shown in Figure 4C and D.
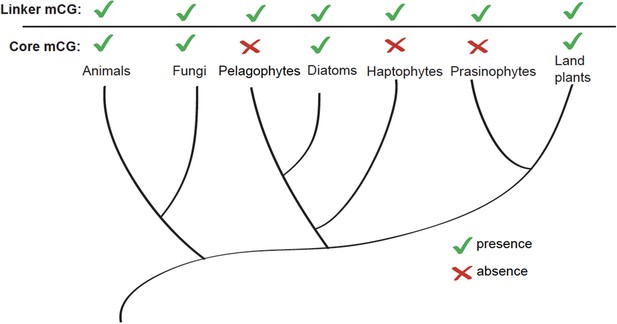
Linker-specific DNA methylation has evolved multiple times.
Phylogenetic tree (Parfrey et al., 2011) illustrating the evolutionary distribution of species that exhibit linker-specific DNA methylation. 'Core' refers to nucleosome core. mCG, CG methylation.

Eukaryotic histone H1 protein sequence alignment in species with and without nucleosomal core DNA methylation.
Multiple sequence alignment of representative linker histone H1 and linker histone-like sequences from across eukaryotes illustrating the absence of the signature winged-helix domain (Kasinsky et al., 2001) in the linker histone-like genes of three of the five marine algae surveyed in (Huff and Zilberman, 2014) (yellow box delineates approximate winged-helix domain). One species (Micromonas pusilla) has a canonical H1 gene. The fifth species (Bathycoccus prasinos) lacks identifiable H1-related genes. Grey shading indicates similar amino acids; identical residues are shaded black. Marine algae are the top four species. See Materials and methods for organism and sequence details.
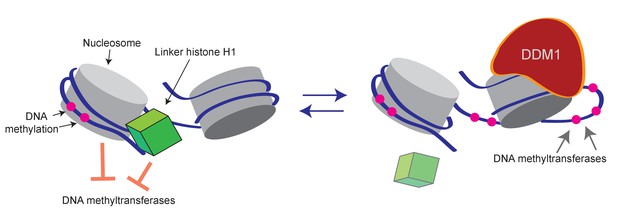
Model of proposed role for H1 and DDM1/Lsh in DNA methylation.
Cartoon depicts a linker histone-bound dinucleosomal fragment of the genome (left) undergoing remodeling by DDM1 (right) to provide methyltransferases access to nucleosomal core DNA.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30674.019





