Neolithic and medieval virus genomes reveal complex evolution of hepatitis B
Figures
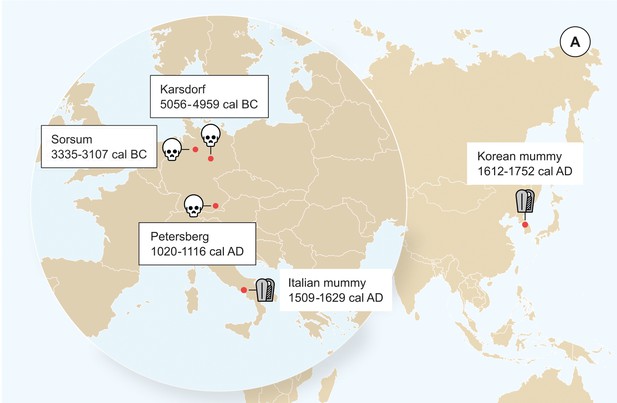
Origin of samples.
Geographic location of the samples from which ancient HBV genomes were isolated. Radiocarbon dates of the specimens is given in two sigma range. Icons indicate the sample material (tooth or mummy). HBV genomes obtained in this study are indicated by black frame.
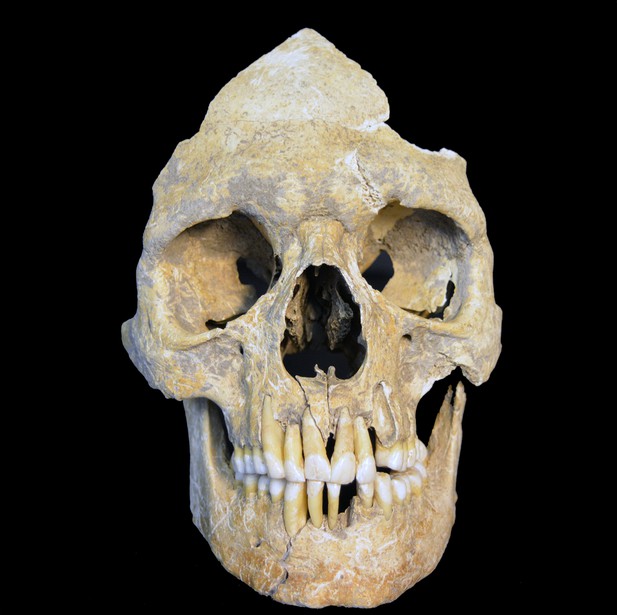
Skull of the investigated Karsdorf individual 537 is from a male with an age at death of around 25–30 years.
https://doi.org/10.7554/eLife.36666.003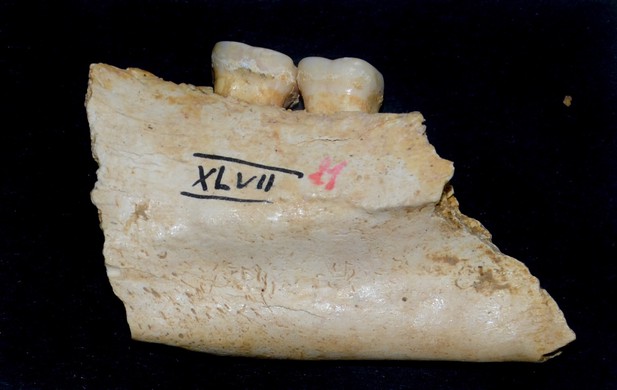
Mandible fragment of the Sorsum individual XLVII 11 analyzed in this study is from a male.
https://doi.org/10.7554/eLife.36666.004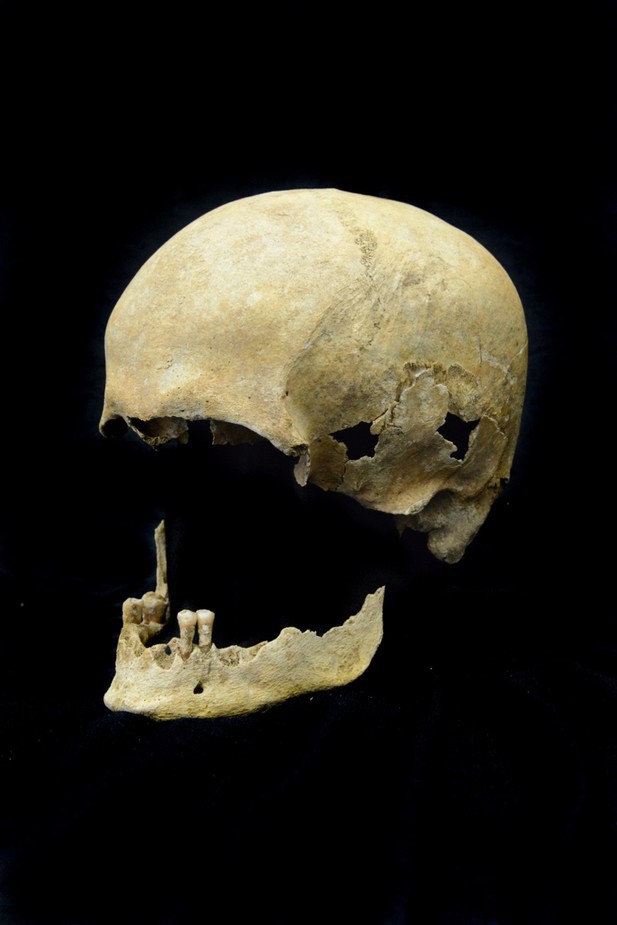
Skull of the analyzed Petersberg individual from grave 820 is from a male with an age at death of around 65–70 years.
https://doi.org/10.7554/eLife.36666.005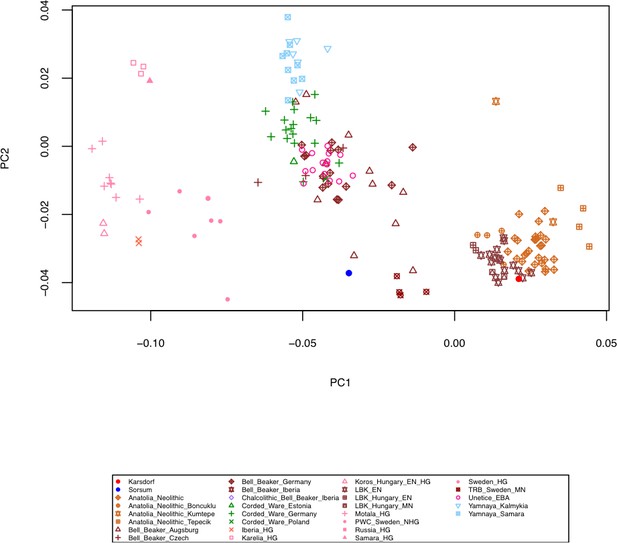
Principal Component Analysis (PCA) of the human Karsdorf and Sorsum samples together with previously published ancient populations projected on 27 modern day West Eurasian populations (not shown) based on a set of 1.23 million SNPs (Mathieson et al., 2015).
https://doi.org/10.7554/eLife.36666.006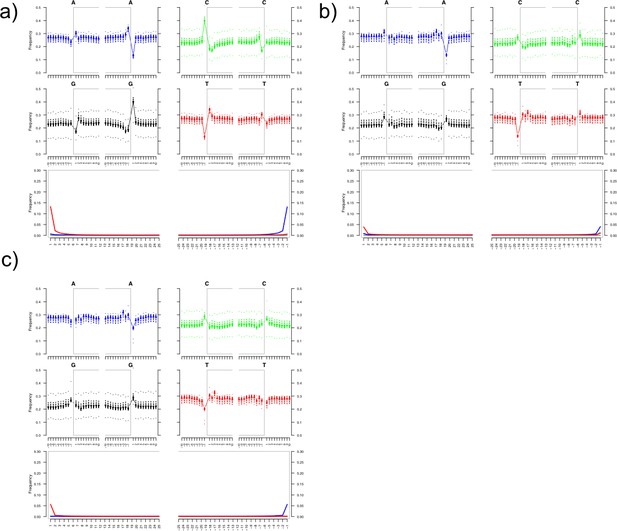
Damage plots showing deamination patterns of hg19-specific reads for the HalfUDG-treated libraries of (a) Karsdorf, (b) Sorsum, (c) Petersberg.
https://doi.org/10.7554/eLife.36666.007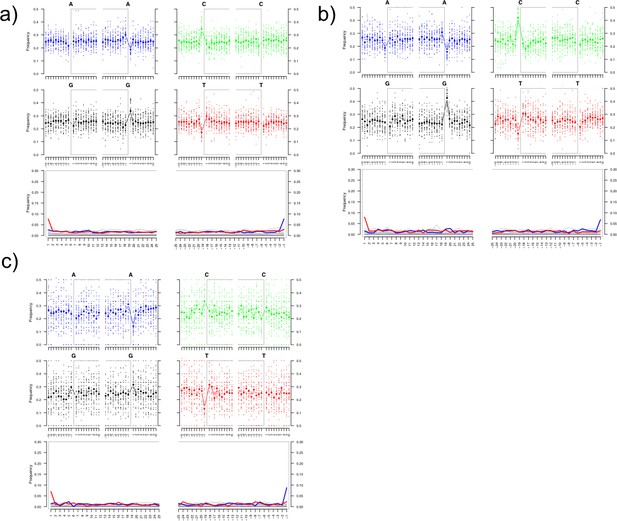
Damage plots showing deamination patterns of HBV-specific reads for the HalfUDG-treated libraries of (a) Karsdorf, (b) Sorsum, (c) Petersberg.
References shown in Supplementary file 1 were used to carry out the alignment.
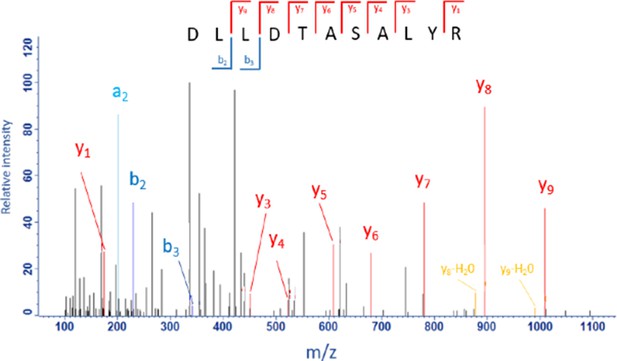
MS/MS spectrum of the proteotypic HBV-peptide DLLDTASALYR from the HBV-protein external core antigen (residues 58–68).
([M + 2 hr]2+): m/z = 1237.6429 Da. Mass accuracy of the precursor peptide = 0.56 ppm.
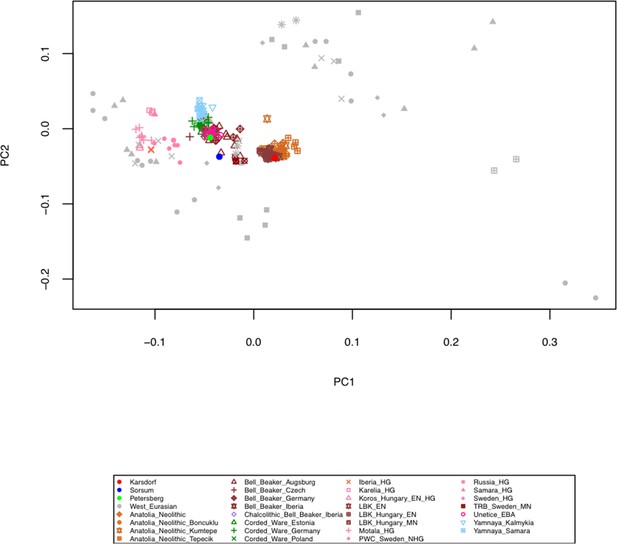
Principal Component Analysis (PCA) of the human Karsdorf and Sorsum samples together with previously published ancient populations projected on 27 modern day West Eurasian populations (shown in gray) based on a set of 1.23 million SNPs (Mathieson et al., 2015).
https://doi.org/10.7554/eLife.36666.010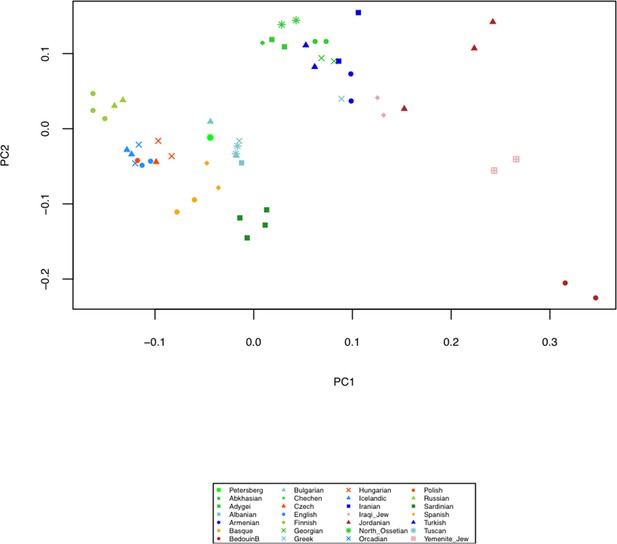
Principal Component Analysis (PCA) of the human Petersberg sample projected on 27 modern day West Eurasian populations based on a set of 1.23 million SNPs (Mathieson et al., 2015).
https://doi.org/10.7554/eLife.36666.011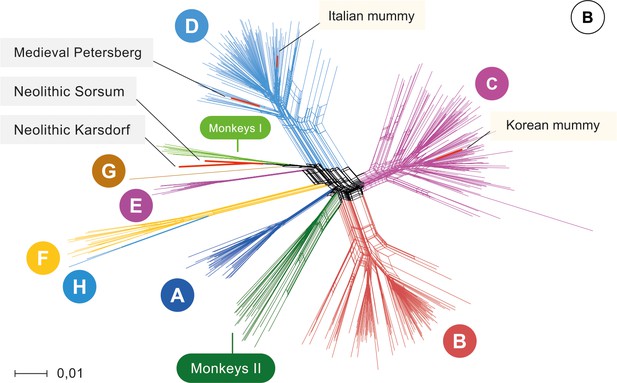
Network.
Network of 493 modern, two published ancient genomes (light yellow box), and three ancient hepatitis B virus (HBV) obtained in this study (grey box). Colors indicate the eight human HBV genotypes (A–H), two monkey genotypes (Monkeys I, African apes and Monkeys II, Asian monkeys) and ancient genomes (red).
-
Figure 2—source data 1
Results of the recombination analysis using the methods RDP, GENECOV, Chimera, MaxChi, BootScan, SiScan, 3Seq within the RDP v4 software package with all modern full reference genomes (n = 493) and five ancient genomes.
- https://doi.org/10.7554/eLife.36666.023
-
Figure 2—source data 2
Multiple sequence alignment of the 493 representative and five ancient HBV genomes.
The multiple sequence alignment was stripped of any sites that had gaps in more than 95%.
- https://doi.org/10.7554/eLife.36666.024
-
Figure 2—source data 3
Maximum-likelihood tree based on the multiple sequence alignment of the 493 representative and five ancient HBV genomes with 2000 replicates.
- https://doi.org/10.7554/eLife.36666.025
-
Figure 2—source data 4
Neighbour-Joining tree based on the multiple sequence alignment of the 493 representative modern and five ancient HBV genomes with 10000 replicates.
- https://doi.org/10.7554/eLife.36666.026
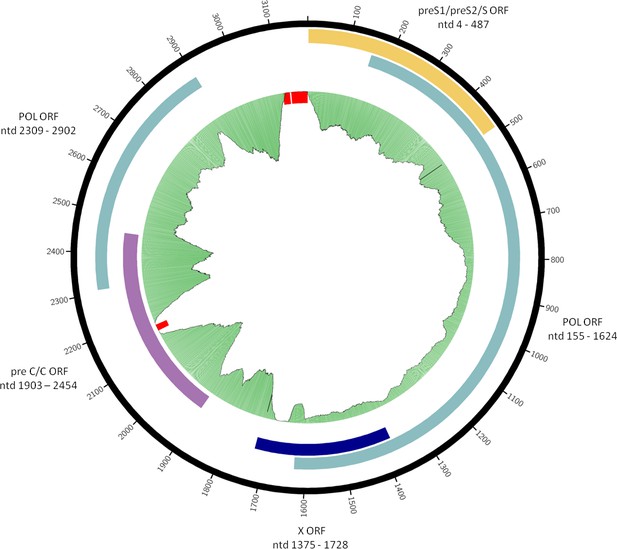
Consensus sequence of the Karsdorf HBV genome.
Organization of overlapping open reading frames and approximate location of single-stranded portion of plus strand are indicated. Gaps in the sequence are marked in red. The green plot depicts the coverage of the re-mapping of raw reads against the consensus. Circular plots were generated using circos-0.69-6 and coverage information from the re-mapping.
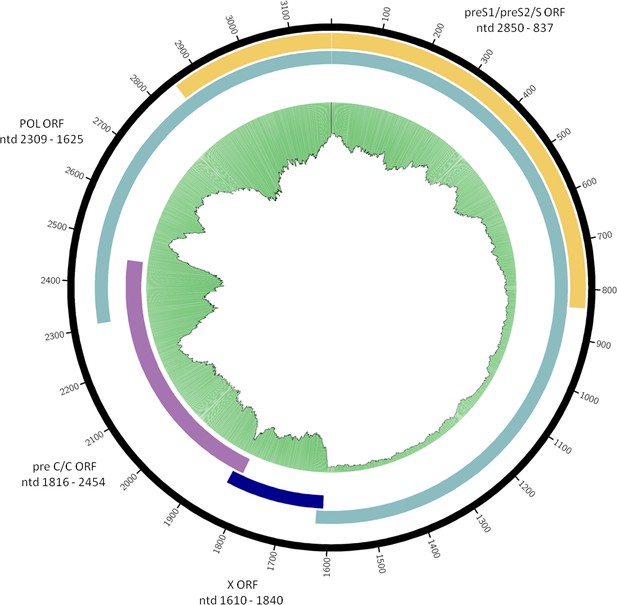
Consensus sequence of the Sorsum HBV genome.
Organization of overlapping open reading frames and approximate location of single-stranded portion of plus strand are indicated. Gaps in the sequence are marked in red. The green plot depicts the coverage of the re-mapping of raw reads against the consensus. Circular plots were generated using circos-0.69-6 and coverage information from the re-mapping.
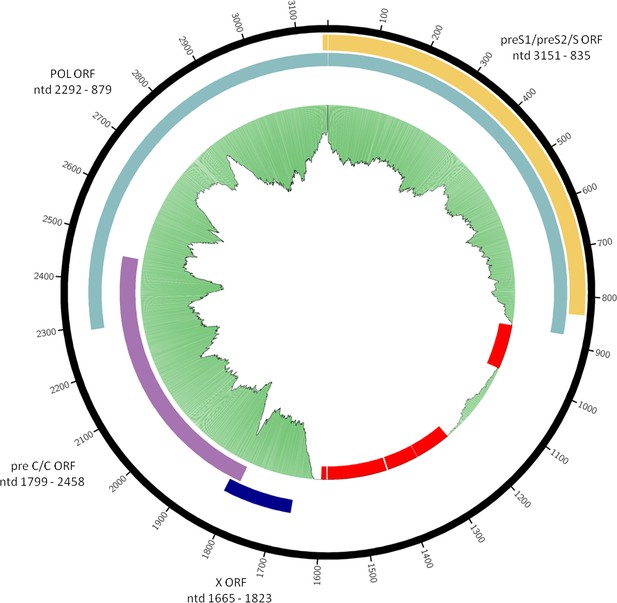
Consensus sequence of the Petersberg HBV genome.
Organization of overlapping open reading frames and approximate location of single-stranded portion of plus strand are indicated. Gaps in the sequence are marked in red. The green plot depicts the coverage of the re-mapping of raw reads against the consensus. Circular plots were generated using circos-0.69-6 and coverage information from the re-mapping.
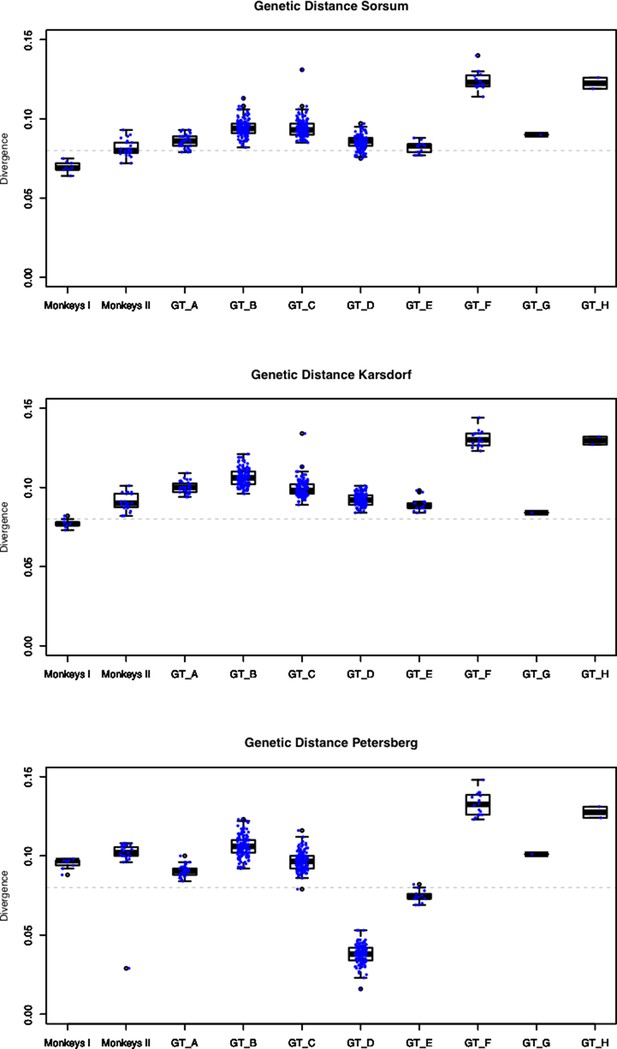
Genetic (hamming) distance of our three ancient HBV genomes compared to all 493 reference genomes.
Gaps or non-called sites (’N') were ignored.
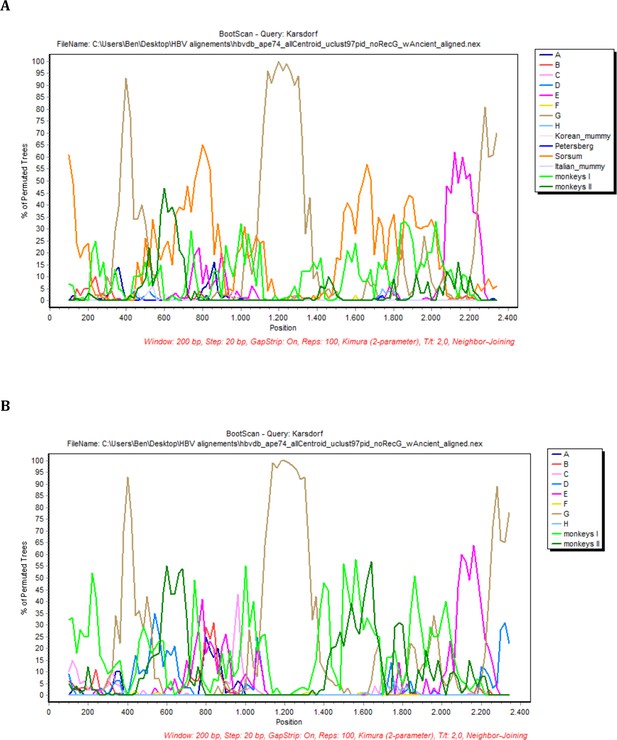
BootScan analysis of the sequence Karsdorf.
In each case, sequence fragments of 200 bases incrementing by 20 bases, 100 bootstrap replicates, were compared with sequence groups of (a) the eight human genotypes, two primate genotypes, and four ancient genomes and (b) the eight human genotypes, two primate genotypes (color coded as described in the legend).
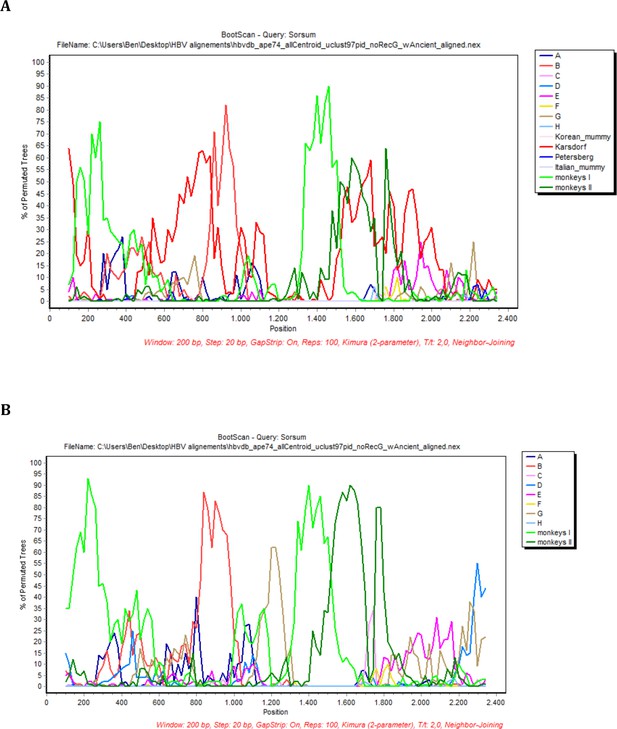
BootScan analysis of the sequence Sorsum.
In each case, sequence fragments of 200 bases incrementing by 20 bases, 100 bootstrap replicates, were compared with sequence groups or 50% consensus sequences of (a) the eight human genotypes, two primate genotypes, and four ancient genomes and (b) the eight human genotypes, two primate genotypes (color coded as described in the legend).
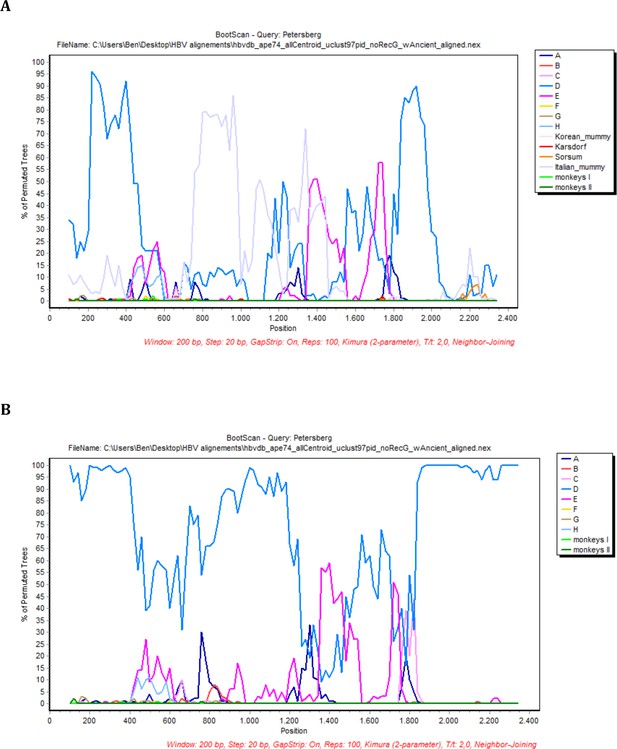
BootScan analysis of the sequence Petersberg.
In each case, sequence fragments of 200 bases incrementing by 20 bases, 100 bootstrap replicates, were compared with sequence groups or 50% consensus sequences of (a) the eight human genotypes, two primate genotypes, and four ancient genomes and (b) the eight human genotypes, two primate genotypes (color coded as described in the legend).
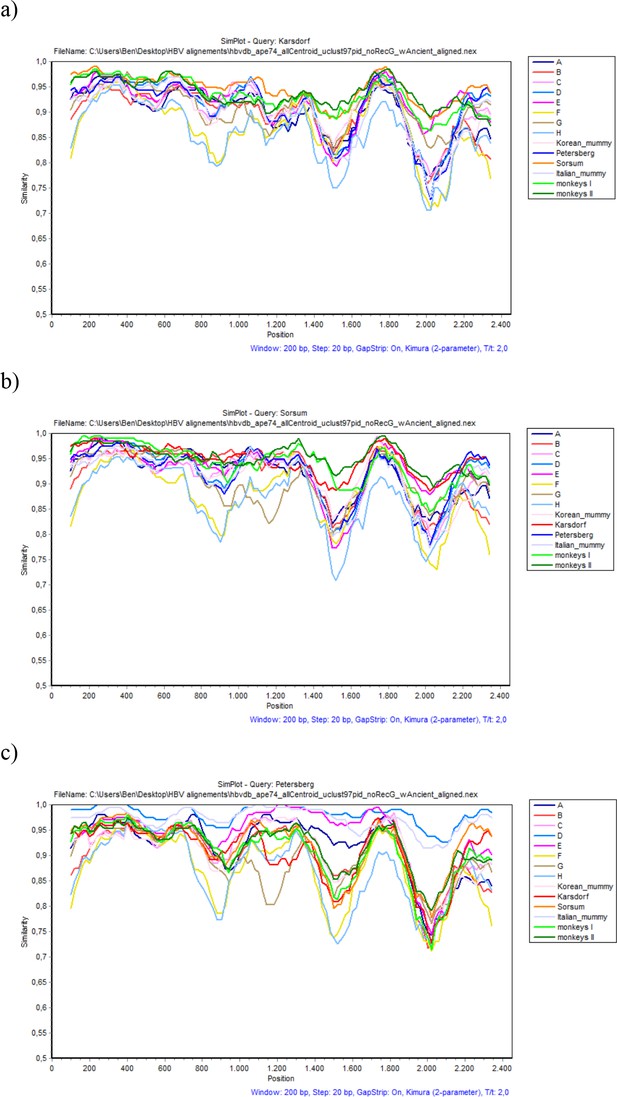
SimPlot analysis of (a) Karsdorf, (b) Sorsum and (c) Petersberg.
In each case, sequence fragments of 200 bases incrementing by 20 bases, 100 bootstrap replicates, were compared with sequence groups of the eight human genotypes, four primate genotypes and four ancient genomes (color coded as described in the legend).
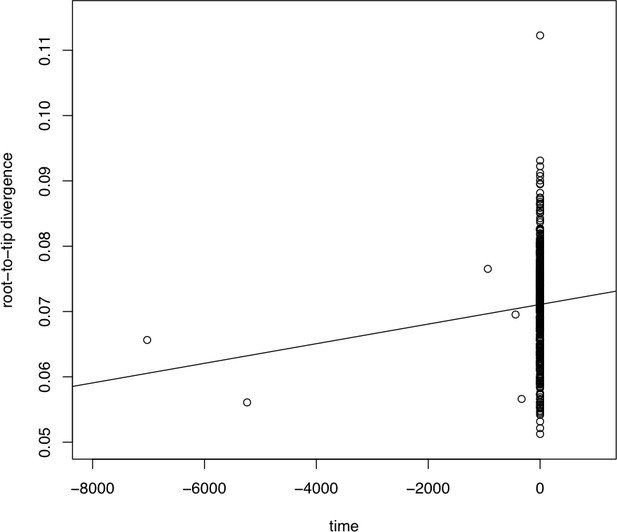
Plot of phylogenetic root-to-tip distance relative to sampling time (TempEst).
Each dot represents one sample.
Tables
Results of the genome reconstruction
https://doi.org/10.7554/eLife.36666.012| *Merged reads | Length of HBV consensus sequence | Mean HBV coverage | Gaps in the consensus sequence at nt position | *Mapped reads HBV | *Mapped reads human | Mean human coverage | Human genomes/HBVgenomes | |
|---|---|---|---|---|---|---|---|---|
| Karsdorf | 386,780,892 | 3183 | 104X | 2157–2175; 3107–3128; 3133–3183 | 10,718 | 122,568,310 | 2.96X | 1: 35.1 |
| Sorsum | 367,574,767 | 3182 | 47X | - | 3249 | 9,856,001 | 1.17X | 1: 40.2 |
| Petersberg | 419,413,082 | 3161 | 46X | 880–1000; 1232–1329; 1331–1415; 1420–1581; 1585–1598 | 2125 | 105,476,677 | 2.88X | 1: 16 |
-
*number.
nt, nucleotide.
Additional files
-
Supplementary file 1
Accession numbers for the reference genomes used in the first alignment step to catch HBV diversity in the sample.
Since monkey HBV strains are not classified into genotypes the column is left blank.
- https://doi.org/10.7554/eLife.36666.027
-
Supplementary file 2
Number of reads mapping against the references shown in Supplementary file 1 before and after duplicate removal.
- https://doi.org/10.7554/eLife.36666.028
-
Supplementary file 3
Number of contigs and combined contig length of the de novo assembly for chosen K-values.
- https://doi.org/10.7554/eLife.36666.029
-
Supplementary file 4
Final consensus length after retrieving gap information from the multiple sequence alignment with Geneious.
- https://doi.org/10.7554/eLife.36666.030
-
Supplementary file 5
Number of reads mapping against hg19 before and after duplicate removal and percentage of the genome where coverage is at least one.
- https://doi.org/10.7554/eLife.36666.031
-
Supplementary file 6
Basic statistics for the mapping against the references shown in table S1.
Shown are mean coverage, mean coverage for the covered region, genome length, number of missing bases and covered bases
- https://doi.org/10.7554/eLife.36666.032
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36666.033





