Topoisomerase VI senses and exploits both DNA crossings and bends to facilitate strand passage
Figures
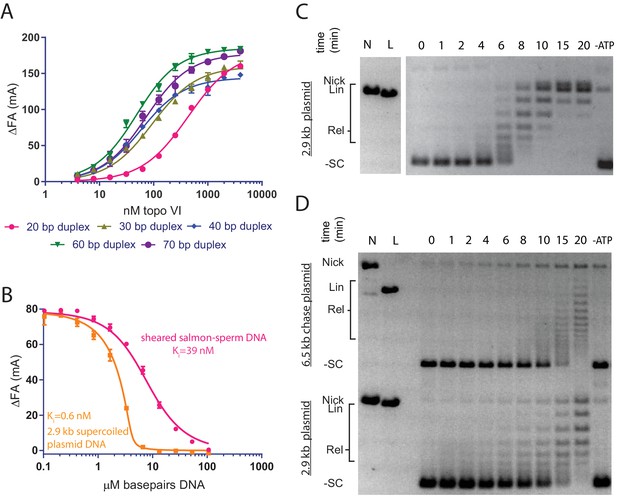
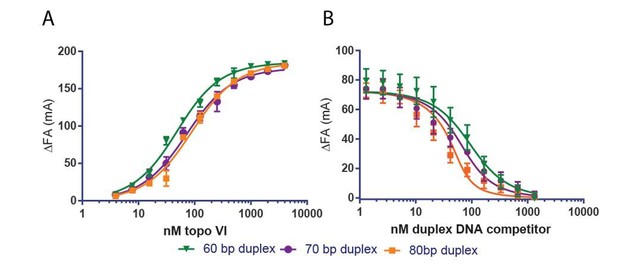
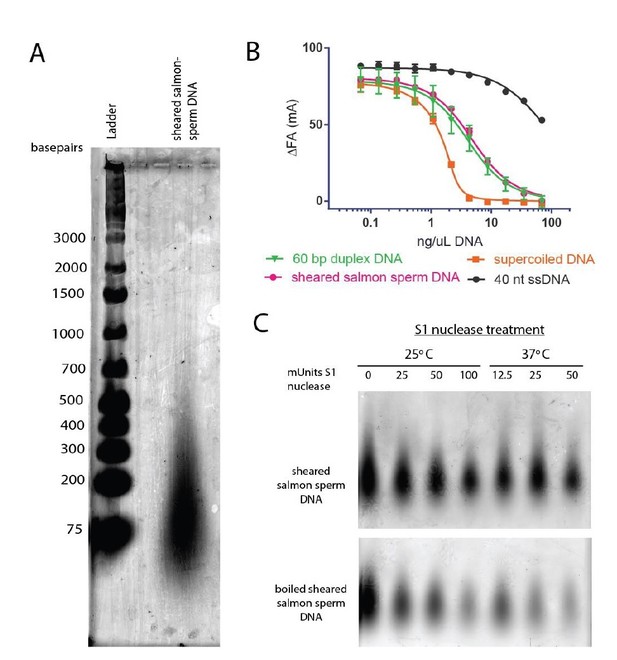
Topo VI binds longer duplexes and preferentially engages features of supercoiled DNA.
(A) Binding of a 20, 30, 40, 60 or 70 bp fluorescein-labeled duplex (20 nM, sequences in Figure 1—source data 1) to topo VI, observed as a change in fluorescence anisotropy (ΔFA) measured in milli-anisotropy units (mA) as a function of enzyme concentration. Points and error bars correspond to the mean and standard deviation of three independent experiments. Curves represent fits to a single-site ligand depletion binding model. Apparent dissociation constants are reported in Figure 1—source data 2. (B) Fluorescence anisotropy experiment assessing the ability of supercoiled DNA and sheared salmon-sperm DNA to compete a fluorescein-labeled 70 bp duplex (20 nM duplex, 1.4 μM bp) from topo VI (100 nM). Non-labeled DNA was titrated from 0.1 μM bp to 106.5 μM bp and competition was observed as a change in fluorescence anisotropy (ΔFA) as measured in milli-anisotropy units (mA). Data are plotted as a function of the base-pair concentration (μM) of competing DNA. Points and error bars correspond to the mean and standard deviation of three independent experiments. Curves represent a fit to a competitive displacement model. Numerical data for (A–B) are reported in Figure 1—source data 3. (C–D) Test of processive supercoil relaxation by topo VI on negatively supercoiled plasmid DNA. Topo VI was pre-incubated in a 1:1.4 ratio to a 2.9 kb negatively supercoiled plasmid (6.7 ng/μL in assay). Reactions were started by addition of either (C) ATP or (D) ATP and a 6.5 kb ‘chase’ plasmid (6.7 ng/μL in assay) to compete for unbound enzyme. Samples were quenched at 0, 1, 2, 4, 6, 8, 10, 15, and 20 min. Each condition was also incubated without ATP for 20 min as a negative control. Plasmid size and topoisomer species are indicated to the left of each gel. For an example of processive supercoil relaxation by a type II topoisomerase, see Figure 1—figure supplement 1.
-
Figure 1—source data 1
Oligonucleotides used for fluorescence anisotropy and FRET experiments.
- https://doi.org/10.7554/eLife.31724.005
-
Figure 1—source data 2
Binding affinities of topo VI for different length duplexes.
- https://doi.org/10.7554/eLife.31724.006
-
Figure 1—source data 3
Numerical data associated with Figure 1.
- https://doi.org/10.7554/eLife.31724.007
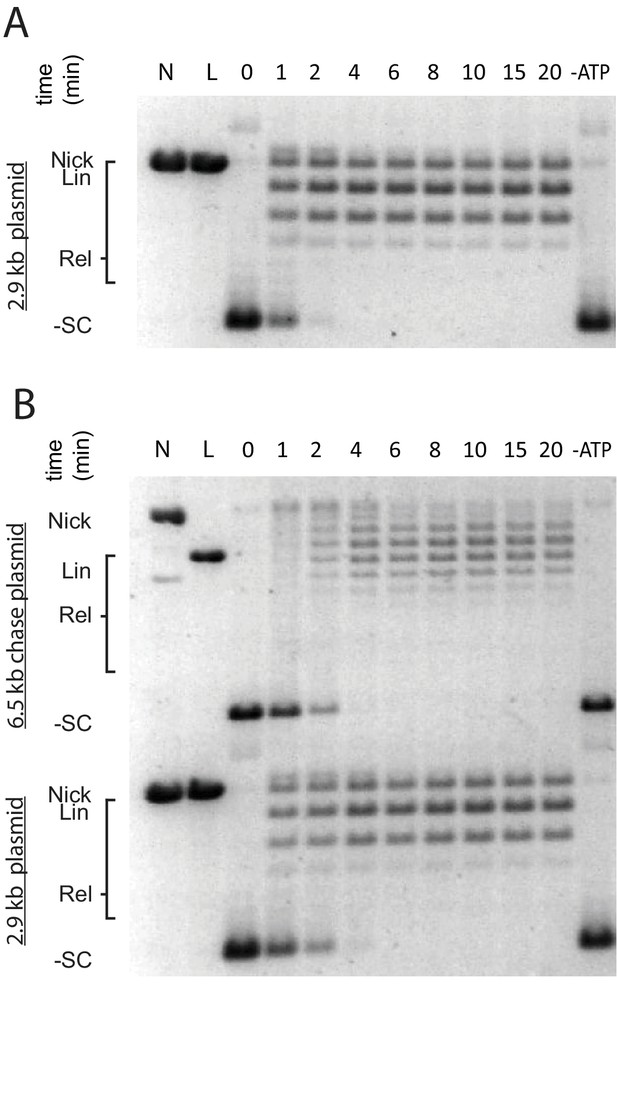
Topo II processively relaxes supercoiled DNA as compared to topo VI.
An example of processive supercoil relaxation by ScTop2ΔCTR, as compared to topo VI in Figure 1C–D. ScTop2ΔCTR was pre-incubated in a 1:1.4 ratio to a 2.9 kb negatively supercoiled plasmid (6.7 ng/uL in assay). Reactions were started by addition of either (A) ATP or (B) ATP and a 6.5 kb ‘chase’ plasmid (6.7 ng/uL in assay) to compete for unbound enzyme. Samples were quenched at 0, 1, 2, 4, 6, 8, 10, 15, and 20 min. Each condition was also incubated without ATP for 20 min as a negative control. Topoisomer species and identification of each plasmid are indicated on the left of each gel.
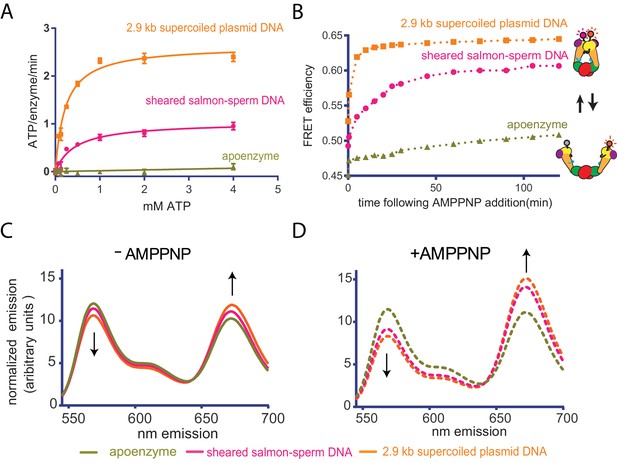
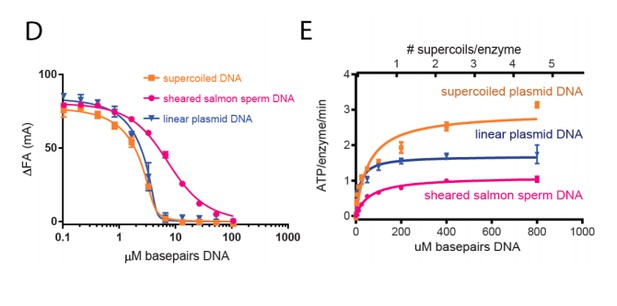
Top6B dimerization and ATP hydrolysis activity are stimulated by supercoiled DNA.
(A) Rate of steady-state ATP hydrolysis catalyzed by topo VI alone, topo VI incubated with 400 μM basepairs of linear sheared salmon-sperm DNA (800:1 basepair to enzyme ratio), or topo VI incubated with 400 μM basepairs of 2.9 kb supercoiled plasmid DNA (800:1 basepair to enzyme ratio) as a function of ATP concentration. Rates were determined spectroscopically using an NADH-coupled assay. Data represent hydrolysis rates after subtracting a small contribution of non-specific ATPase activity from assays performed with an ATPase-deficient topo VI construct (Figure 2—figure supplement 1). Points and bars correspond to the mean and standard error of the mean of three independent experiments. Curves represent fits to a Michealis-Menten kinetics model reported in Figure 2—source data 1. (B) The ratiometric FRET efficiency of Alexa555/Alexa647-labeled topo VIcyslite-155C (see Figure 2—figure supplement 2) was monitored over time following the addition of AMPPNP for enzyme alone, enzyme bound to supercoiled DNA, or enzyme bound to short linear (sheared salmon-sperm) DNA. Numerical data for (A–B) are reported in Figure 2—source data 2. (C–D) Example fluorescence emission spectra produced by 530 nm excitation of Alexa555/Alexa647-labeled topo VIcyslite-155C assessing the conformation of the Top6B ATPase domain in the absence of nucleotide ((C), solid lines) and 120 min following addition of AMPPNP ((D), dashed lines). Spectral emission was normalized by total emission from 545 nm to 700 nm. This behavior contrasts that of a model type IIA topoisomerase, ScTop2 (see Figure 2—figure supplement 3).
-
Figure 2—source data 1
Apparent kinetic parameters for ATP hydrolysis by topo VI.
- https://doi.org/10.7554/eLife.31724.012
-
Figure 2—source data 2
Numerical data associated with Figure 2.
- https://doi.org/10.7554/eLife.31724.013
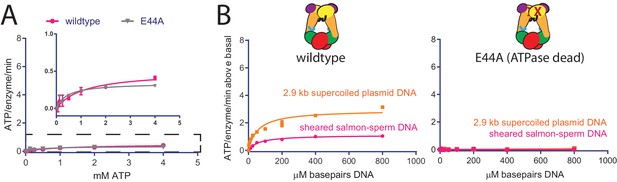
Determination of contaminating ATPase activity levels present in topo VI preparations using a hydrolysis-deficient mutant.
(A) Rate of steady-state ATP hydrolysis measured as a function of ATP concentration for topo VI alone and an ATPase-deficient construct of topo VI (Top6BE44A). Rates were determined spectroscopically using an NADH-coupled assay (Materials and methods). Points and error bars correspond to the mean and standard deviation of three independent experiments. Measured rates for Top6BE44A were used to estimate the topo VI-specific ATPase rates presented in Figure 2A. (B) Stimulation of ATP hydrolysis above basal levels catalyzed by wildtype topo VI (left) or the Top6BE44A ATPase-deficient construct (right) as a function of the basepair concentration (µM) of sheared salmon-sperm, DNA (pink), or a 2.9 kb supercoiled plasmid (orange). ATP was held at 2 mM, and rates were determined spectroscopically using an NADH coupled assay. Points and error bars correspond to the mean and standard deviation of three independent experiments. Numerical data for (A–B) are reported in Figure 2—source data 1.
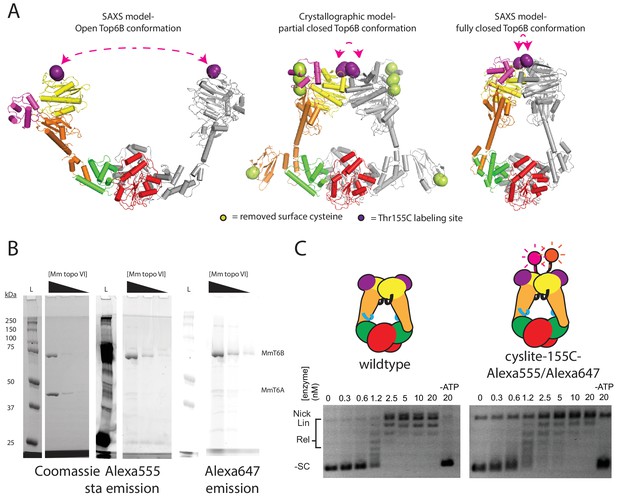
Design and production of a FRET pair-labeled topo VI to report on B-subunit conformation.
(A) Three different conformations of the topo VI holoenzyme, an open SAXS model of S. shibatae topo VI, a partially closed crystallographic model of M. mazei topo VI, and a fully closed SAXS model of Ss topo VI are shown with Thr155 displayed in purple sticks and the distance between Thr155 dimer pairs shown by pink dashed arrows. Native cysteine residues mutated to alanine or serine residues to remove off-target labeling sites are shown as yellow-green sticks on the Mm topo VI crystal structure. (B) Purified topo VIcyslite-155C-labeled with Alexa555 and Alexa647, separated on SDS-PAGE and stained for protein with coomassie blue (left) after scanning for Alexa555 emission (middle), and Alexa647 emission (right). A dilution series of enzyme (1 μg, 0.1 μg, 0.01 μg) was run, along with a standards ladder (lane marked L, molecular weights in kilodaltons labeled on left). Positions of MmTop6B and MmTop6A are labeled on the right. (C) Supercoil relaxation activity of FRET pair-labeled topo VI compared to wild type as a function of enzyme concentration. For the enzyme titrations (0.3–20 nM in two-fold steps, marked above), each experiment proceeded for 30 min prior to quenching and contained 3.5 nM plasmid (10.2 μM bp DNA). Topoisomer products were separated on agarose gel with the position of supercoiled and relaxed topoisomers marked on the left. Cartoons denote the construct used in each activity assay. This enzyme was used for Top6B dimerization experiments reported in Figure 2B–D.
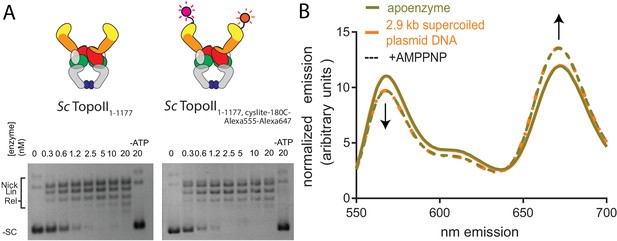
AMPPNP is sufficient to close the ATP gate of ScTop2.
(A) Supercoil relaxation activity of the FRET pair-labeled ScTop2ΔCTR-cyslite-180C was compared to wild type as a function of enzyme concentration. For the enzyme titrations (0.3–20 nM in two-fold steps, marked above), each experiment proceeded for 30 min prior to quenching and contained 3.5 nM plasmid (10.2 μM bp DNA). Topoisomer products were separated on agarose gel with the position of supercoiled and relaxed topoisomers marked on the left. Cartoons denote the construct used. (B) Fluorescence emission spectra produced by 530 nm excitation of Alexa555/Alexa647-labeled ScTop2ΔCTR-cyslite-180C show that in contrast to the behavior of topo VI reported in Figure 2B–D, addition of AMPPNP (dashed line spectra), is sufficient to dimerize the ATP gate as assessed by the relative increase in acceptor emission and decrease in donor emission as compared to controls without nucleotide (solid line spectra). Presence (orange spectra) or absence (goldenrod spectra) of supercoiled DNA does not appreciably modify the conformational state of the ATPase domain as accessed by FRET. Spectral emission was normalized by total emission from 545 nm to 700 nm.
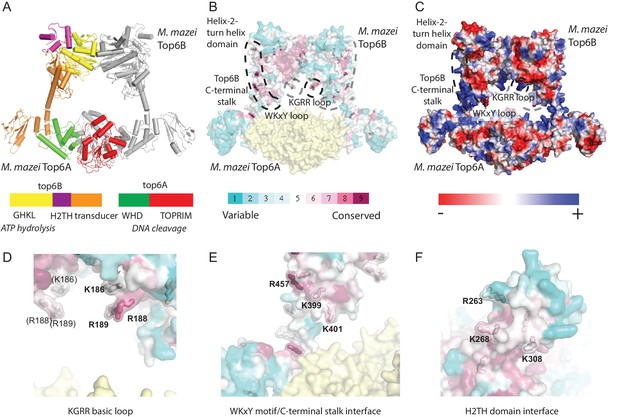
Identification of potential DNA-binding elements in Top6B.
(A) Primary and tertiary structure [Protein Data Bank (PDB) ID: 2Q2E] of the M. mazei topo VI heterotetramer. Domains for one Top6A-Top6B heterodimer are colored as shown in the primary structure and the partner Top6A-Top6B heterodimer is shown in grey. Catalytic function is denoted in italics under primary structure. (B) Mapping of sequence conservation in Top6B based on a PSI-BLAST multiple sequence alignment. Conserved surface-exposed arginine and lysine residues (ConSurf score of ≥6) are shown as sticks. Coloration from cyan to magenta denotes variable to conserved. Top6A is represented in yellow. (C) Electrostatic surface representation of topo VI. A conserved basic loop in the T-segment storage cavity, and a conserved basic interface stretching from the WKxY motif and C-terminal stalk of Top6B to the Helix-2-turn-helix (H2TH) domain are labeled. (D) View of the KGRR basic loop motif. (E) View of the C-terminal stalk/WKxY interface. (F) View of the H2TH DNA-binding interface, rotated 90° towards the point of view as compared to A–C. See Figure 3—figure supplement 1 for a more detailed rationale for the functional importance of this interface. In (D–F), residues mutated to alanine or to glutamate for functional studies are labeled. Mutations to these interfaces produced well-behaved functional mutants (see Figure 3—figure supplement 2).
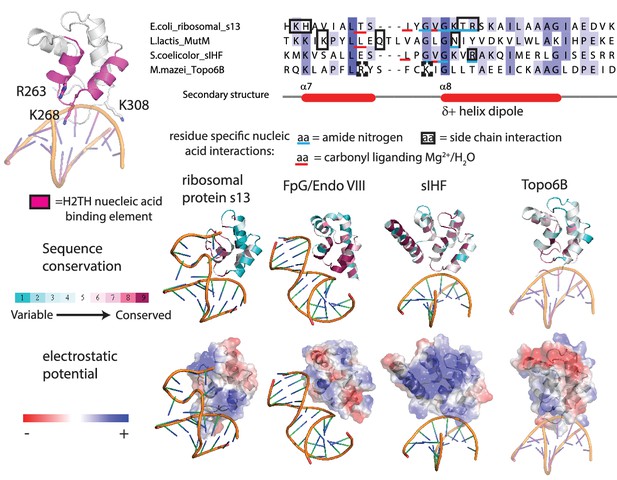
Comparison between H2TH domain homologs predicts a DNA-binding interface in Top6B.
Representation of the H2TH domain with the nucleic-acid-binding element colored pink, potential DNA-binding residues labeled, and DNA model based on a DNA-bound sIHF structure (top left). Structural comparison reveals ribosomal s13 (PDB ID = 4YBB), MutM (PDB ID = 1TDZ) and sIHF (PDB ID = 4ITQ) bind DNA or RNA through interactions with main chain amide nitrogens (blue underline) or bridging H2O or Mg2+ liganded by main chain carbonyl oxygens (red underline). Side chain interactions (black box) are poorly conserved (top right). Nevertheless, the nucleic-acid-binding interface (middle) and electrostatic surface (bottom) is maintained within each family and aided identification of the H2TH point substitutions shown in Figure 3F.
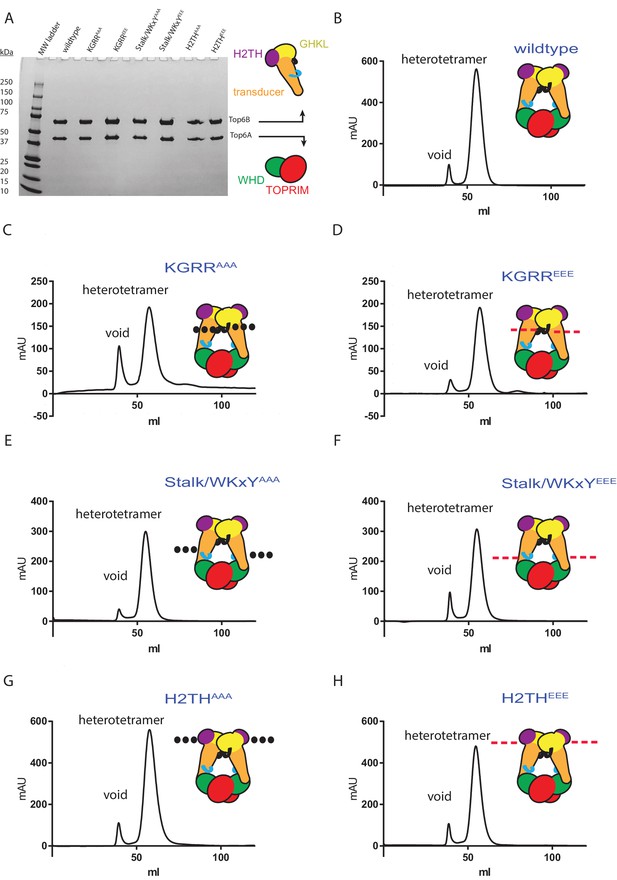
Topo VI functional mutants show similar solution properties to wild-type enzyme as judged by gel filtration.
(A) Purity of constructs used in this study (Figures 1–2 and 4–7) was assessed by running ~4 μg of each construct on SDS-PAGE (4%–20% gradient) and staining with coomassie blue. Molecular weight standards (left) and subunit identity (right) are labeled. (B–H) Size exclusion chromatographs for wild type (B), KGRRAAA (C), KGRREEE (D), Stalk/WKxYAAA (E), Stalk/WKxYEEE (F), H2THAAA (G), and H2THEEE (H) topo VI constructs. Void and heterotetramer peaks are noted for each chromatograph. The placement and nature of mutations in each construct are depicted in the cartoons above each chromatograph (‘•••' - AAA; ‘---' – EEE). Each protein preparation gives rise to a monodisperse peak eluting at the same expected volume.
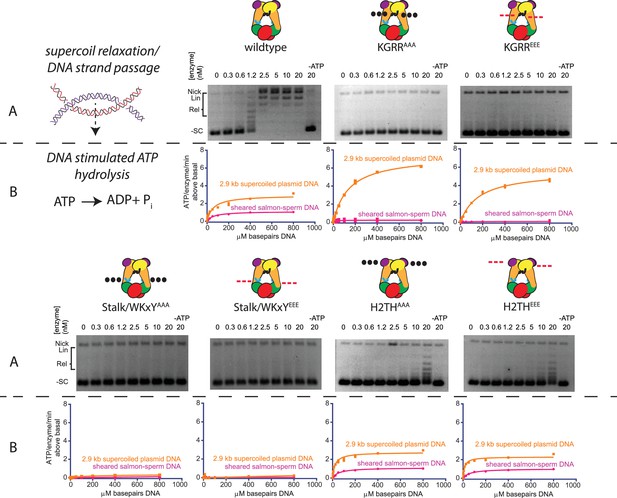
Effect of neutralization and charge reversal mutations to the KGRR loop, Stalk/WKxY region, or H2TH DNA-binding interface on supercoil relaxation activity and ATP hydrolysis by topo VI.
(A) Activity of mutant topo VI constructs for relaxing supercoiled DNA compared to wild type as a function of enzyme concentration. For the enzyme titrations (0.3–20 nM in two-fold steps), each assay proceeded for 30 min prior to quenching with EDTA and SDS and contained 3.5 nM plasmid (10.2 μM bp DNA). Similar behavior for each mutant may be observed by timecourse (Figure 4—figure supplement 1). The placement and nature of mutations in each construct are depicted in the cartoons above each titration (‘•••' - AAA; ‘- - -' – EEE). (B) The rate of steady-state ATP hydrolysis above basal levels (Figure 4—figure supplement 2) catalyzed by wild-type topo VI compared to mutant topo VI constructs, plotted as a function of the basepair concentration (µM) of sheared salmon-sperm DNA (pink), or a 2.9 kb supercoiled plasmid DNA (orange). ATP was held at 2 mM, and rates were determined spectroscopically using an NADH-coupled assay. Points and bars correspond to the mean and standard deviation of three independent experiments. Curves represent a fit to a Michealis-Menten type kinetics model reported in Figure 4—source data 1. Numerical data are reported in Figure 4—source data 2.
-
Figure 4—source data 1
Kinetic parameters for DNA-dependent stimulation of topo VI ATPase activity.
- https://doi.org/10.7554/eLife.31724.020
-
Figure 4—source data 2
Numerical data associated with Figure 4.
- https://doi.org/10.7554/eLife.31724.021
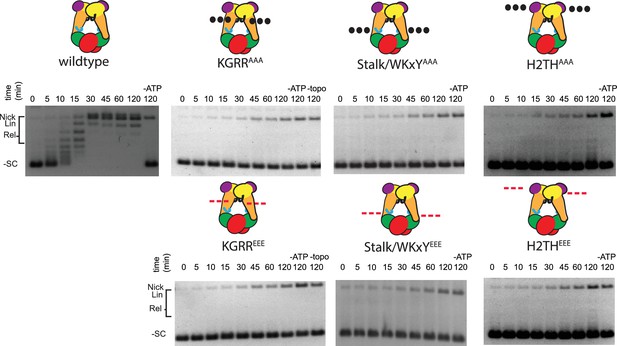
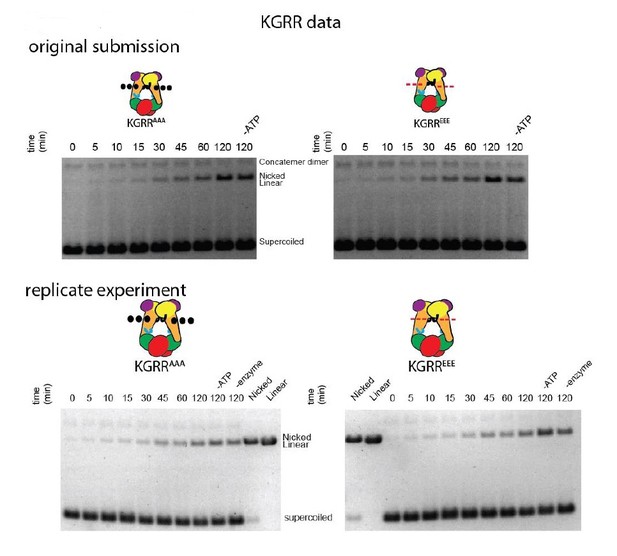
Supercoil relaxation activity of topo VI mutants as a function of time.
Strand passage activity of mutant topo VI constructs for relaxing supercoiled DNA as compared to wild type are shown as a function of time and in relation to enzyme titrations reported in Figure 4A. In each time course experiment, 2.5 nM of enzyme processed 3.5 nM of negatively supercoiled plasmid (10.2 μM bp) and was quenched with EDTA and SDS at the indicated time (0–120 min). A no-ATP control (-ATP) was taken after a 120 min incubation period for each variant. A no-enzyme control (-topo) was also taken and run with the timecourse experiments for the KGRR constructs. The placement and nature of mutations in each construct are depicted in the cartoons above each timecourse (‘•••' - AAA; ‘---' – EEE).
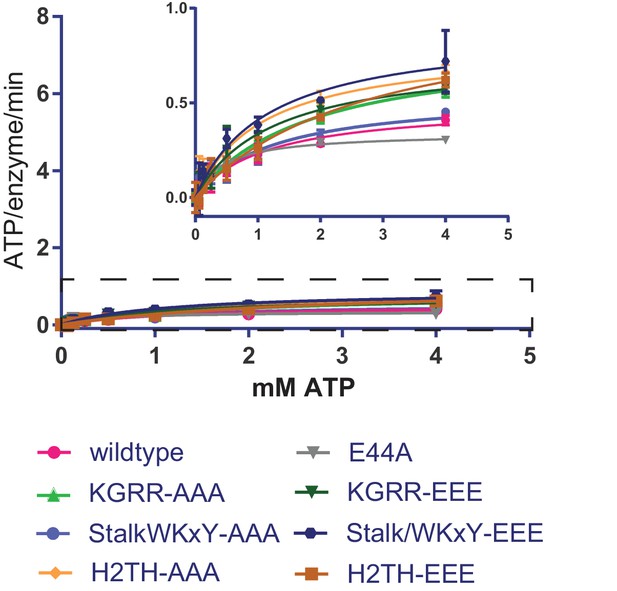
Comparison of basal ATPase activities for topo VI mutants.
Rate of steady-state ATP hydrolysis measured as a function of ATP concentration for each topo VI mutant in the absence of DNA, as related to Figure 4B. Rates were determined spectroscopically using an NADH-coupled assay. Points and error bars correspond to the mean and standard deviation of three independent experiments. Numerical data are reported in Figure 4—source data 2.
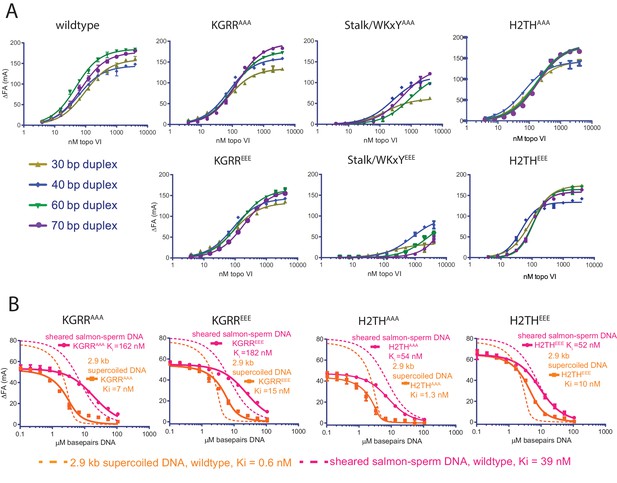
Effect of neutralization and charge reversal mutations to Top6B on DNA binding affinity and preferential engagement of supercoiled DNA.
(A) Binding of a 30, 40, 60, or 70 bp fluorescein-labeled duplex (20 nM) to topo VI mutant constructs. Binding was observed as a change in fluorescence anisotropy (ΔFA) and measured in milli-anisotropy units (mA) as a function of enzyme concentration. Points and error bars correspond to the mean and standard deviation of three independent experiments. For the H2THEEE mutant, curves represent fits to a Hill-type cooperative binding model. All other curves represent fits to a single site ligand depletion binding model. Binding isotherms for the wildtype enzyme are reproduced from Figure 1A for reference. Apparent dissociation constants are reported in Figure 5—source data 1. (B) Binding assay assessing the ability of supercoiled DNA and sheared salmon-sperm DNA to compete a fluorescein-labeled 70 bp duplex (20 nM duplex, 0.14 μM bp) from 100 nM H2THAAA, H2THEEE, KGRRAAA or KGRREEE, topo VI enzyme. Non-labeled DNA was titrated from 0.1 μM bp to 106.5 μM bp with competition observed as a change in fluorescence anisotropy (ΔFA) measured in milli-anisotropy units (mA). Data are plotted as a function of the basepair concentration (µM) of competitor DNA. Points and error bars correspond to the mean and standard deviation of three independent experiments. Curves represent a fit to an explicit competitive displacement model (Figure 5—source data 2). Dashed curves corresponding to the competitive binding data for wildtype enzyme (Figure 1B) are shown for reference. Numerical data are reported in Figure 5—source data 3.
-
Figure 5—source data 1
Binding affinities of Top6B mutants for different length duplexes.
- https://doi.org/10.7554/eLife.31724.024
-
Figure 5—source data 2
Affinities of H2TH and KGRR mutants for supercoiled and sheared salmon-sperm DNA as compared to wild type presented in Figure 1.
- https://doi.org/10.7554/eLife.31724.025
-
Figure 5—source data 3
Numerical data associated with Figure 5.
- https://doi.org/10.7554/eLife.31724.026
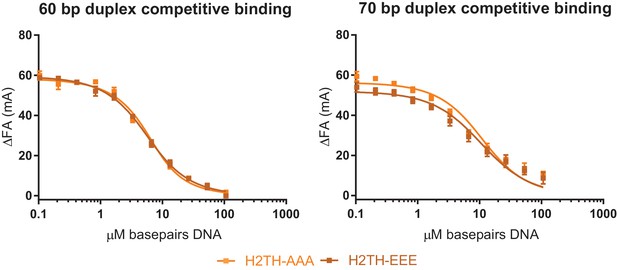
Affinities of H2THAAA and H2THEEE topo VI mutants for short, defined duplexes as determined by competitive binding.
Experiments assessing the ability of an unlabeled 60 bp duplex (left) or unlabeled 70 bp duplex (right) to compete with a fluorescein-labeled 70 bp duplex (20 nM duplex, 0.14 μM bp) for binding to 100 nM H2THAAA (light orange) or 100 nM H2THEEE (dark orange) topo VI, as related to Figure 5A. Non-labeled DNA was titrated from 0.1 μM bp to 106.5 μM bp with competition observed as a change in fluorescence anisotropy (ΔFA) measured in milli-anisotropy units (mA). Data are plotted as a function of the basepair concentration (µM) of competitor DNA. Points and bars correspond to the mean and standard deviation of three independent experiments. Curves represent a fit to an explicit competitive displacement model. Numerical data are reported in Figure 5—source data 3.
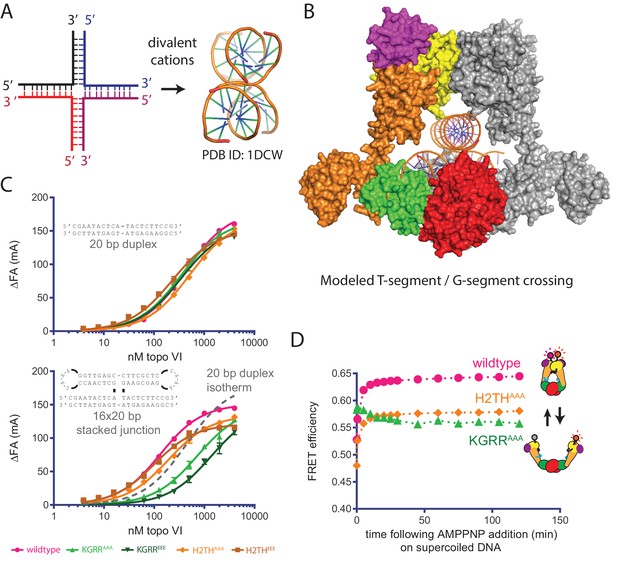
Effect of neutralization and charge reversal mutations to the KGRR loop or H2TH domain on DNA crossing affinity and Top6B dimerization.
(A) A four-way junction folds into a stacked-X structure in the presence of divalent cations (PDB ID 1DCW) (Eichman et al., 2000). (B) Modeling of a prospective G-segment and T-segment DNA into a previously published structure of M. mazei topo VI (PDB ID 2Q2E)(Corbett et al., 2007). Domains are colored as in Figure 3A. The juxtaposition of the two DNAs in this intermediate closely mimic the stacked-X junction structure in Figure 6A. (C) Binding of a 20 bp fluorescein-labeled duplex (top) or 20 bp by 16 bp fluorescein-labeled stacked junction substrate (bottom, both 20 nM) to topo VI or mutant constructs. Binding was observed as a change in fluorescence anisotropy (ΔFA) and measured in milli-anisotropy units (mA) as a function of enzyme concentration. Points and bars correspond to the mean and error of three independent experiments. Curves represent fits to a single site ligand depletion binding model. In the plot of the enzyme-stacked junction binding isotherms, the fit of wildtype topo VI binding to the 20 bp duplex is displayed (---) for reference. Apparent dissociation constants are reported in Figure 6—source data 1. (D) Change in ratiometric FRET efficiency for the indicated Alexa555/647-labeled topo VI constructs incubated with supercoiled DNA was monitored over time following the addition of AMPPNP. As further detailed in Figure 6—figure supplement 1, incubation with supercoiled DNA alone increases the FRET efficiency for each construct. Numerical data are reported in Figure 6—source data 2.
-
Figure 6—source data 1
Affinities of wildtype, H2TH and KGRR mutants for stacked junction DNA.
- https://doi.org/10.7554/eLife.31724.029
-
Figure 6—source data 2
Numerical data associated with Figure 6.
- https://doi.org/10.7554/eLife.31724.030
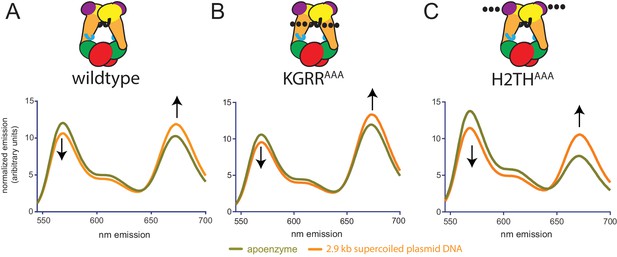
Conformational response of H2THAAA and KGRRAAA constructs on different substrates as determined by FRET in the absence of nucleotide.
Comparison of the fluorescence emission spectra produced by 530 nm excitation of Alexa555/Alexa647-labeled (A) TopoVIcyslite-155C, (B) KGRRAAA, cyslite-155C and (C) H2THAAA, cyslite-155C alone and on supercoiled DNA, as related to Figure 6D. For all three constructs, supercoiled DNA modulates the conformation of the Topo6B ATPase domain in the absence of nucleotide as assessed by FRET. Spectral emission was normalized by total emission from 545 nm to 700 nm.
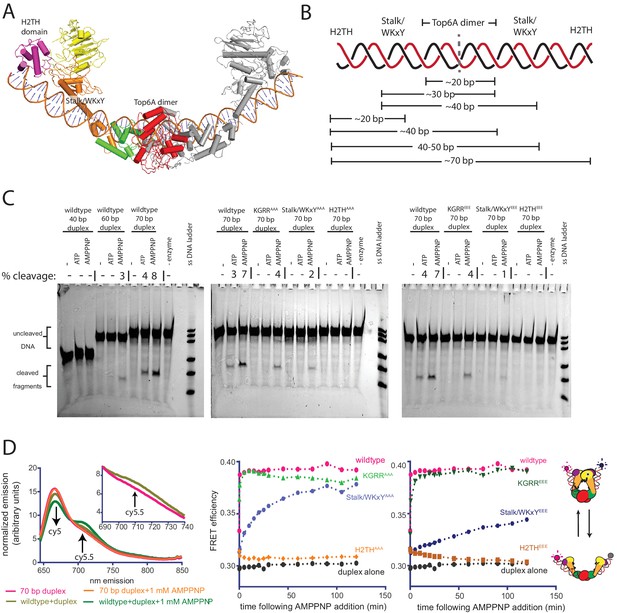
Topo VI requires H2TH-mediated, nucleotide-dependent bending of a 70 bp duplex G-segment to induce cleavage.
(A) Model of a 70 bp bent duplex which spans dimer-related H2TH domains through the TOPRIM/Winged Helix Domain cleavage site of Top6A (using a previously published SAXS model of S. shibatae topo VI with Top6B in an open conformation (Corbett et al., 2007) see also PDB ID: 2ZBK for a similar conformation, stabilized by the inhibitor radicicol (Graille et al., 2008)). Domains are colored as in Figure 3A. DNA was modeled with a continuous bend using web 3DNA (Zheng et al., 2009). (B) Schematic of the estimated duplex lengths needed to span across the H2TH, Stalk/WKxY, and Top6A dimer DNA-binding regions, using the G-segment path modeled in Figure 7A. Note that the Stalk/WKxY region may allow for the asymmetric binding of DNA in different registers, accounting for the jump in affinity seen between 20 and 30 bp DNA duplexes. (C) Nucleotide-dependent cleavage of fluorescein-labeled DNA duplexes by topo VI and mutant constructs. Length-dependent cleavage by wildtype (left), cleavage of a 70 bp duplex by basic-to-neutral mutants (middle), and cleavage by basic-to-acidic mutants (right) was tested. Cleavage reactions containing a 2:1 ratio of enzyme:duplex were run on denaturing PAGE to separate reaction products, and were visualized using a laser gel scanner. Enzyme construct (wildtype, KGRRAAA, KGRREEE, Stalk/WKxYAAA, Stalk/WKxYEEE, H2THAAA, or H2THEEE), duplex length (40 bp, 60 bp, or 70 bp), and addition of 1 mM ATP or 1 mM AMPPNP is noted above each lane. A no enzyme control containing 1 mM AMPPNP and a single strand DNA ladder consisting of 20, 30, 40, 60, 70, and 80 nt oligonucleotides were run for reference. Where present, the percentage of cleavage product relative to intact DNA is quantified above the lane. (D) Nucleotide-dependent bending of a Cy5/Cy5.5-labeled 70 bp duplex was assessed using bulk FRET. Fluorescence emission spectra (left) produced by 630 nm excitation of the Cy5-Cy5.5-labeled DNA show an increase in cy5.5 emission in the presence of topo VI (left, inset) and AMPPNP, but not in the presence of AMPPNP alone. Spectral emission was normalized by total emission from 645 nm to 850 nm. Ratiometric FRET efficiency was monitored over time upon addition of AMPPNP for the noted basic-to-neutral mutant (middle) or basic-to-acidic topo VI mutant (right). Wildtype and duplex alone are shown in each case for comparison. Figure 7—figure supplement 1 confirms FRET changes arise from DNA bending. Figure 7—figure supplement 2 further considers the gate closure activity of KGRRAAA on the 70 bp duplex substrate. Numerical data are reported in Figure 7—source data 1.
-
Figure 7—source data 1
Numerical data associated with Figure 7.
- https://doi.org/10.7554/eLife.31724.034
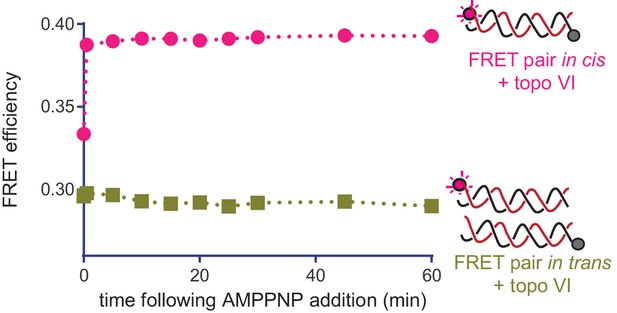
The AMPPNP-dependent FRET increase observed for a labeled 70 bp duplex arises from DNA bending by topo VI.
Change in the ratiometric FRET efficiency between Cy5 and Cy5.5 fluorophores placed on opposite ends of the same 70 bp duplex (pink) compared to two separate duplexes (goldenrod) in the presence of a 2:1 molar excess of topo VI, followed as a function of time after the addition of AMPPNP. An increase in FRET is only observed when the fluorophores are on the same DNA, verifying the FRET responses reported in Figure 7D reflect intramolecular bending, rather than the simultaneous capture of two duplexes. Numerical data are reported in Figure 7—source data 1.
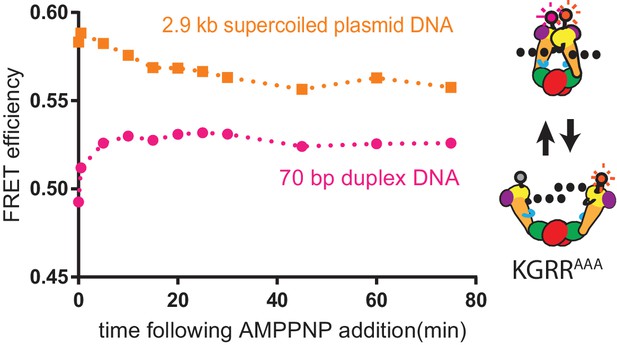
AMPPNP-dependent conformational response of KGRRAAA in bending assay conditions bound to excess 70 bp duplex or supercoiled DNA.
Change in ratiometric FRET efficiency for the Alexa555/647-labeled KGRRAAA construct incubated with either excess 70 bp duplex or supercoiled DNA was monitored over time after the addition of AMPPNP. AMPPNP promotes Top6B dimerization when KGRRAAA is bound to the 70 bp duplex, but does not stabilize gate closure to the extent of supercoiled DNA alone, reconciling the Top6B dimerization (Figure 6B) and bending assay (Figure 7D) data for this mutant. Numerical data are reported in Figure 7—source data 1.
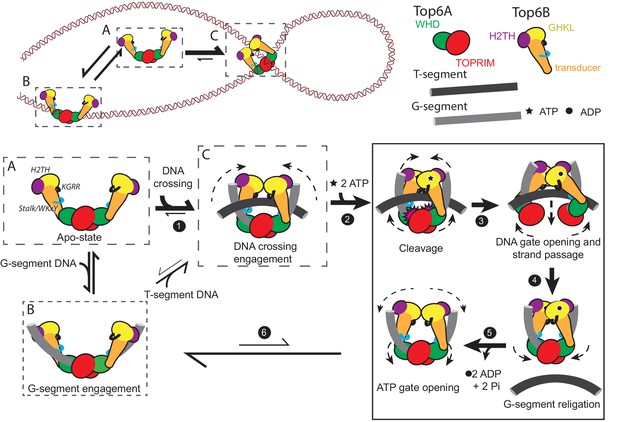
A new model for the Type IIB topoisomerase catalytic cycle.
Free topo VI (A) binds to linear DNA (B), but preferentially engages DNA crossings (C). Binding to a hooked DNA crossing (C) by the KGRR loop and Stalk/WKxY region (step 1) induces a conformational change that presets Top6B dimerization. From this state ATP binding (step 2) introduces H2TH-dependent G-segment DNA bending and shifts the catalytic tyrosines on the Winged Helix Domain (WHD) into a cleavage-competent conformation, thus committing the enzyme to strand passage and ATP hydrolysis. While the KGRR/T-segment interaction stabilizes Top6B dimerization, T-segment capture potentiates DNA-gate opening by introducing strain in the storage cavity (step 3). T-segment release allows for DNA-gate closure and G-segment religation (step 4). Without a DNA crossing to stabilize Top6B closure, ADP and Pi are released, the WHDs relax to an inactive conformation (step 5) and Top6B returns to a relaxed, open conformation (step 6). From this G-segment bound state (B) topo VI tends to dissociate from DNA (to state A), but will infrequently capture another T-segment, regenerating a DNA crossing (C). The mechanistic implications of this model for meiotic recombination systems are considered in Figure 8—figure supplement 1.
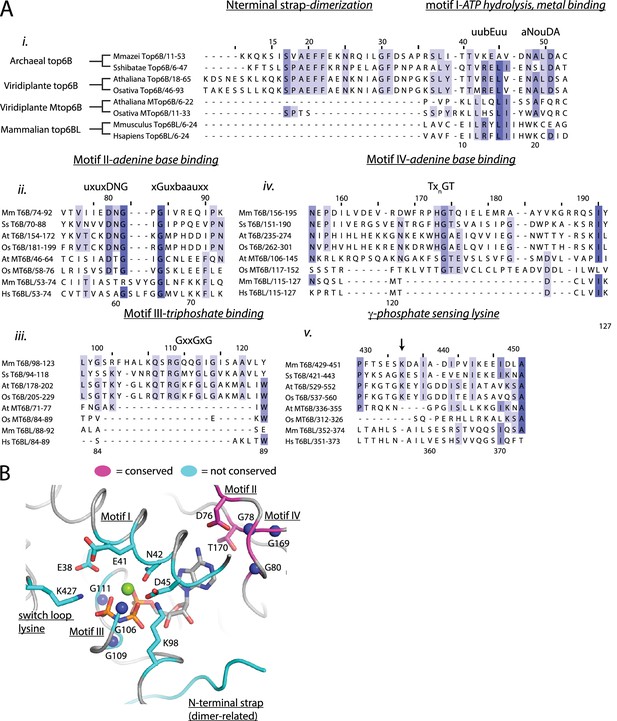
Conservation of canonical GHKL elements between Top6B, MTop6B, and Top6BL.
(A) Alignment of archaeal Top6B (M. mazei, S. shibatae), Viridiplante Top6B (A. thaliana, O. sativa), Viridiplante Mtop6B (A. thaliana, O. sativa), and mammalian Top6BL (Mus musculus, Homo sapiens) comparing motifs important for ATP hydrolysis. These include (i) the N-terminal dimerization strap and motif I, metal binding and ATP hydrolysis; (ii) motif II, adenine binding and base discrimination; (iii) motif III, triphosphate binding; (iv) motif IV, adenine binding and (v)transducer domain γ-phosphate sensor loop (Dutta and Inouye, 2000; Schoeffler and Berger, 2008; Thomsen and Berger, 2008). Numbering above the alignment corresponds to M. mazei Top6B, numbering below the alignments corresponds to H. sapiens Top6BL. (B) Ribbon representation of the ATP binding pocket of S. shibatae Top6B bound to AMPPNP (PDB ID = 1M × 0 [Corbett and Berger, 2003]) with the sidechains of residues involved in hydrolysis (E38), metal binding (N42), triphosphate binding (G106, G109, G111, K98), adenine binding (D76, T170, G78, G80, G168), phosphate sensing (K427), or binding pocket integrity (E41, D45) shown as sticks. Nitrogen atoms of denoted main chain glycine residues are shown as blue spheres. Residues generally conserved in the meiotic Top6B homologs are shown in magenta and poorly conserved residues are shown in cyan. Motifs I-IV and the switch lysine residue are labeled. Residues are numbered according to S. shibatae Top6B.
Tables
Summary of topo VI functional activities and mutant effects.
https://doi.org/10.7554/eLife.31724.037| Assay | Wildtype activity | Effect of mutation to: | ||
|---|---|---|---|---|
| KGRR loop | Stalk/WKxY | H2TH | ||
| Supercoil relaxation | distributive strand passage activity | kills strand passage | kills strand passage | greatly impairs strand passage |
| ATPase activity | ||||
| -on linear DNA | stimulates above basal activity | no activity | no activity | ~wildtype activity |
| -on supercoiled DNA | stimulates more than linear DNA | increased activity, futile cycling | no activity | ~wildtype activity, futile cycling |
| DNA binding | ||||
| -short duplexes | affinity increases from 20 bp to 40 bp in length | moderately impairs binding for longer duplexes | greatly impairs binding | slightly impairs binding for longer duplexes |
| -sheared salmon-sperm DNA | similar affinity as for 40–70 bp duplexes | moderately impairs binding | N.D. | slightly impairs binding |
| -supercoiled DNA | increased affinity compared to linear DNA | moderately impairs binding | N.D. | moderately impairs binding |
| -stacked junction | tighter binding than to duplex | greatly impairs binding | N.D. | ~wildtype affinity |
| Top6B dimerization | ||||
| -on short DNA duplexes | DNA promotes closure AMPPNP promotes further closure | loss of substrate promoted closure AMPPNP promotes some closure | N.D. | N.D. |
| -on supercoiled DNA | promotes greater closure than linear DNA AMPPNP promotes further closure | supercoiled DNA promotes closure loss of AMPPNP promoted closure | N.D. | weaker substrate dependent closure than wildtype AMPPNP promotes further closure |
| Short duplex cleavage | AMPPNP promotes cleavage on 60 and 70 bp duplexes | similar to wildtype | greatly impairs cleavage | no cleavage |
| Short duplex bending | AMPPNP promotes bending | similar to wildtype | greatly slows bending | no bending |
Summary of enzyme, DNA and nucleotide conditions by type of experiment.
https://doi.org/10.7554/eLife.31724.038| Assay | [enzyme] | [nucleotide] | [DNA] |
|---|---|---|---|
| DNA binding | 0, 3.9–4000 nM | N/A | 20 nM (0.4–1.4 μM bp) probe duplex |
| Competitive binding | 100 nM | N/A | 20 nM (1.4 μM bp) probe duplex 0, 0.1–106 μM bp DNA competitor/ 0, 0.3–36 nM plasmid |
| Supercoil relaxation titration timecourse/chase | 0, 0.3-20 nM 2.5 nM | 1 mM ATP 1 mM ATP | 2.9 kb primary plasmid- 10.2 μM bp DNA/3.5 nM plasmid 6.5 kb chase plasmid- 10.2 μM bp DNA/1.6 nM plasmid |
| ATP hydrolysis- ATP titration DNA titration | 500 nM 500 nM | 0, 0.06–4 mM ATP 2 mM ATP | 400 μM bp DNA/136 nM plasmid 0, 3.1–800 μM bp DNA/ 0, 1–273 nM plasmid |
| Top6B dimerization | 200 nM | 1 mM AMPPNP | 100 μM bp DNA/34 nM plasmid |
| Short duplex cleavage | 200 nM | 1 mM ATP or AMPPNP | 100 nM duplex (7 μM bp DNA) |
| Short duplex bending | 200 nM | 1 mM AMPPNP | 100 nM duplex (7 μM bp DNA) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Methanosarcina Mazei) | Top6A | N/A | NCBI Gene ID: 1480760 | |
| Gene (Methanosarcina Mazei) | Top6B | N/A | NCBI Gene ID: 1480759 | |
| Strain, strain background (E. coli) | BL21(DE3)-RIL | QB3-MacroLab | ||
| Strain, strain background (E. coli) | XL1-Blue | QB3-MacroLab | ||
| Recombinant DNA reagent | M. mazei Top6AB expression vector | PMID: 17603498 | ||
| Recombinant DNA reagent | M. mazei Top6AB-KGRRAAA expression vector | this paper | Construct generated by introduction of point mutations: K186A, R188A, and R189A to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-KGRREEE expression vector | this paper | Construct generated by introduction of point mutations: K186E, R188E, and R189E to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-Stalk/ WKxYAAA expression vector | this paper | Construct generated by introduction of point mutations: K399A, K401A, and R457A to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-Stalk/ WKxYEEE expression vector | this paper | Construct generated by introduction of point mutations: K399E, K401E, and R457E to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-H2THAAA expression vector | this paper | Construct generated by introduction of point mutations: R263A, K268A, and K308A to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-H2THEEE expression vector | this paper | Construct generated by introduction of point mutations: R263E, K268E, and K308E to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-cyslite- 155C expression vector | this paper | Construct generated by introduction of point mutations: T155C, C267S, C278A, C316A, and C550A to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-KGRRAAA cyslite-155C expression vector | this paper | Construct generated by introduction of point mutations: K186A, R188A, and R189A to Top6B gene on M. Mazei Top6AB-cyslite-155C expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-H2THAAA cyslite-155C expression vector | this paper | Construct generated by introduction of point mutations: R263A, K268A, and K308A to Top6B gene on M. Mazei Top6AB-cyslite-155C expression vector | |
| Recombinant DNA reagent | M. mazei Top6AB-E44A expression vector | this paper | Construct generated by introduction of point mutations: E44A to Top6B gene on M. Mazei Top6AB expression vector | |
| Recombinant DNA reagent | pSG483 (plasmid DNA) | PMID: 16023670 | 2.9 kb plasmid used as supercoiled substrate | |
| Sequence-based reagent (13 oligonucleotides) | See Figure 1—source data 1 | Integrated DNA Technologies | ||
| Chemical compound, drug | salmon sperm DNA, sheared | Thermo Fisher Scientfic | ThermoFisher:AM9680 | |
| Chemical compound, drug | Alexa Fluor 555 C2 Maleimide | Thermo Fisher Scientfic | ThermoFisher:A20346 | |
| Chemical compound, drug | Alexa Fluor 647 C2 Maleimide | Thermo Fisher Scientfic | ThermoFisher:A20347 | |
| Software, algorithm | ConSurf Server | PMID: 20478830 | RRID:SCR_002320 | |
| Software, algorithm | w3DNA server | PMID: 19474339 | ||
| Software, algorithm | PyMol | Schrödinger, LLC | RRID:SCR_000305 | |
| Software, algorithm | Prism 7 | Graphpad Software | RRID:SCR_015807 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31724.039





