The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets
Figures
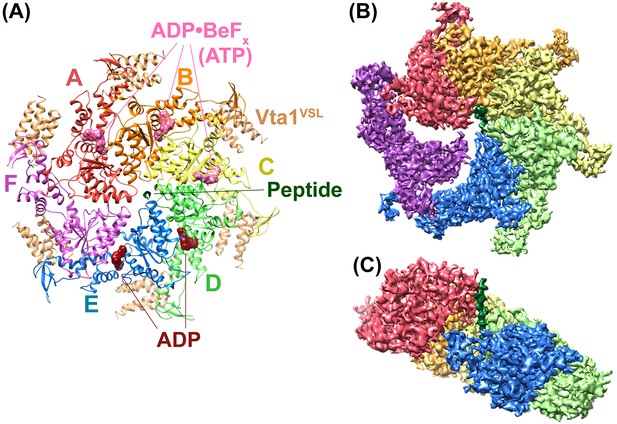
Overall structure of the Vps4 complex.
(A) Ribbon representation of the complex viewed from the ‘top’ N-terminal side of Vps4 and N-terminal end of the peptide. (B) Similar orientation as panel A showing a segmented map contoured around Vps4 and peptide. (C) Same as panel B viewed from the side with density for subunit F removed for clarity.

Cryo-EM of the Vps4 complex.
(A) Representative cryo-EM micrograph of Vps4101-437-Hcp1 particles. (BC) Representative 2D class averages, (B) before and (C) after Hcp1 signal subtraction. (D) Gold-standard FSC of the Hcp1-subtracted particle reconstructions on independent (odd:even particles) halves of the data (blue) and FSC between the refined model and the density map (orange). (E) Cross-validation of refined model (see Materials and methods). (F) Angular distribution plot based on orientation assignments in RELION and visualized in UCSF Chimera. Cylinders scaled (low to high) and colored (blue to red) proportional to number of particles in the assigned orientation. (G) Local resolution estimates determined by ResMap (Kucukelbir et al., 2014).
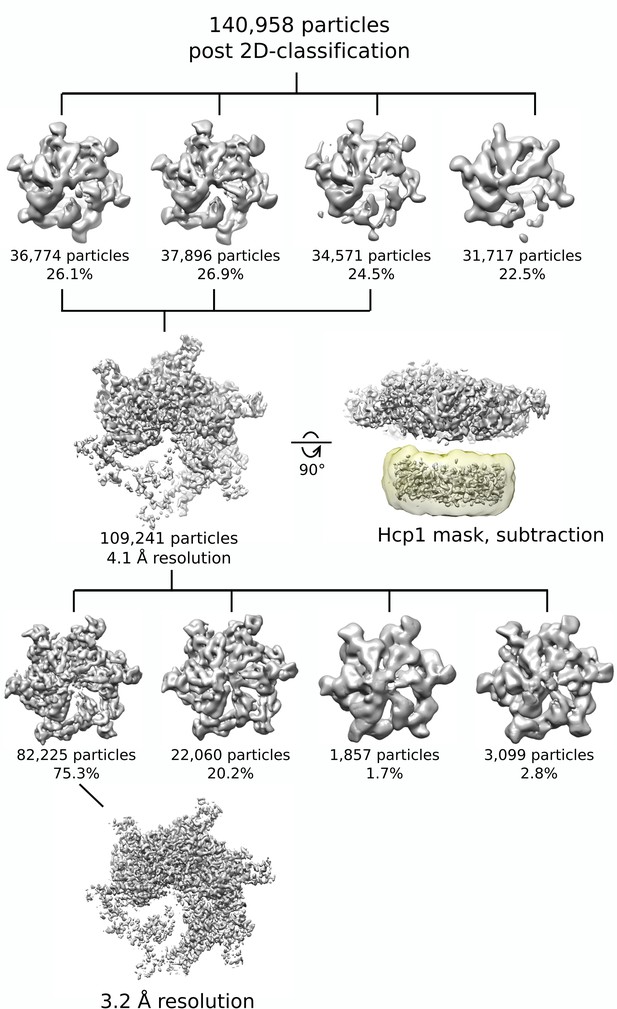
Classification and signal-subtraction scheme for the Vps4 complex.
140,958 particles were input for 3D classification. 109,241 particles were sorted into classes with good Vps4 features and used to generate a consensus reconstruction of the entire Vps4101-437-Hcp1 complex at 4.1 Å resolution. Signal subtraction of Hcp1 was performed using a previously described strategy (see Materials and methods). An additional round of 3D classification was performed using the Hcp1-subtracted particles. 82,225 particles were sorted into a single class with high-resolution Vps4 features and used to generate the final 3.2 Å Vps4 reconstruction.
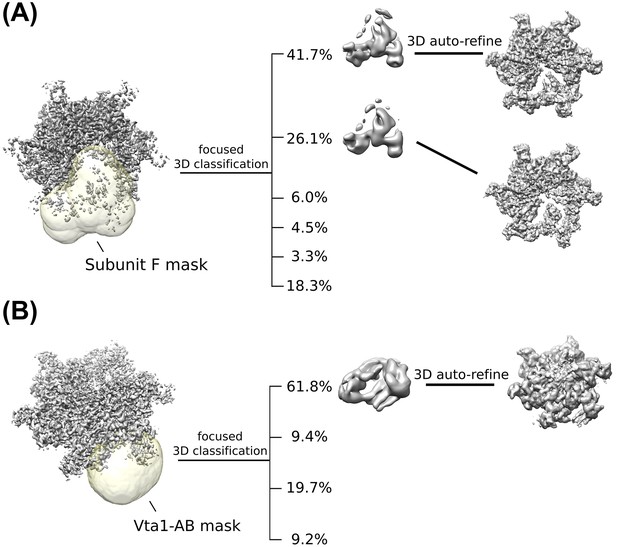
Focused classification of Vps4 subunit F and Vta1.
(A) Representative masking scheme for subunit F. Custom masks were generated for structurally heterogeneous features and focused 3D classification was performed using the masks. Classes with good features were used to isolate particles for additional rounds of RELION auto-refinement. (B) Representative masking scheme for Vta1.

Surface Representation.
Similar orientation to Figure 1A. Shows the gaps between the subunit F large ATPase domain and its neighboring subunits, and the highly solvated channel between subunit F and the peptide.
Representative density.
Charge density map shown over the β-sheet of the large domain of subunit B.
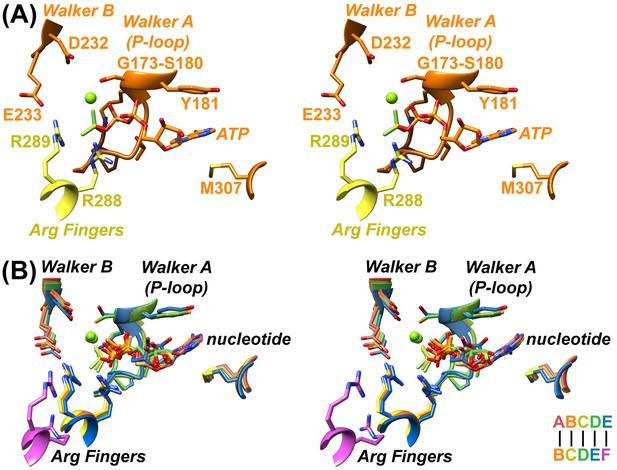
Nucleotide coordination and subunit interfaces.
(A) Stereoview of a representative ADP·BeFx coordination shown at subunit B (BC interface). Subunits color-coded as in Figure 1. (B) Stereoview of nucleotide-binding sites at subunits A, B, C, D, and E following superposition on the large domains of the first subunit at each interface.
Nucleotide densities.
Density shown with the refined models of ADP·BeFx (subunits A-C), and ADP (subunits D and E). A Mg2+ ion is modeled in subunits A-D.
Coordination of ADP·BeFx (ATP) at a representative subunit.
Coordination of ADP·BeFx is shown at subunit B and the interface with subunit C.
Comparison of nucleotide coordination at subunits A, B, and C.
Pairs of Vps4 subunits (AB, BC, and CD) are superposed by overlap on the P loop residues of the first subunit.
Comparison of nucleotide coordination at subunits A and D.
Same as Figure 2—video 4 but showing overlap of the ADP·BeFx at subunit A and the ADP at subunit D.
Comparison of nucleotide coordination at subunits A and E.
Same as Figure 2—video 4 but showing overlap of the ATP/ADP·BeFx at subunit A and the ADP at subunit E.
Interface at a representative subunit pair in the Vps4 helix.
The interface between subunits A and B is highlighted. It comprises three regions: large domain to large domain, small domain to large domain, and nucleotide-mediated.
Comparison of AB, BC, and CD interfaces.
Overlap was performed on the large ATPase domain of the first subunit in each pair.
Comparison of AB and DE interfaces.
Overlap was performed on the large ATPase domain of the first subunit in each pair. Subunit E is rotated by 8° but maintains very similar contacts with the preceding subunit and relative position of pore loops 1 and 2.
Conservation of the interface between small and large domains.
Small domain – large domain interfaces following overlap of subunit pairs by superposition on the large domain of the second subunit. In all cases, the surface formed by residues M348, I351, and W388 of the first subunit contacts the surface formed by residues L151, F155, L158, F159, and R163 of the second subunit.
Similarity between the small domain - large domain interfaces and the major contacts in Vps4 crystal structures.
The cryo-EM AB (same as BC, CD, DE, FA) interface and the EF interface are overlapped. Major contacts in crystal structures of archaeal Vps4 proteins overlap closely with the AB interface. Major contacts in crystal structures of eukaryotic Vps4 proteins overlap with the EF interface, except for two of the mouse Vps4 contacts, which are intermediate between the AB and EF interfaces.
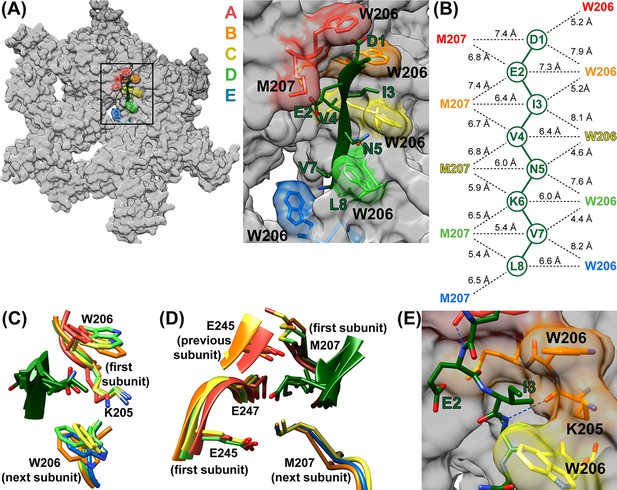
ESCRT-III peptide conformation and coordination.
(A) Left – tilted view of a surface representation showing how the pore loop residues form an array of class I and II binding pockets through the hexamer pore. W206 and M207 from subunits A-E are highlighted. Right – close up of the pore region. (B) Distances between Cα atoms of the peptide and pore loop 1 W206 and M207 indicate equivalent binding in the different class I and class II pockets. (C) Superposition of the four Class I pockets following superposition on Cα atoms of the class I pocket residues of subunits A and B. (D) Superposition of the four class II pockets following superposition on Cα atoms of the class II pocket residues of subunits A and B. (E) The H-bond seen between the NH of even-numbered ESCRT-III residues and the K205 CO of Vps4 subunits A-D – here centered on the bond between ESCRT-III V4 and subunit B. The bond between E2 and subunit A is also visible.
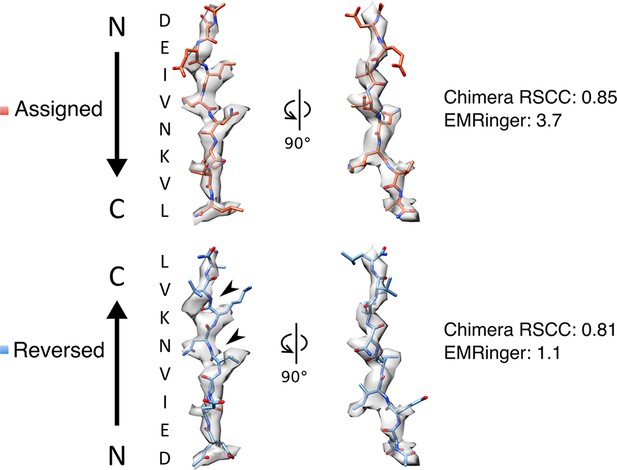
Fit of peptide to density when refined in the assigned and reversed orientations.
Visual inspection shows that the assigned peptide orientation is a better fit to the map than the inverted orientation. Arrowheads indicate notably poor agreement between model and map in the inverted orientation. Chimera RSCC and EMRinger scores also support the assigned orientation.
Charge density map at the ESCRT-III peptide and residues of pore loop 1 and 2.
https://doi.org/10.7554/eLife.31324.022The ESCRT-III peptide spirals around the helix axis.
https://doi.org/10.7554/eLife.31324.023Class I side chain binding pockets.
https://doi.org/10.7554/eLife.31324.024Class II side chain binding pockets.
https://doi.org/10.7554/eLife.31324.025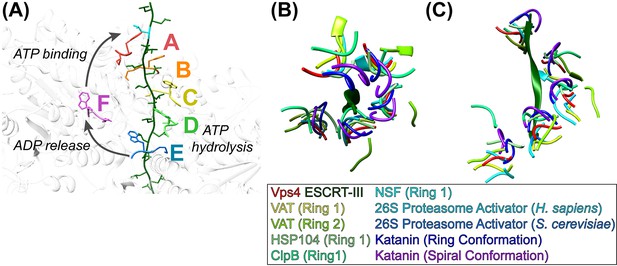
Mechanism of translocation.
(A) Proposed mechanism of ESCRT-III translocation by Vps4. W206 and M207 residues of the six Vps4 subunits are shown, with the peptide passing through the Vps4 hexamer. The peptide model was constructed by changing the side chains to leucine without adjusting the main chain, and building out in the N and C directions by overlapping copies of the peptide model. The proposed mechanism envisions that Vps4 progresses through states A to E while bound to successive dipeptides of its substrate. ATP hydrolysis at subunit D destabilizes the DE interface and promotes displacement of subunit E toward the transitioning subunit F configuration, which allows displacement of ADP. Subsequent ATP binding allows subunit F to pack against subunit A, bind to the next dipeptide of ESCRT-III, and assume the subunit A configuration. (BC) Conservation of helical pore loop structure in AAA ATPases. Overlap on the large ATPases of multiple AAA ATPase structures gives a similar helical arrangement of pore loop 1 residues from five subunits. (B) Top and (C) side views are shown of the ESCRT-III peptide (green) and Vps4 pore loop 1 (red) with the equivalent residues of: VAT (Ripstein et al., 2017) (pdbid 5vca), HSP104 (Gates et al., 2017) (5vjh), NSF (Zhao et al., 2015) (3j94), human 26S proteasome (Huang et al., 2016) (5gjr), yeast 26S proteasome (Wehmer et al., 2017) (5mp9), katanin (Zehr et al., 2017) (5wc0, 5wcb).
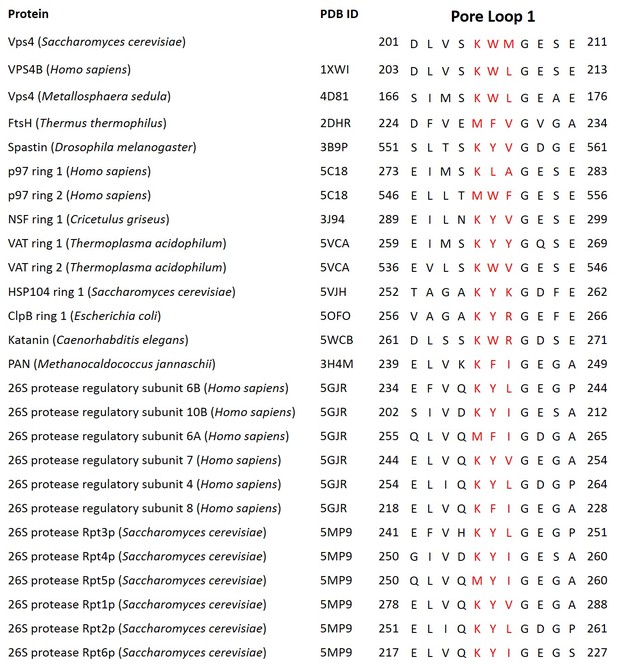
Structure-based alignment of pore loop 1 sequences.
Pore loop 1 residues that contact the ESCRT-III peptide (red font) are shown with four flanking residues on either side.
Model of translocation.
Translocation of a peptide modeled by interpolation between the six Vps4 subunit states seen in the cryo-EM structure. The pore loop 1 W206 and M207 side chains are highlighted. The peptide residue highlighted in cyan corresponds to the odd-numbered residue that will be bound in the class 1 pocket that will be formed as subunit F packs against subunit A. The transitioning subunit (initially F) moves between the extreme positions apparent from focused classification without change in the other subunits. Other changes are coordinated in a linear interpolation with all parts of the complex, and imply that two Vps4 subunits are disengaged from substrate for much of the transition.
Comparison with other classic clade AAA+ ATPases.
All structures are superimposed on the large ATPase domains of the five most helical subunits. Subsequent positions of pore loop 1 residues are shown for all six subunits in each structure, and then for the 5 most helical subunits.
Tables
Reconstruction, Refinement, and Model Statistics of Vps4.
https://doi.org/10.7554/eLife.31324.008| Reconstruction | |
|---|---|
| Particle images | 82,225 |
| Resolution (0.143 FSC) (Å) | 3.2 |
| Map sharpening B-factor (Å2) | −125 |
| EMDB accession number | EMD-8887 |
| Model refinement and validation of Vps4 subunits A-E | |
| PDB accession number | 6BMF |
| Resolution used for refinement (Å) | 3.2 |
| Number of atoms | 11033 |
| RMSD | |
| Bond length (Å) | 0.01 |
| Bond angles (°) | 0.18 |
| Ramachandran | |
| Favored (%) | 89.5 |
| Allowed (%) | 10.5 |
| Outlier (%) | 0 |
| Validation scores | |
| Molprobity score/percentile (%) | 1.83 (100%) |
| Clashscore/percentile (%) | 5.02 (100%) |
| EMRinger score | 2.04 |
Additional files
-
Supplementary file 1
MolProbity report.
This is for the parts of the model that were defined in charge density at a resolution that justified refinement (Subunits A-E, nucleotides, ESCRT-III peptide). Data in Table 1 are based on this report.
- https://doi.org/10.7554/eLife.31324.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31324.031





