Coupling chemosensory array formation and localization
Figures
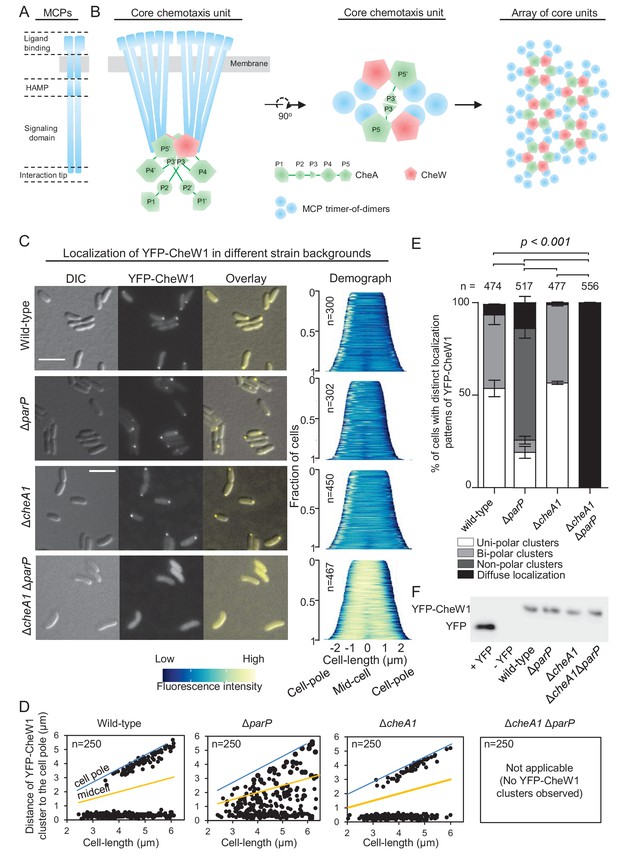
ParP contributes to signaling array formation.
(A) Schematic of the domain architecture of the MCP dimer. (B) Schematic showing the structure of chemotaxis core units and how these units assemble into signaling arrays. (C) Fluorescence microscopy images showing the intracellular localization of YFP-CheW1 in wild-type and indicated V. cholerae mutant backgrounds. Demographs show the fluorescence intensity of YFP-CheW1 along the cell length in a population of V. cholerae cells relative to cell length. Scale bars represents 5 µm. (D) Graphs depicting the distance of YFP-CheW1 foci from the cell pole as a function of cell length. (E) Bar graph showing the percentage of cells with distinct YFP-CheW1 localization patterns in the indicated V. cholerae strain backgrounds. Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. (F) Immunoblot using JL8 anti-YFP antibodies to detect the presence of YFP and YFP-CheW1 in V. cholerae strains imaged in (C). As a positive control, a strain expressing YFP from plasmid pMF390 was included (+YFP). A strain not expressing YFP (- YFP) was included as a negative control.
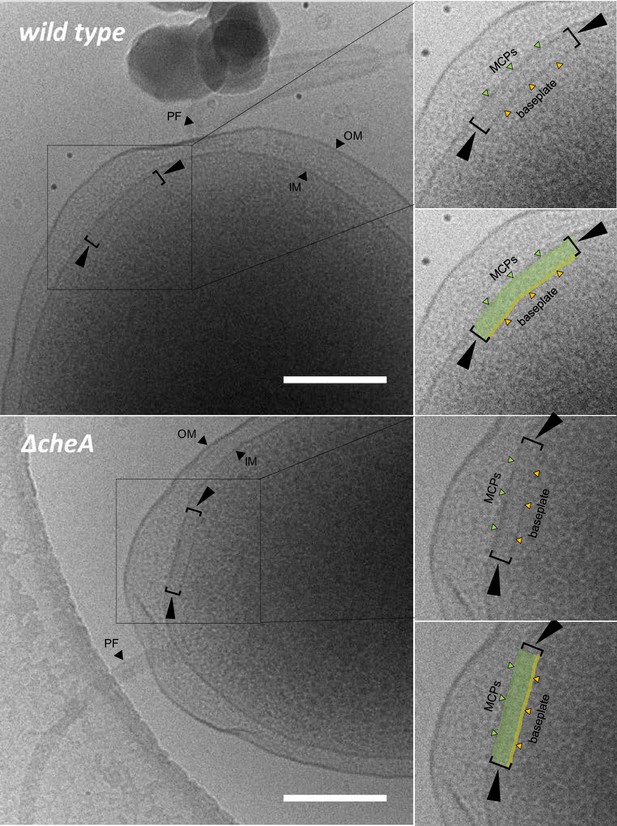
Chemotaxis arrays form in the absence of CheA.
Cryo-EM images of the cell pole of wild-type and ΔcheA V. cholerae cells, showing the presence of chemotaxis signaling arrays in both strain backgrounds. Signaling arrays are indicated by brackets. PF: polar flagellum; OM: outer membrane; IM: inner membrane. Green highlighted regions indicate MCP arrays and yellow highlighted regions indicate the baseplate of the signaling arrays. Scale bar represents 200 nm.
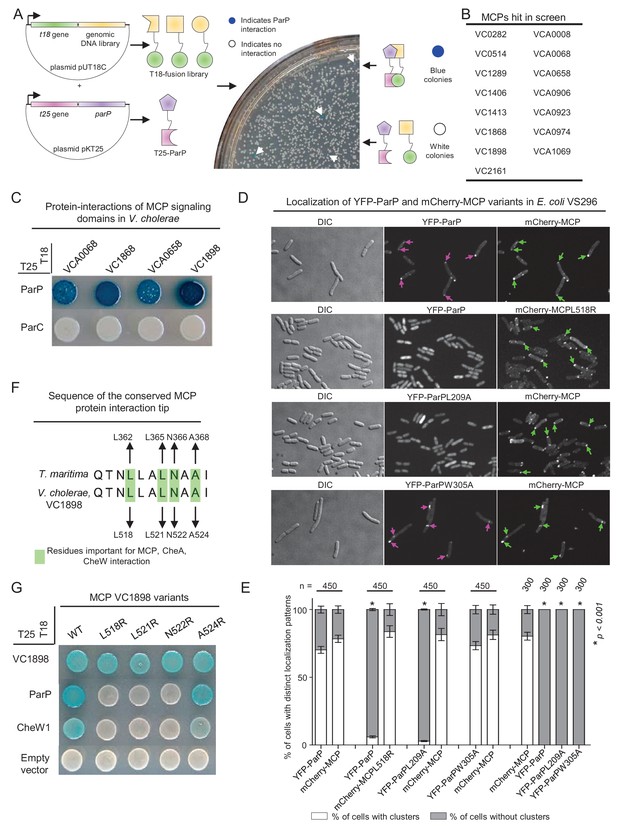
ParP interacts with the protein interaction tip of methyl-accepting-chemotaxis proteins.
(A) To screen for ParP interaction factors, E. coli strain BTH101 carrying plasmid pAK08 (encoding T25-ParP) was transformed with the plasmid library and spread on indicator plates. Bacterial colonies encoding a candidate ParP interaction partner turned blue. Blue colonies were picked and the chromosomal DNA inserted in the pUT18 vector was identified by sequencing. (B) Summary of the MCP proteins identified as ParP interaction partners in the bacterial-two-hybrid (BACTH) screen. (C) BACTH experiment assaying for protein interactions of the V. cholerae MCP signaling domains (SD) of MCPs VCA0068, VC1868, VCA0658, and VC1898 with ParP and ParC. Blue coloration of bacterial colonies indicates an interaction. (D) Fluorescence microscopy of YFP-ParP and mCherry-MCP VC1898 (mCherry-MCP) variants in E. coli strain VS296. Purple arrows indicate clusters of YFP-ParP variants. Green arrows indicate clusters of mCherry-MCP. (E) Bar graphs indicate the percentage of cells with clusters of YFP-ParP variants and mCherry-MCP in E. coli VS296. Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. Asterisks indicate p<0.001 compared to VS296 co-expressing wild-type YFP-ParP with mCherry-MCP. (F) Alignment of the conserved protein interaction tips of MCP TM1143 from T. maritima and MCP VC1898 of V. cholerae. Highlighted V. cholerae amino acids were chosen as candidates for amino acid substitution. (G) BACTH experiment assaying for protein interactions of V. cholerae MCP VC1898 and its variants.
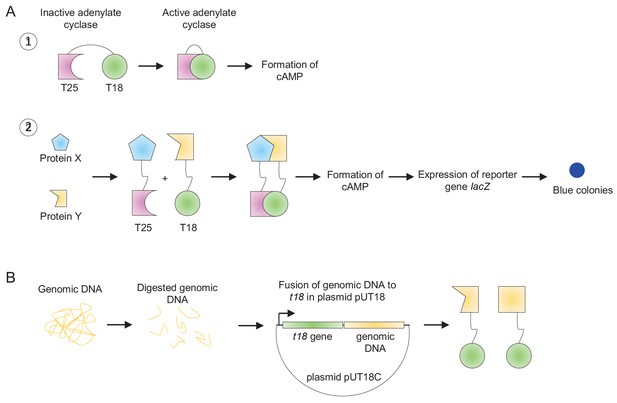
Bacterial-two-hybrid screen for identification of ParP interaction partners.
(A) Schematic showing the principle behind bacterial-two-hybrid assays for protein-protein interactions; 1) adenylate cyclase consists of two domain, T25 and T18, which come together to form an active enzyme resulting in the formation of cAMP; 2) To assay for protein interaction, the T25 and T18 fragments of the adenylate cyclase are split in two separate domains. Subsequently the two domains are fused to the two proteins, X and Y, of interest. If X and Y interact, they bring T25 and T18 in close proximity, which results in the formation of a functional adenylate cyclase and in turn blue colonies on indicator plates. (B) Construction of a library of chromosomal DNA fused to the gene encoding the T18 fragment. Chromosomal DNA is purified and digested with rare cutting restriction enzymes. DNA fragments in the size 1 kb-5kb are purified in inserted into the multiple cloning site of plasmid pUT18 and transformed into E. coli strain DH5αλpir. Bacterial colonies are pooled and a plasmid DNA purified, resulting in a library of plasmid DNA carrying chromosomal DNA fused to the T18 fragment.
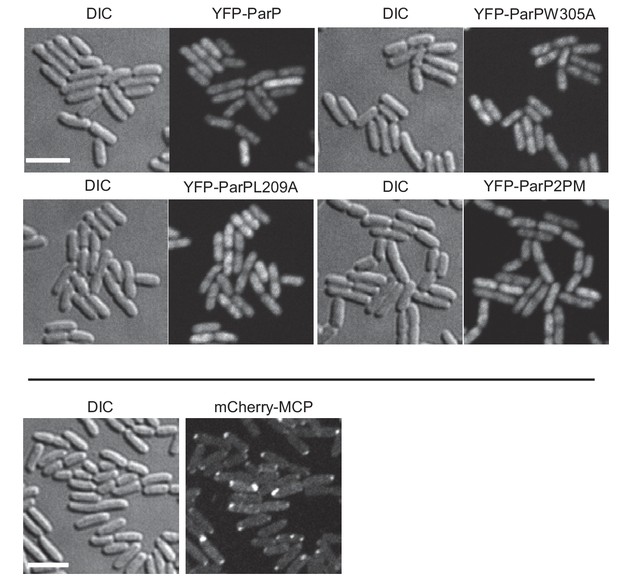
YFP-ParP is diffusely localized to the cytoplasm in E. coli.
Fluorescence microscopy of YFP-ParP variants in E. coli strain VS296. All variants are diffusely localized in the cytoplasm.

Alignment of the MCP protein interaction tip of MCPs from V. cholerae.
Alignment of Thermotoga maritima MCP TM1143 domain responsible for interaction with CheW and CheA-P5 and all predicted MCPs from V. cholerae. Residues in green highlight the residues in the MCP TM1143 from T. maritima that have been shown to be important for mediating interaction to CheW and CheA-P5: L362, L365, N366, and A368. These residues are conserved in all of but two of the MCPs of V. cholerae and in all of the MCPs indentified in the BACTH screen.
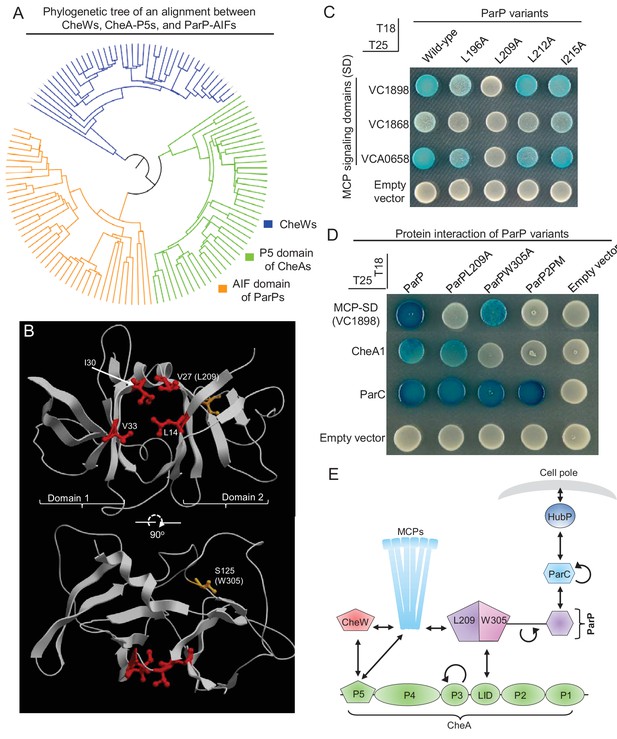
Two distinct interaction interfaces of ParP-AIF mediate its interaction with MCP and CheA respectively.
(A) Phylogenetic tree of the SH3-like domains of CheW, CheA-P5 and ParP-AIF proteins. (B) Structure of Thermotoga maritima MSB8 CheW (PDB 3UR1, [Briegel et al., 2012]). CheW consists of two subdomains (1 and 2) responsible for interaction with subdomains 2 and 1 of the P5 domain of CheA. The junction between the two subdomains consists of branched hydrophobic residues (amino acids highlighted in red) that form a groove where CheW interacts with the MCP signaling domain helix. The corresponding amino acid L209 of ParP-AIF is noted in parentheses. The amino acid (S125) corresponding to the position of W305 in ParP-AIF is highlighted in orange. (C) BACTH experiment assaying interaction between ParP variants carrying amino acid substitutions in the predicted MCP binding pocket and MCP proteins VC1898, VC1868, and VCA0658. (D) BACTH experiment assaying interaction between ParP variants and MCP VC1898, CheA1, and ParC. (E) Schematic depicting ParP’s three interaction interfaces, which enable the protein to interact with MCPs, CheA and ParC. L209 and W305 refer to amino acids important for interaction with MCP-SD and CheA respectively, within the two interaction interfaces of ParP-AIF.
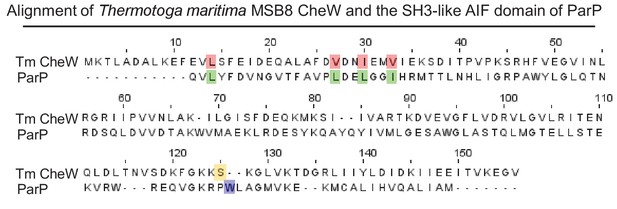
ParP contains a C-terminal SH3-like domain.
Sequence alignment between T. maritima CheW and ParP-AIF. Amino acid alignment of T. maritima MSB8 CheW and the ParP-AIF domain. Numbers refer to amino acid position in T. maritima MSB8 CheW. Red: amino acids required for interaction between CheW and the MCP signaling domain and the corresponding amino acids in ParP-AIF. Purple: highlights W305 in ParP-AIF responsible for interaction with CheA. The amino acid with the corresponding position (S125) in T. maritima MSB8 CheW is highlighted in yellow.
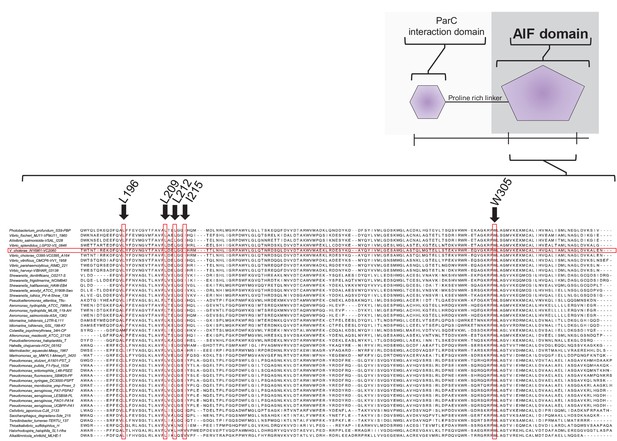
Residues responsible of MCP and CheA interaction are highly conserved amongst ParP proteins.
Sequence alignment of the C-terminal AIF-domain of ParP proteins.
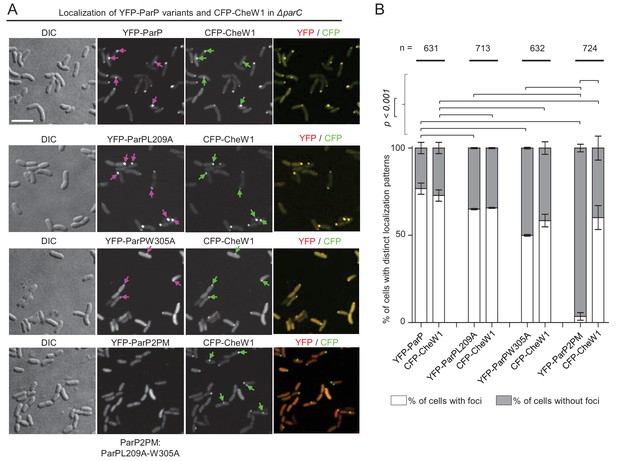
ParP’s interaction with MCPs or CheA is required for its association with signaling arrays.
(A) Fluorescence microscopy showing the intracellular localization of YFP-ParP variants and CFP-CheW1 in V. cholerae ΔparC. Scale bar represents 5 µm. Purple arrows indicate clusters of YFP-ParP variants. Green arrows indicate CFP-CheW1 clusters and pink arrows indicate YFP-ParP clusters. (B) Bar graphs indicate percentage of cells with foci of YFP-ParP variants and of CFP-CheW1 in V. cholerae ΔparC. Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. (A–B) ParP2PM refers to a ParP variant carrying both the L209A and W305A amino acid substitutions.
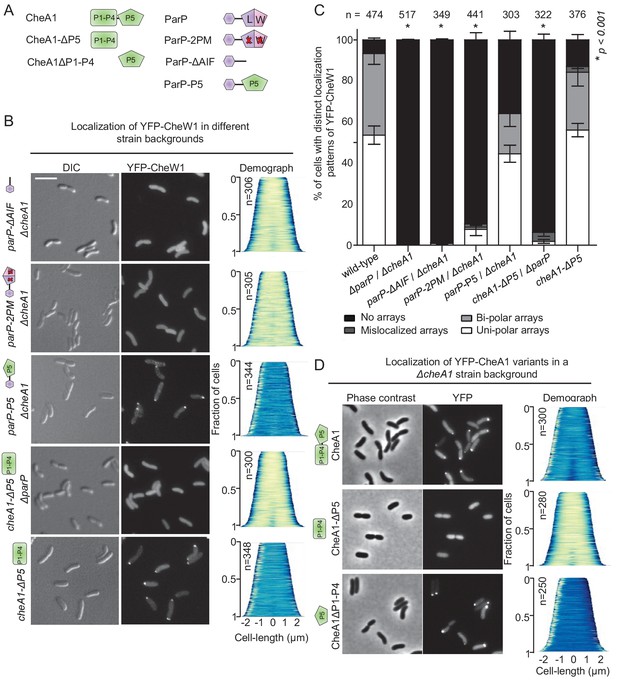
The ParP AIF domain and CheA-P5 promote formation of signaling arrays.
(A) Schematic depicting the various CheA and ParP variants analyzed. (B) Fluorescence microscopy showing the intracellular localization of YFP-CheW1 in the indicated V. cholerae strain backgrounds. Scale bar represents 5 µm. (C) Bar graph showing the percentage of cells with distinct YFP-CheW1 localization patterns in the indicated V. cholerae strain backgrounds. Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. Asterisks indicate p<0.001 compared to wild-type. (D) Fluorescence microscopy showing the intracellular localization of full-length and truncated versions of CheA1 fused to YFP in a ΔcheA1 strain background. CheA: full-length CheA protein; CheA-(P1–P4): truncated version of CheA consisting of domain P1 to P4; CheA-P5: truncated version of CheA only consisting of the P5 domain.
YFP-CheW1 protein is stable in the analyzed V. cholerae strain backgrounds.
Immunoblot using JL8 anti-YFP antibodies to detect the presence of YFP and YFP-CheW1 in the indicated V. cholerae strains imaged in Figure 5B. As a positive control, a strain expressing YFP from plasmid pMF390 was included (+YFP). A strain not expressing YFP (- YFP) was included as a negative control.
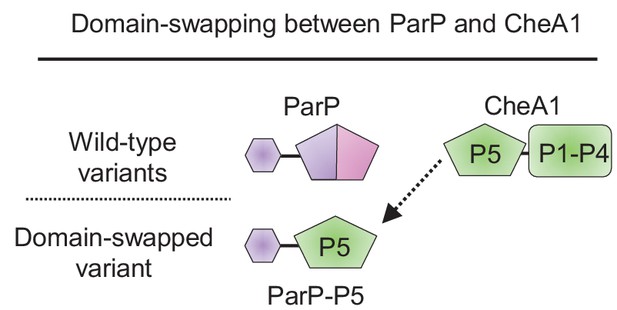
Domain swapping.
Schematic showing the design of the chimeric protein ParP-P5 by domain swapping of the AIF-domain of ParP with the P5-domain of CheA. Demographs show the fluorescence intensity of YFP-CheW1 along the cell length in a population of V. cholerae cells relative to cell length.
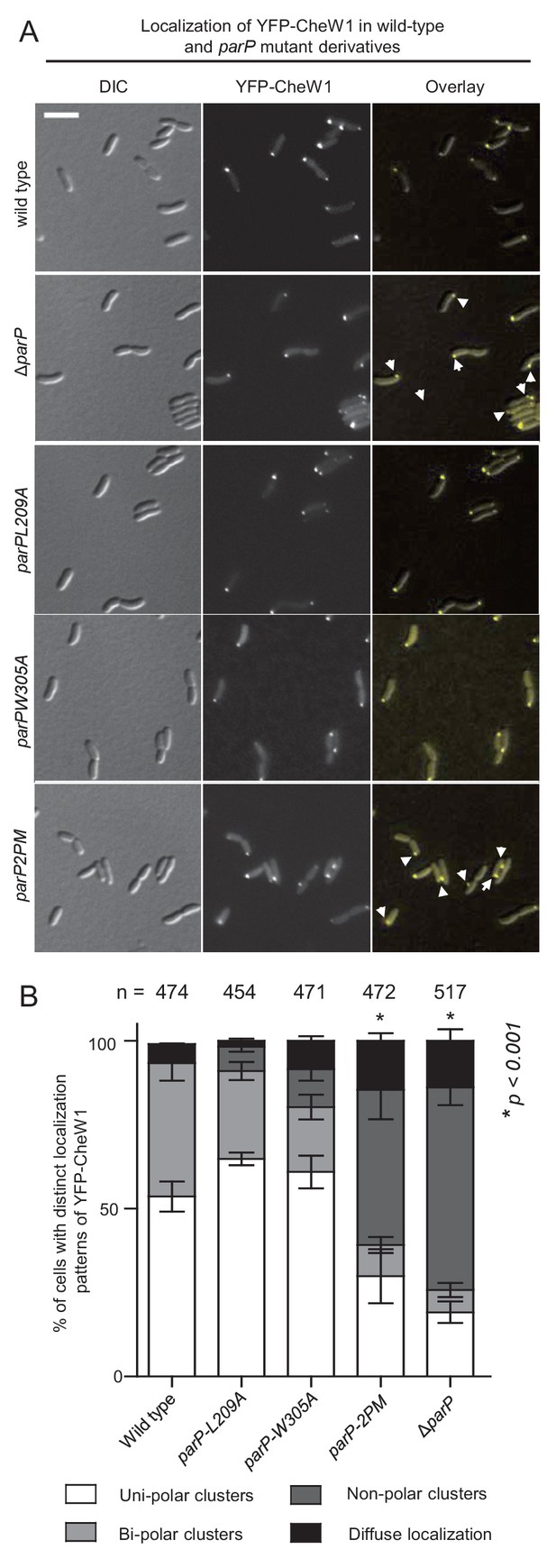
Interactions between ParP, MCPs, and CheA ensure proper polar localization and inheritance of signaling arrays.
(A) Fluorescence microscopy images showing the intracellular localization of YFP-CheW1 in wild-type and different V. cholerae parP mutant backgrounds. Arrows indicate non-polar clusters of YFP-CheW1. Scale bar represents 5 µm. (B) Bar graph showing percentage of cells with distinct localization patterns of YFP-CheW1 in wild-type and different V. cholerae mutant backgrounds. Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. Asterisks indicate p<0.001 compared wild-type.
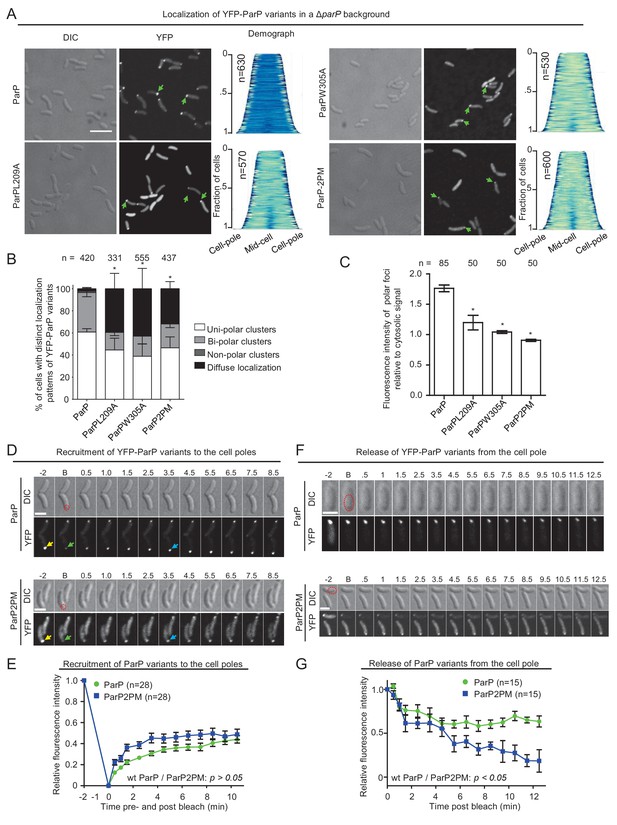
Integration of ParP within signaling arrays stabilizes recruitment of ParP to the cell pole.
(A) Fluorescence microscopy images showing the intracellular localization of YFP-ParP variants in a V. cholerae ΔparP background. Demographs show the fluorescence intensity of YFP along the cell length in a population of V. cholerae cells relative to cell length. Scale bar represents 5 µm. Green arrows indicate polarly localized ParP clusters. (B) Bar graph showing the percentage of cells with distinct YFP-ParP localization patterns in the indicated V. cholerae strain backgrounds. (C) Bar graph showing the fluorescence intensity of polar ParP foci relative to cytosolic signal. (B–C) Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. Asterisks indicate p<0.001 compared to wild-type. (D) Fluorescence-recovery-after-photobleaching (FRAP) experiment of YFP-ParP and YFP-ParP2PM clusters at the cell poles showing that clusters recover post-bleaching. Numbers indicate minutes pre- and post-bleach. ‘B’ indicates bleaching. The red dashed circle shows the bleached region. Yellow arrows indicate the pre-bleach cluster, green arrows indicate the bleached cluster. Blue arrows indicate clusters with recovered YFP signal. (E) Graph depicting the fluorescence intensity of YFP-ParP and YFP-ParP2PM pre- and post-bleach at the bleached cell pole relative to the initial intensity at the pole pre-bleach during time-lapse series. The average recovery from 28 distinct cells is shown. Error-bars indicate standard error mean (SEM). (F) Release of YFP-ParP and YFP-ParP2PM from the cell pole post-bleach of the cytoplasmic signal. Numbers indicate minutes pre- and post-bleach. ‘B’ indicates bleaching. The red dashed circle shows the bleached region. (G) Graph depicting the fluorescence intensity of polar YFP-ParP and YFP-ParP2PM clusters post-bleach relative to the initial intensity during time-lapse series. The average of 15 distinct cells is shown. Error-bars indicate standard error mean (SEM).
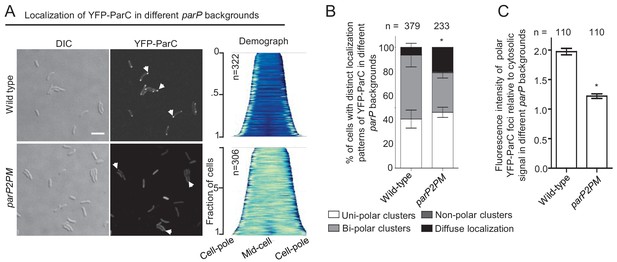
Integration of ParP within signaling arrays is required for proper polar localization of ParC.
(A) Fluorescence microscopy images showing the intracellular localization of YFP-ParC in wild-type and parP2PM V. cholerae. Demographs show the fluorescence intensity of YFP along the cell length in a population of V. cholerae cells relative to cell length. Scale bar represents 5 µm. White arrows indicate polarly localized ParC clusters. (B) Bar graph showing the percentage of cells with distinct YFP-ParC localization patterns in the indicated V. cholerae strain backgrounds. (C) Bar graph showing the fluorescence intensity of polar ParC foci relative to cytosolic signal. (B–C) Error bars indicate standard error of the mean (SEM). The n-value indicates the total number of cells analyzed from three independent experiments. Asterisks indicate p<0.001 compared wild-type.
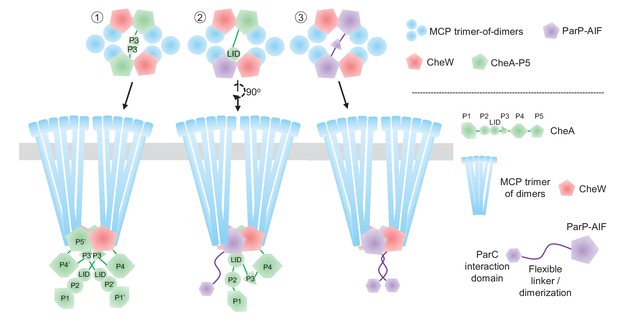
Model of ParP’s integration within the chemotactic core unit.
Schematic model of potential ParP interactions in the chemotaxis core unit – discussed in detail in the main text.
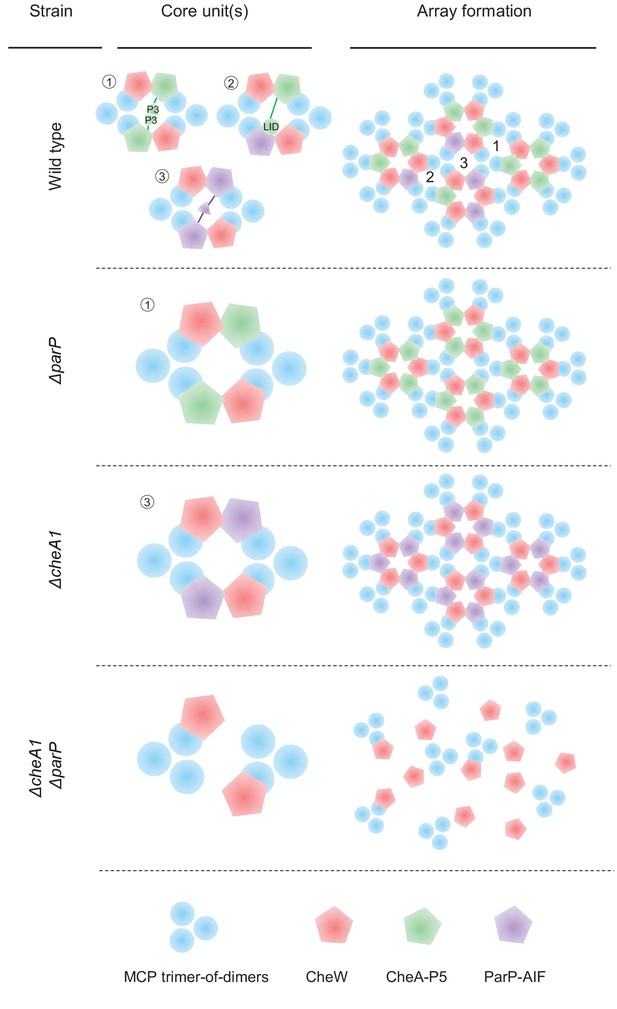
Chemotaxis core-unit- and array structure in different strain backgrounds.
In wild-type cells three distinct core unit occurs and the arrays might consist of all three types of core units. In ΔparP and ΔcheA deletion backgrounds the arrays consist of type #1 and type #3 core units only, respectively. In the absence of both ParP and CheA, core units, and in consequence signaling arrays, no longer form.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information | |
|---|---|---|---|---|---|
| Gene (Vibrio cholerae) | parP | N/A | NCBI-GeneID: 2613440 | ||
| Gene (V.cholerae) | cheA | N/A | NCBI-GeneID: 2613443 | ||
| Gene (V. cholerae) | vca0068 | N/A | NCBI-GeneID: 2612100 | ||
| Gene (V. cholerae) | vc1868 | N/A | NCBI-GeneID: 2613622 | ||
| Gene (V.cholerae) | cheW | N/A | NCBI-GeneID: 2613439 | ||
| Gene (V. cholerae) | parC | N/A | NCBI-GeneID: 2613441 | ||
| Gene (V. cholerae) | mcp; vc1898 | N/A | NCBI-GeneID: 2613527 | ||
| Gene (V. cholerae) | vca0658 | N/A | NCBI-GeneID: 2612769 | ||
| Strain (V. cholerae N16961) | wild type | Clinical isolate | NCBI-Taxonomy ID: 243277 | For complete strain list see Supplementary file 1 | |
| Antibody | Mouse monoclonal anti-GFP | Clontech Laboratories, Inc. (USA) | Cat#: 632381 | ||
| Recombinant DNA reagent | See Supplementary file 2 for Plasmid list | ||||
| Sequence-based reagents | Oligonucleotides | This paper | See Supplementary file 1 for Primer list | Eurofins MWG Operon (Ebersberg) | |
| NucleoSpin Gel and PCR Clean-up kit | N/A | Macherey-Nagel (Düren) | Ref.: 740609.250 | ||
| NucleoSpin Plasmid Kit | N/A | Macherey-Nagel (Düren) | Ref.: 740588.250 | ||
| Metamorph v7.5 | N/A | Molecular Devices (Union City, CA) | |||
| ImageJ-Fiji | N/A | http://rsbweb.nih.gov/ij | |||
| R studio version 3.0.1 | N/A | http://www.rstudio.com/ | |||
| GraphPad Prism version 6.07 | N/A | GraphPad Software (La Jolla CA) | |||
| NIS-Elements Software AR 4.60.00 (Nikon) | N/A | LIM (Prague) | ` | ||
Additional files
-
Supplementary file 1
Table S1 - strain and plasmid list.
Table listing the strains and plasmids used in the study.
- https://doi.org/10.7554/eLife.31058.021
-
Supplementary file 2
Table S2 - primer list.
Table listing the primer names and sequences used in the study.
- https://doi.org/10.7554/eLife.31058.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31058.023





