Hypoxia-inducible factor cell non-autonomously regulates C. elegans stress responses and behavior via a nuclear receptor
Figures
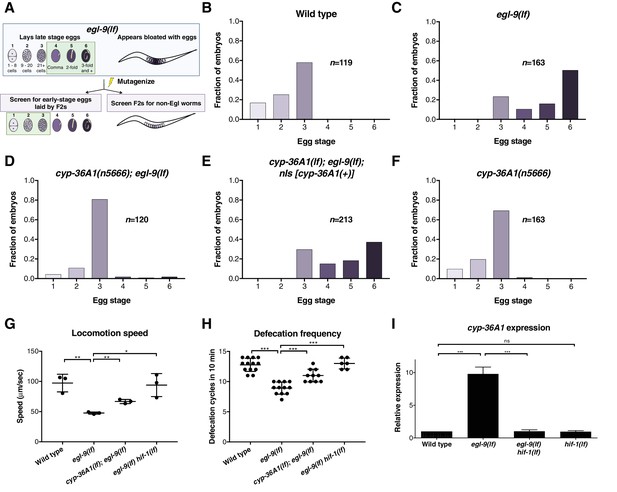
The cytochrome P450 gene cyp-36A1 is an effector of the hypoxia-response pathway that regulates behavior.
(A) Schematic of the screen design. Stages of embryonic development adapted from Ringstad and Horvitz (2008) and Paquin et al. (2016). (B–F) Distribution of stages of eggs newly laid by adult hermaphrodites, used as a proxy for egg retention time in utero, of animals of the indicated genotypes. (B) Stages of eggs laid by wild-type animals. (C) egl-9 loss-of-function (lf) mutants laid later stage eggs than the wild type (p<0.001, Chi-square test with Holm-Bonferroni correction). (D) cyp-36A1(n5666) suppressed the egg-laying defect of egl-9(lf) mutants (p<0.001). (E) The cyp-36A1(+) transgene, which contains wild-type cyp-36A1, rescued the suppression of the egg-laying defect observed in cyp-36A1(n5666); egl-9(lf) mutants (p<0.001). (F) cyp-36A1(n5666) mutants displayed wild-type egg laying (p>0.05). (G) egl-9(lf) mutants were defective in locomotion rate, and this defect was suppressed by hif-1(lf) and cyp-36A1(lf) mutations. Mean ± SD of n = 3 biological replicates, *p<0.05, **p<0.01 considered significant (Student’s t-test with Holm-Bonferroni correction). (H) egl-9(lf) mutants were defective in defecation rate, and this defect was suppressed by hif-1(lf) and cyp-36A1(lf). Mean ± SD of n ≥ 6 animals, ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction). (I) Relative expression of cyp-36A1 mRNA in the wild type, egl-9(lf), egl-9(lf) hif-1(lf), and hif-1(lf) mutants, measured by qRT-PCR and normalized to the expression of the large ribosomal subunit rpl-32. Mean ± SD of n = 3 biological replicates, ***p<0.001 considered significant. ns, not significant (Student’s t-test with Holm-Bonferroni correction). Alleles used for (B–F) were cyp-36A1(n5666), egl-9(n586), and nIs674 (nIs [cyp-36A1(+)]), and all strains used in (B–F) contained the nIs470 (Pcysl-2::gfp) transgene. Alleles used for (G–I) were egl-9(sa307), hif-1(ia4), and cyp-36A1(gk824636).
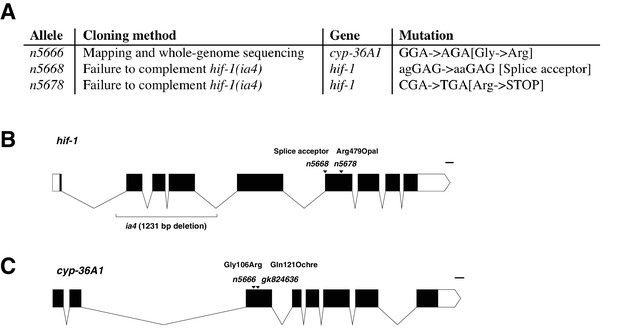
A screen for suppressors of the egl-9(lf) egg-laying defect.
(A) Summary of three cloned alleles from a suppressor screen of the egl-9(lf) egg-defect. (B) Gene diagram of hif-1; isoform F38A6.3b is shown. The ia4 deletion allele was generated by deletion screening (Jiang et al., 2001). Scale bar, 100 bases. (C) Gene diagram of cyp-36A1. The gk824636 allele is from the Million Mutation Project (Thompson et al., 2013). Scale bar, 100 bases.
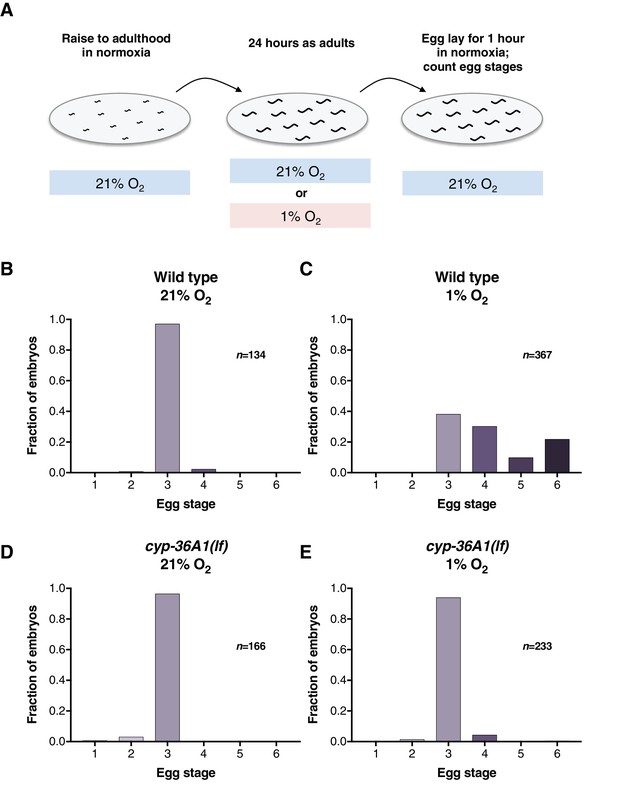
cyp-36A1(lf) suppresses the egg-laying defect of hypoxia-exposed animals.
(A) Schematic depicting experimental design. (B–E) Distribution of stages of eggs laid by adult hermaphrodites. (B) Stages of eggs laid by wild-type animals grown in normoxia (21% O2). (C) Animals exposed to hypoxia (1% O2) for 24 hr as adults laid later stage eggs than those in normoxia (p<0.001, Chi-square test with Holm-Bonferroni correction). (D) cyp-36A1(gk824636) mutants grown in normoxia displayed wild-type egg laying (p>0.05). (E) cyp-36A1(gk824636) mutants exposed to 1% O2 laid eggs at earlier stages than wild-type animals in the same condition (p<0.001).
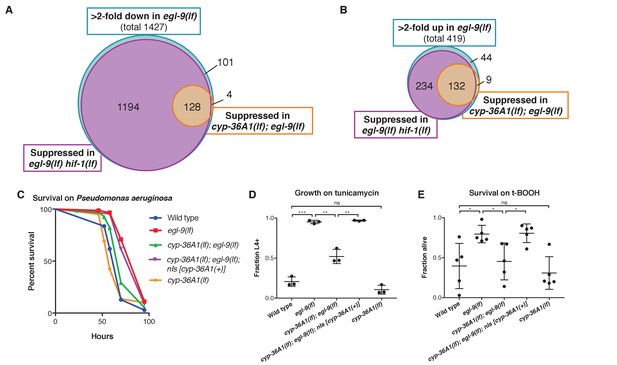
CYP-36A1 acts downstream of HIF-1 to regulate gene expression changes and stress responses.
(A) Blue circle: Genes that were at least twofold downregulated in egl-9(lf) mutants. Purple and orange circles: Subset of egl-9(lf)-downregulated genes that were significantly upregulated in egl-9(lf) hif-1(lf) double mutants (purple) or cyp-36A1(lf); egl-9(lf) double mutants (orange) vs. egl-9(lf) single mutants. (B) Blue circle: Genes that were at least twofold upregulated in egl-9(lf) mutants. Purple and orange circles: Subset of egl-9(lf)-upregulated genes that were significantly downregulated in egl-9(lf) hif-1(lf) double mutants (purple) or cyp-36A1(lf); egl-9(lf) double mutants (orange) vs. egl-9(lf) single mutants. Significance for all comparisons in (A) and (B) was based on two biological replicates and determined by the Benjamini-Hochberg procedure with a false-discovery rate of 0.05. (C) Survival of animals grown from the L4 larval stage on the pathogen Pseudomonas aeruginosa. Wild type (n = 86 animals) vs. egl-9(lf) (n = 154) p<0.001; egl-9(lf) vs. cyp-36A1(lf); egl-9(lf) (n = 83), p<0.001; cyp-36A1(lf); egl-9(lf) vs. cyp-36A1(lf); egl-9(lf); nIs [cyp-36A1(+)] (n = 97), p<0.001; wild type vs. cyp-36A1(lf) (n = 61) p<0.05, as determined by the log-rank (Mantel-Cox) test, correcting for multiple comparisons with the Holm-Bonferroni method. See figure supplement for replicate data. (D) Survival of animals to the L4 larval stage or later after growth for three days from the L1 larval stage on plates containing 5 μg/ml tunicamycin. Mean ± SD of n = 3 biological replicates. **p<0.01, ***p<0.001 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction). (E) Survival of animals exposed to 7.5 mM tert-butyl hydroperoxide for 10 hr as young adults. Mean ± SD of n = 5 biological replicates. *p<0.05 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction). Alleles used for (A) and (B) were egl-9(sa307), hif-1(ia4), and cyp-36A1(gk824636). Alleles used for (C–E) were egl-9(n586), cyp-36A1(n5666), and nIs674 (nIs [cyp-36A1(+)]), and all strains used in (C–E) contained the nIs470 (Pcysl-2::gfp) transgene.
-
Figure 2—source data 1
RNA-seq GO enrichment analysis.
Gene ontology enrichment analysis terms for genes more than twofold upregulated in egl-9 mutants and significantly suppressed by hif-1(lf) and cyp-36A1(lf).
- https://doi.org/10.7554/eLife.36828.007
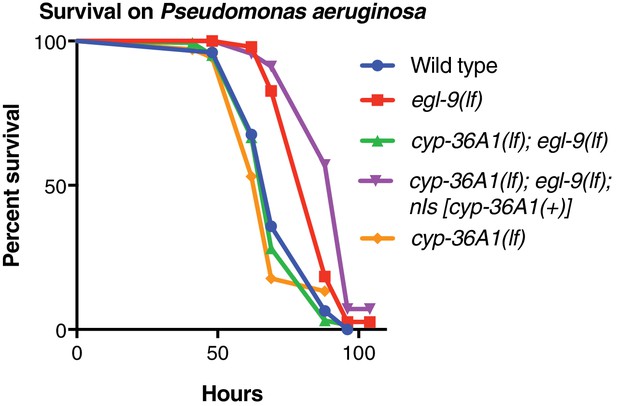
Replicate data for survival on Pseudomonas aeruginosa.
Replicate data for Figure 2C. Wild type (n = 150) vs. egl-9(lf) (n = 50), p<0.001; egl-9(lf) vs. cyp-36A1(lf); egl-9(lf) (n = 163), p<0.001; cyp-36A1(lf); egl-9(lf) vs. cyp-36A1(lf); egl-9(lf); nIs [cyp-36A1(+)] (n = 51), p<0.001; wild type vs. cyp-36A1(lf) (n = 35), p>0.05, as determined by the log-rank (Mantel-Cox) test, correcting for multiple comparisons with the Holm-Bonferroni method.
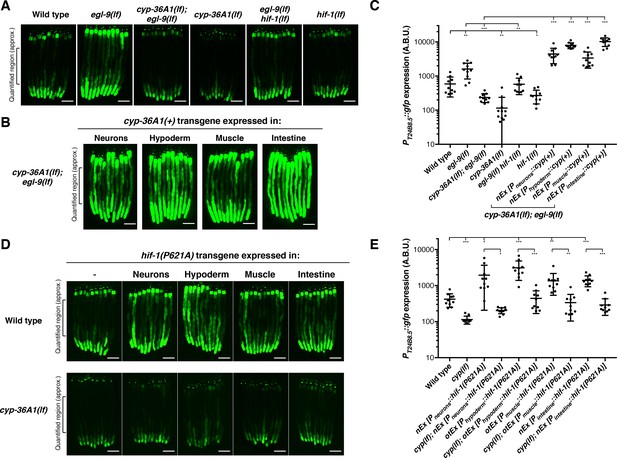
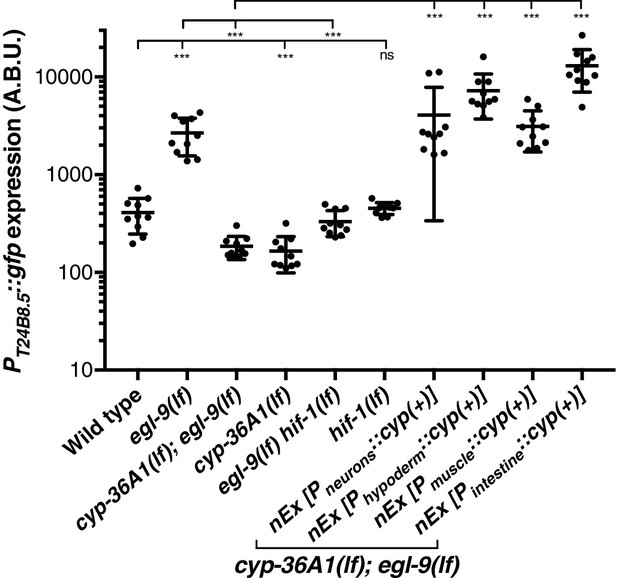
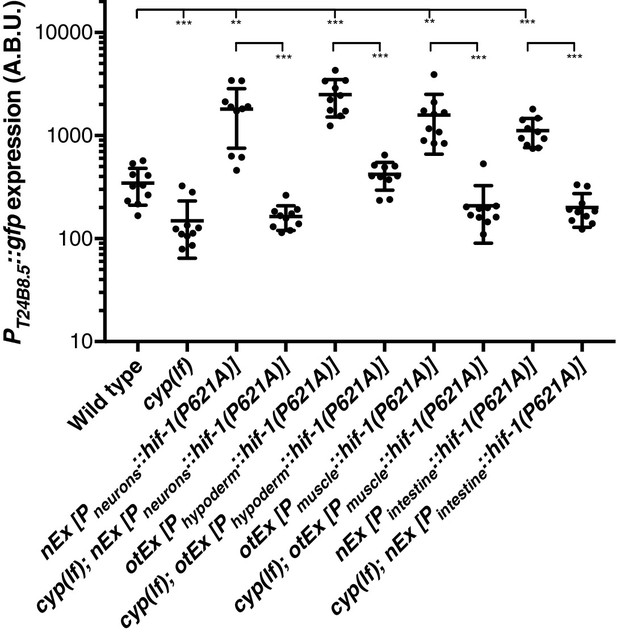
HIF-1 and CYP-36A1 cell non-autonomously regulate expression of a stress-responsive gene.
(A) PT24B8.5::gfp expression of the indicated genotypes (n = 10 animals per image). Scale bars, 100 μm. (B) Expression of cyp-36A1(+) specifically in neurons, hypoderm, muscle, or intestine of cyp-36A1(lf); egl-9(lf) animals increased expression of PT24B8.5::gfp in intestine (n = 10 animals per image). Scale bars, 100 μm. (C) Quantification of fluorescence intensity for (A) and (B), measured as average intensity for a 300 μm section of the intestine in the midbody of each animal, as indicated. *p<0.05, **p<0.01, ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction). Mean ± SD of n = 10 animals. See figure supplement for replicate data. (D) Expression of hif-1(P621A), which encodes a stable variant of HIF-1 (Pocock and Hobert, 2008), specifically in neurons, hypoderm, muscle, or intestine increased expression of PT24B8.5::gfp in intestine; increased expression was suppressed by cyp-36A1(lf) (n = 10 animals per image). Scale bars, 100 μm. (E) Quantification of fluorescence intensity for (D), measured as average intensity for a 300 μm section of the intestine in the midbody of each animal, as indicated. *p<0.05, ***p<0.01, ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction). Mean ± SD of n = 10 animals. See figure supplement for replicate data. Alleles used were egl-9(sa307), hif-1(ia4), cyp-36A1(gk824636), nEx2699[Pneurons::hif-1(P621A)], otEx3156 [Phypoderm::hif-1(P621A)], otEx3165 [Pmuscle::hif-1(P621A)], nEx2860 [Pintestine:: hif-1(P621A)], nEx2853 [Pneurons::cyp(+)], nEx2856 [Phypoderm::cyp(+)], nEx2859 [Pmuscle::cyp(+)], and nEx2849 [Pintestine::cyp(+)]. All strains contained the agIs219 (PT24B8.5::gfp) transgene.
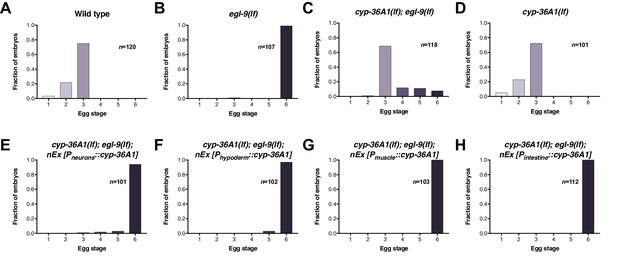
cyp-36A1 expression in multiple tissues rescues the egg-laying phenotype of cyp-36A1(lf); egl-9(lf).
(A–H) Distribution of stages of eggs laid by adult hermaphrodites of the indicated genotypes. All strains contained the agIs219 (PT24B8.5::gfp) transgene. (A) Stages of eggs laid by wild-type animals. (B) egl-9(sa307) animals laid later stage eggs than wild type (p<0.001, Chi-square test with Holm-Bonferroni correction). (C) cyp-36A1(gk824636) suppressed the egg-laying defect of egl-9(sa307) mutants (p<0.001). (D) Stages of eggs laid by cyp-36A1(gk824636) single mutants were not significantly different from those laid by wild-type animals (p>0.05). (E–H) Expression of cyp-36A1(+) specifically in neurons, hypoderm, muscle, or intestine of cyp-36A1(gk824636); egl-9(sa307) animals all rescued the suppression by cyp-36A1(lf) of the egg-laying defect of egl-9(lf) (p<0.001).
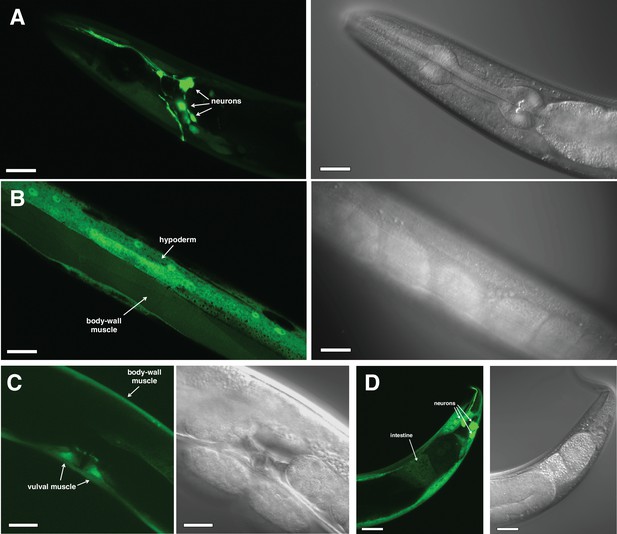
cyp-36A1 is expressed in many tissues.
(A–D) Paired fluorescent (left in each panel) and Nomarski (right in each panel) micrographs showing expression of a transcriptional Pcyp-36A1::gfp reporter (nIs682) in an adult worm. Expression was observed in the head (A), midbody (B and C), and tail (D), including in neurons, body-wall muscle, vulval muscle, intestine, and hypoderm, as indicated. Scale bars, 20 μm.
Replicate data for PT24B8.5::gfp reporter expression.
Replicate data for Figure 3C. Quantification of n = 10 animals. ***p<0.001 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction).
Replicate data for PT24B8.5::gfp reporter expression.
Replicate data for Figure 3E. Quantification of n = 10 animals. **p<0.01, ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction).
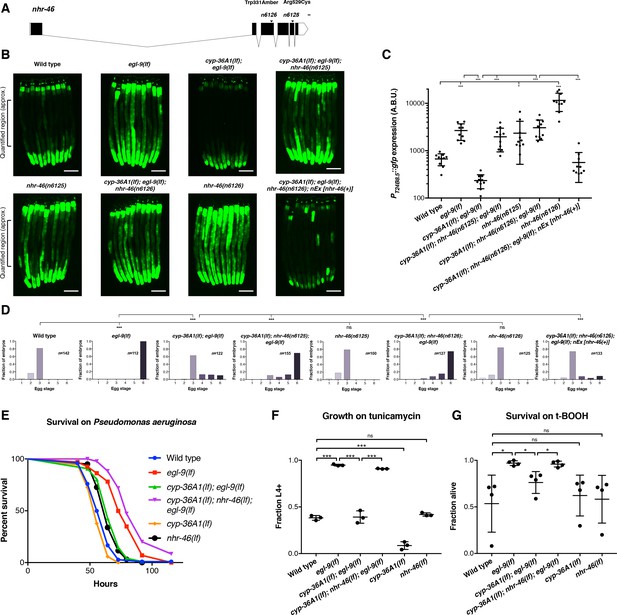
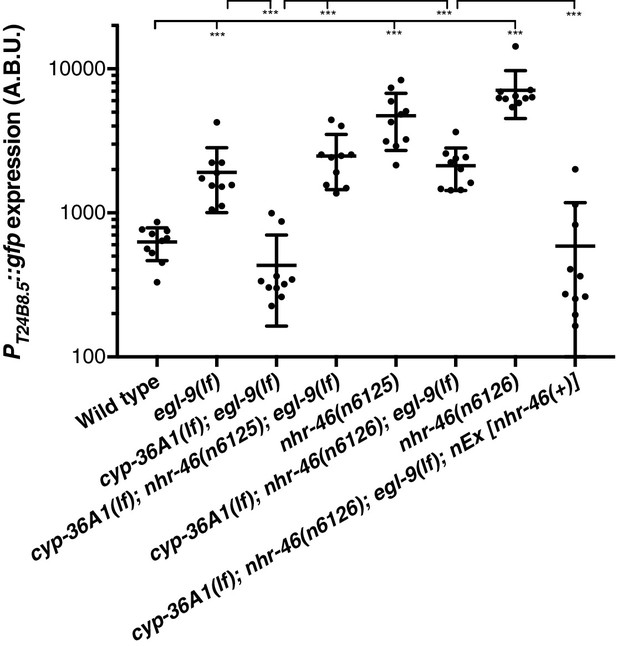
The nuclear hormone receptor NHR-46 acts downstream of CYP-36A1.
(A) nhr-46 gene diagram; isoform C45E5.6b is shown. n6125 and n6126 are in the NHR-46 ligand-binding domain. Scale bar, 100 bases. (B) PT24B8.5::gfp fluorescence of the indicated genotypes (n = 10 animals per image). Scale bars, 100 μm. (C) Quantification of fluorescence intensity for (B), measured as average intensity for a 300 μm section of the intestine in the midbody of each animal, as indicated. *p<0.05, ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction). Mean ±SD of n = 10 animals. See figure supplement for replicate data. (D) Distribution of stages of eggs newly laid by adult hermaphrodites of the indicated genotypes. ***p<0.001 considered significant. ns (p>0.05), not significant (Chi-square test with Holm-Bonferroni correction). (E) Survival of animals grown from the L4 larval stage on the pathogen Pseudomonas aeruginosa. Wild type (n = 129) vs. egl-9(lf) (n = 67) p<0.001; egl-9(lf) vs. cyp-36A1(lf); egl-9(lf) (n = 129), p<0.001; cyp-36A1(lf); egl-9(lf) vs. cyp-36A1(lf); nhr-46(lf); egl-9(lf) (n = 94), p<0.001; wild type vs. cyp-36A1(lf) (n = 106), p<0.05; wild type vs. nhr-46(lf) (n = 121), p<0.001, as determined by the log-rank (Mantel-Cox) test, correcting for multiple comparisons with the Holm-Bonferroni method. egl-9(lf) allele was egl-9(n586). See figure supplement for replicate data. (F) Survival of animals to the L4 larval stage or later after growth for three days from the L1 larval stage on plates containing 5 μg/ml tunicamycin. Mean ± SD of n = 3 biological replicates. ***p<0.001 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction). (G) Survival of animals exposed to 7.5 mM tert-butyl hydroperoxide for 10 hr as young adults. Mean ± SD of n = 4 biological replicates. *p<0.05 considered significant. ns, not significant (Student’s t-test with Holm-Bonferroni correction). Alleles used for (B–G) were egl-9(sa307), cyp-36A1(gk824636), nhr-46(n6126), and nEx2586 (nEx [nhr-46(+)]) except where otherwise noted. All strains in (B), (C), (D), (F), and (G) contained the agIs219 (PT24B8.5::gfp) transgene.
Replicate data for PT24B8.5::gfp reporter expression.
Replicate data for Figure 4C. Quantification of n = 10 animals. ***p<0.001 considered significant (Student’s t-test with Holm-Bonferroni correction).
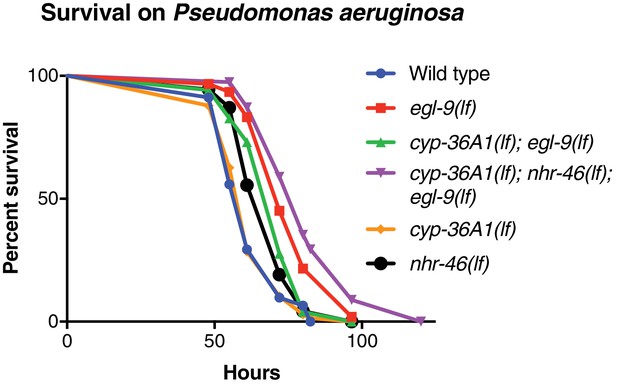
Replicate data for survival on Pseudomonas aeruginosa.
Replicate data for Figure 4E. Wild type (n = 34) vs. egl-9(n586) (n = 61), p<0.001; egl-9(lf) vs. cyp-36A1(lf); egl-9(lf) (n = 52), p<0.05; cyp-36A1(lf); egl-9(lf) vs. cyp-36A1(lf); nhr-46(lf); egl-9(lf) (n = 39), p<0.001; wild type vs. cyp-36A1(lf) (n = 91), p>0.05; wild type vs. nhr-46(lf) (n = 54), p<0.05, as determined by the log-rank (Mantel-Cox) test, correcting for multiple comparisons with the Holm-Bonferroni method.
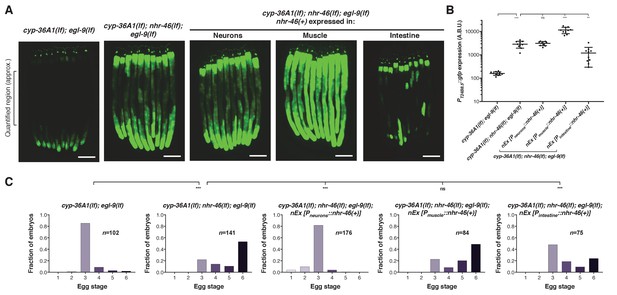
NHR-46 acts in different tissues to regulate T24B8.5 expression and egg laying.
(A) Expression of nhr-46(+) in the intestine but not in neurons or muscle rescued the high PT24B8.5::gfp expression in the intestine of cyp-36A1(lf); nhr-46(lf); egl-9(lf) mutants (n = 10 animals per image). Scale bars, 100 μm. (B) Quantification of fluorescence intensity for (A), measured as average intensity for a 300 μm section of the intestine in the midbody of each animal, as indicated. **p<0.01, ***p<0.001 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction). Mean ±SD of n = 10 animals. See figure supplement for replicate data. (C) Distribution of stages of eggs laid by adult hermaphrodites. Expression of nhr-46(+) in the intestine partially rescued the egg-laying defect of cyp-36A1(lf); nhr-46(lf); egl-9(lf) mutants. Neuronal nhr-46(+) expression fully rescued the egg-laying defect. Expression of nhr-46(+) in muscle did not rescue the egg-laying defect of cyp-36A1(lf); nhr-46(lf); egl-9(lf) mutants. ***p<0.001 considered significant. ns (p>0.05), not significant (Chi-square test with Holm-Bonferroni correction). Alleles used were egl-9(sa307), cyp-36A1(gk824636), nhr-46(n6126), nEx2713 [Pneurons::nhr-46(+)], nEx2715 [Pmuscle::nhr-46(+)], and nEx2864 [Pintestine::nhr-46(+)]. All strains contained the agIs219 (PT24B8.5::gfp) transgene.
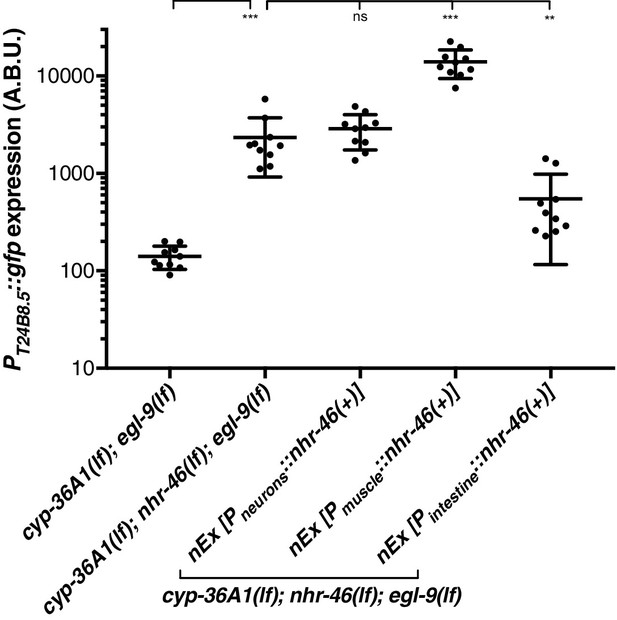
Replicate data for PT24B8.5::gfp reporter expression.
Replicate data for Figure 5B. Quantification of n = 10 animals. **p<0.01, ***p<0.001 considered significant. ns (p>0.05), not significant (Student’s t-test with Holm-Bonferroni correction).
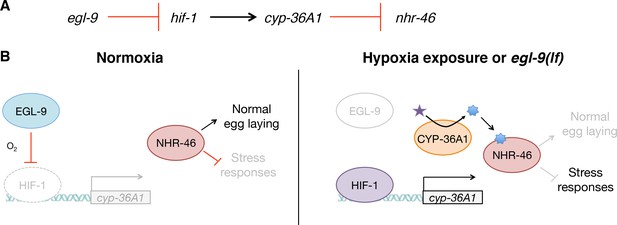
Model for the regulation of physiology and behavior by NHR-46 and CYP-36A1.
(A) The genetic pathway in which egl-9 inhibits hif-1, which activates cyp-36A1, which in turn inhibits nhr-46. (B) Model for how CYP-36A1 and NHR-46 function downstream of HIF-1. We suggest that CYP-36A1, which is transcriptionally upregulated by HIF-1, generates a hormone that binds NHR-46, thereby promoting transcriptional and physiological changes. See text for details.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditis elegans) | AU78 | Dennis Kim | agIs9 III | |
| Strain, strain background (C. elegans) | CB6088 | Jonathan Hodgkin | egl-9(sa307) V hif-1(ia4) V | |
| Strain, strain background (C. elegans) | JT307 | Creg Darby | egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT20483 | Dengke Ma/Bob Horvitz | nIs470 IV | |
| Strain, strain background (C. elegans) | MT22836 | this paper | cyp-36A1(gk824636) I ; egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT23218 | this paper | nIs682 X | |
| Strain, strain background (C. elegans) | MT24164 | this paper | cyp-36A1(gk824636) I ; agIs9 III | |
| Strain, strain background (C. elegans) | MT24165 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT24166 | this paper | agIs9 III ; egl-9(sa307) V hif-1(ia4) V | |
| Strain, strain background (C. elegans) | MT24167 | this paper | agIs9 III ; egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT24177 | this paper | agIs9 III ; hif-1(ia4) V | |
| Strain, strain background (C. elegans) | MT24179 | this paper | cyp-36A1(n5666) I ; (nIs470) IV | |
| Strain, strain background (C. elegans) | MT24520 | this paper | cyp-36A1(n5666) I ; (nIs470) IV ; egl-9(n586ts) V ; nIs674 | |
| Strain, strain background (C. elegans) | MT24622 | this paper | cyp-36A1(n5666) I ; (nIs470) IV ; egl-9(n586ts) V | |
| Strain, strain background (C. elegans) | MT24684 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6126) IV ; egl-9(sa307) V ; nEx2586 | |
| Strain, strain background (C. elegans) | MT24690 | this paper | agIs9 III ; otEx3165 | |
| Strain, strain background (C. elegans) | MT24692 | this paper | agIs9 III ; otEx3156 | |
| Strain, strain background (C. elegans) | MT24911 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6126) IV ; egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT25047 | this paper | agIs9 III ; nEx2699 | |
| Strain, strain background (C. elegans) | MT25048 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nEx2699 | |
| Strain, strain background (C. elegans) | MT25050 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; otEx3156 | |
| Strain, strain background (C. elegans) | MT25051 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; otEx3165 | |
| Strain, strain background (C. elegans) | MT25104 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6126) IV ; egl-9(sa307) V ; nEx2713 | |
| Strain, strain background (C. elegans) | MT25107 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6126) IV ; egl-9(sa307) V ; nEx2715 | |
| Strain, strain background (C. elegans) | MT25196 | this paper | agIs9 III ; nhr-46(n6125) IV | |
| Strain, strain background (C. elegans) | MT25197 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6125) IV ; egl-9(sa307) V | |
| Strain, strain background (C. elegans) | MT25211 | this paper | cyp-36A1(gk824636) I | |
| Strain, strain background (C. elegans) | MT25212 | this paper | cyp-36A1(gk824636) I ; egl-9(n586ts) V | |
| Strain, strain background (C. elegans) | MT25213 | this paper | cyp-36A1(gk824636) I ; nhr-46(n6126) IV ; egl-9(n586ts) V | |
| Strain, strain background (C. elegans) | MT25214 | this paper | nhr-46(n6126) IV | |
| Strain, strain background (C. elegans) | MT25215 | this paper | egl-9(n586ts) V | |
| Strain, strain background (C. elegans) | MT25596 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; egl-9(sa307) V ; nEx2853 | |
| Strain, strain background (C. elegans) | MT25599 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; egl-9(sa307) V ; nEx2856 | |
| Strain, strain background (C. elegans) | MT25601 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; egl-9(sa307) V ; nEx2849 | |
| Strain, strain background (C. elegans) | MT25605 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; egl-9(sa307) V ; nEx2859 | |
| Strain, strain background (C. elegans) | MT25606 | this paper | agIs9 III ; nEx2860 | |
| Strain, strain background (C. elegans) | MT25610 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nhr-46(n6126) IV ; egl-9(sa307) V ; nEx2864 | |
| Strain, strain background (C. elegans) | MT25611 | this paper | cyp-36A1(gk824636) I ; agIs9 III ; nEx2860 | |
| Strain, strain background (C. elegans) | MT25627 | this paper | agIs9 III ; nhr-46(n6126) IV | |
| Strain, strain background (C. elegans) | ZG31 | Huaqi Jiang/Jo Anne Powell-Coffman | hif-1(ia4) V |
Additional files
-
Supplementary file 1
Genes that were significantly differentially expressed between wild type and egl-9(lf) and at least twofold downregulated in egl-9(lf) vs. wild type.
- https://doi.org/10.7554/eLife.36828.019
-
Supplementary file 2
Genes that were significantly differentially expressed between wild type and egl-9(lf) and at least twofold upregulated in egl-9(lf) vs. wild type.
- https://doi.org/10.7554/eLife.36828.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36828.021





