Thioredoxin shapes the C. elegans sensory response to Pseudomonas produced nitric oxide
Figures
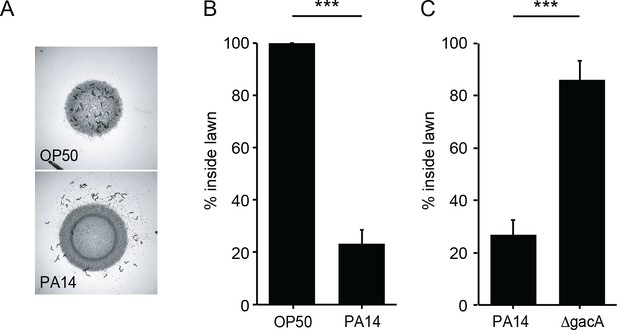
C. elegans avoids PA14, but not OP50 or ΔgacA.
(A) Foraging behavior of wild-type animals after 8 hr exposure to E. coli OP50 (top) and P. aeruginosa PA14 (bottom) lawns is shown. (B) Lawn occupancy of wild-type animals on OP50 and PA14 after 8 hr are compared. (C) Lawn occupancy of wild-type animals on PA14 vs. ΔgacA after 8 hr. ***p<0.001 (Student’s t-test). Values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM.
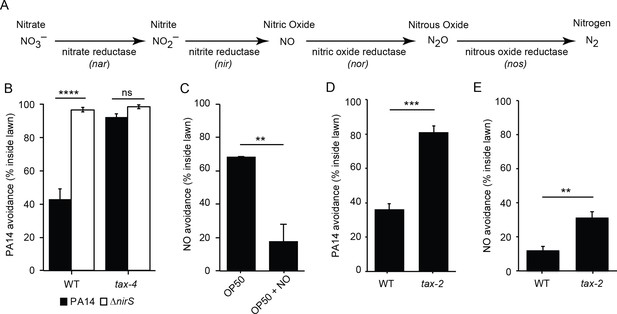
Bacterial NO is required for PA14 avoidance.
(A) The NO biosynthetic pathway in P. aeruginosa is shown. (B) Avoidance of PA14 ΔnirS lawns was dramatically reduced compared to wild-type PA14. By contrast, neither wild-type nor nirS mutants elicited lawn avoidance in tax-4 mutant worms. ****p<0.0001, ns: not significant, (one-way ANOVA, Tukey’s multiple comparisons test) (F = 66.7, p<0.0001). (C) E. coli OP50 (OP50) lawns supplemented with the NO donor DPTA NONOate elicited C. elegans avoidance after a 30 min exposure. (D–E) The mutation tax-2(p671) decreased PA14 (D) and NO donor (E) avoidance. (C–E) **p<0.01 and ***p<0.001 (Student’s t-test). Values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM.
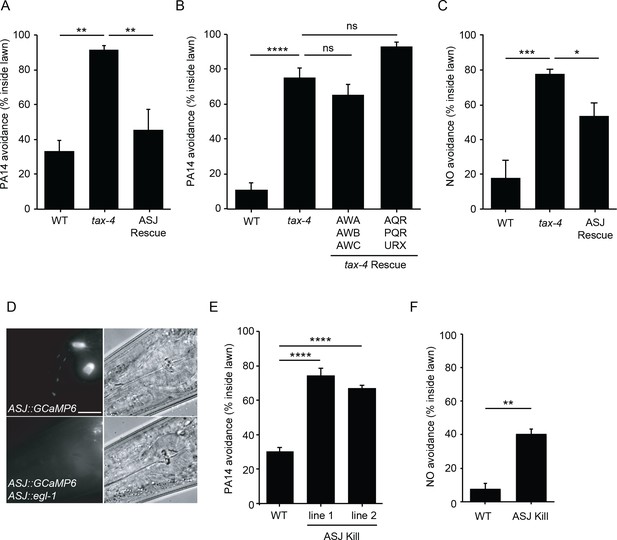
ASJ mediates both PA14 and NO avoidance behavior.
(A, B, C) The mutation tax-4(p678) abolished PA14 (A, B) and NO donor (C) avoidance. A transgene expressing tax-4 selectively in ASJ (A, C), but not in AWA/AWB/AWC or AQR/PQR/URX (B) rescued both PA14 and NO donor avoidance defects. (D) Fluorescence (left) and Nomarski (right) images of head region of wild-type animals expressing ptrx-1::GCaMP6.0 (top), or ptrx-1::GCaMP6.0 with pssu-1::egl-1 (bottom). Scale bar indicates 5 μm. (E, F) ASJ-ablated animals exhibited significantly decreased PA14 (E) and NO donor (F) avoidance. (A, B, C, E) *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, ns: not significant (one-way ANOVA, Tukey’s multiple comparisons test) (A, F = 14.61, p=0.0015; B, F = 53.71, p<0.0001; C, F = 14.06, p=0.0012; E, F = 69.88, p<0.0001). (F) **p<0.01 (Student’s t-test). Values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM. tax-4 transgenes are as follows: ASJ (trx-1 promoter), AWA/AWB/AWC (odr-3 promoter), AQR/PQR/URX (gcy-36 promoter).
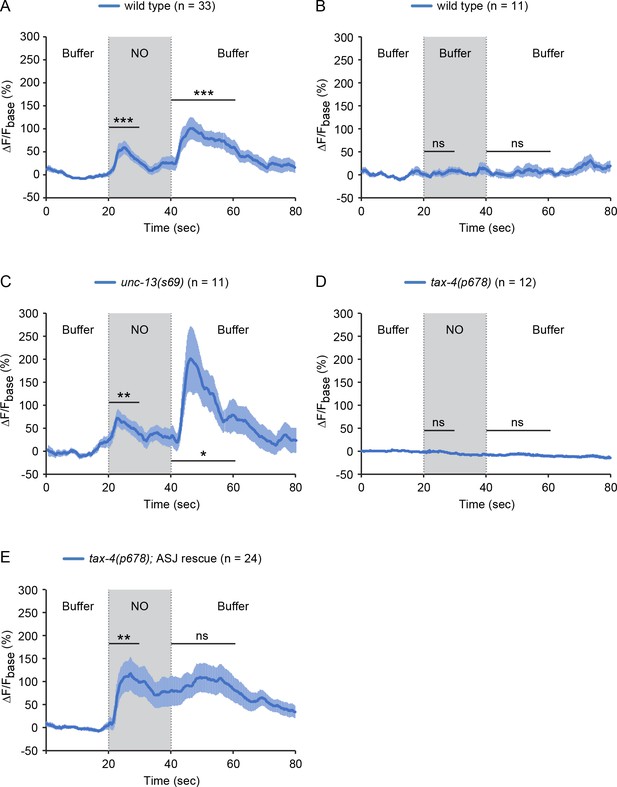
The sensory neurons ASJ respond to NO.
(A, B) ASJ neurons respond to the onset and removal of NO with increased GCaMP6 signal (A) but do not respond when switched between control buffer (B). (C) Blocking synaptic transmission (in unc-13 mutants) had little effect on ASJ responses to NO. (D, E) The tax-4(p678) mutation abolished the ASJ response to the onset and the removal of NO (D). Expressing a wild-type tax-4 cDNA in ASJ rescued ASJ response to the onset of NO stimulation (E). Mean (solid line) and SEM (shaded area) of GCaMP fluorescence are shown. Wilcoxon signed rank test for data that were not normally distributed (the response to NO onset in A, E and the response to NO removal in A, B, D, E); paired Student’s t-test for normally distributed data (the response to NO onset in B-D and the response to NO removal in C). ***p<0.001, **p<0.01, *p<0.05, ns: not significant (Materials and methods).
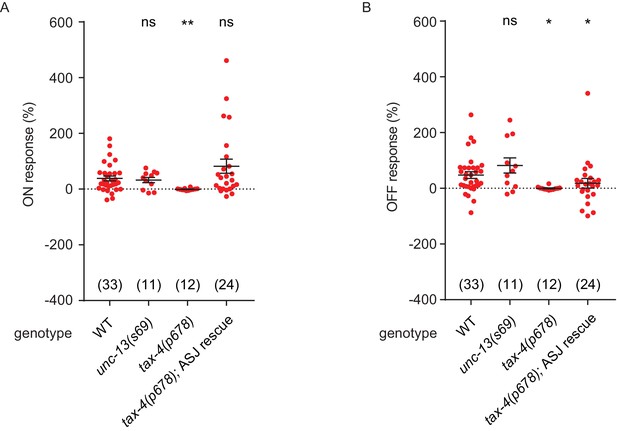
NO response amplitudes for data in Figure 4.
(A, B) NO evoked ON (A) and OFF (B) response amplitudes (scatter plots and mean ± SEM) are shown for recordings in Figure 4. ON and OFF response amplitudes are defined in the Methods. For statistical analysis, responses of all mutant genotypes in Figures 4–7 were compared with a common set of wild type controls using Kruskal–Wallis one-way analysis of variance. Significant differences from wild type are indicated as follows: *p<0.05; **p<0.01; ns, not significant. The number of animals analyzed is indicated for each genotype.
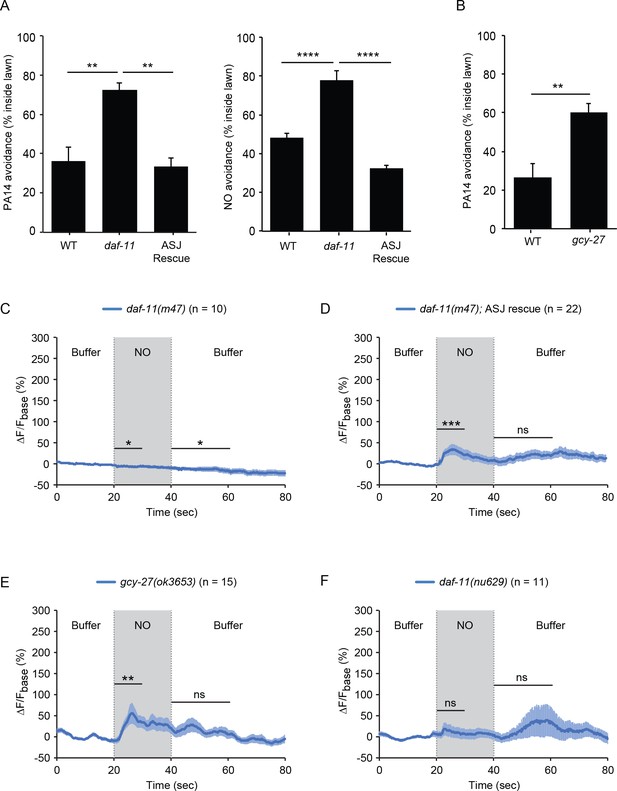
The rGCs DAF-11 and GCY-27 are required for ASJ responses to NO.
(A) The daf-11(m47) mutation abolished PA14 (left) and NO donor (right) avoidance, both of which were reinstated by a transgene that expresses daf-11 in the ASJ neurons. **p<0.01, and ****p<0.0001 (one-way ANOVA, Tukey’s multiple comparisons test) (left: F = 12.83, p=0.0046; right: F = 43.05, p<0.0001). (B) The gcy-27(ok3653) mutation decreased PA14 avoidance. **p<0.01 (Student’s t-test). For (A–B), values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM. (C, D) The daf-11(m47) mutation abolished the NO-evoked calcium transients in the ASJ neurons (C). Expressing a wild-type daf-11 cDNA in ASJ rescued the NO-evoked ON response (D). Note, NO-onset and removal slightly suppressed the ASJ GCaMP6 signal in daf-11(m47) mutants. (E) The gcy-27(ok3653) mutation eliminated the ASJ OFF response to NO. (F) The daf-11(nu629ΔHNOBA) mutation abolished the ASJ ON and OFF responses to NO. For (A and B), values represent means of four independent experiments, with 40 animals analyzed in each replicate. For (C–F), mean (solid line) and SEM (shaded area) GCaMP fluorescence are shown. Wilcoxon signed rank test for data that were not normally distributed (for the response to NO onset in D-F and for the response to NO removal in D, F); paired Student’s t-test for normally distributed data (for the response to NO onset in C and for the response to NO removal in C, E). ***p<0.001, **p<0.01, *p<0.05, ns: not significant (Materials and methods).
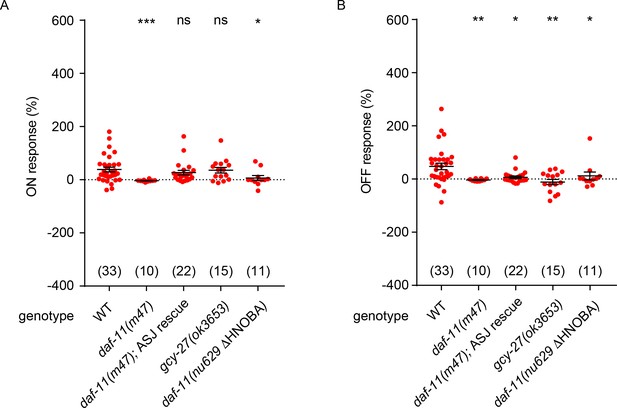
NO response amplitudes for data in Figure 5.
(A, B) NO evoked ON (A) and OFF (B) response amplitudes (scatter plots and mean ± SEM) are shown for recordings in Figure 5. ON and OFF response amplitudes are defined in the Methods. For statistical analysis, responses of all mutant genotypes in Figures 4–7 were compared with a common set of wild type controls using Kruskal–Wallis one-way analysis of variance. Significant differences from wild type are indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ns, not significant. The number of animals analyzed is indicated for each genotype.
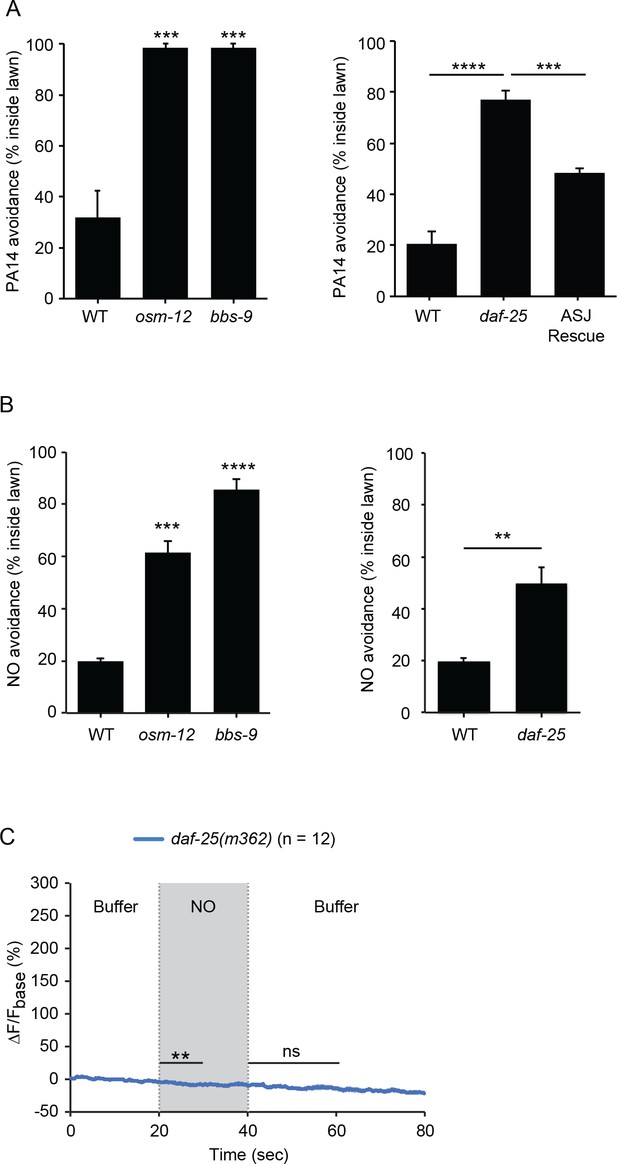
C. elegans senses NO as an external cue.
(A, B) osm-12(n1606), bbs-9(gk471) and daf-25(m362) mutants were defective for PA14 (A) and NO donor (B) avoidance. Expressing a wild-type daf-25 cDNA in ASJ partially rescued the daf-25 mutant PA14 avoidance defect (A). (A) ***p<0.001, and ****p<0.0001 (one-way ANOVA, Tukey’s multiple comparisons test) (left panel: F = 33.77, p=0.0005; right panel: F = 50.09, p<0.0001). (B) Left panel, ***p<0.001, and ****p<0.0001 (one-way ANOVA, Tukey’s multiple comparisons test) (F = 77.17, p<0.0001). Right panel, **p<0.01 (Student’s t-test). For (A and B), values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM. The daf-25 ASJ transgene was expressed by the ssu-1 promoter. (C) daf-25(m362) mutants lacked ASJ responses to the onset and removal of NO. Mean (solid line) and SEM (shaded area) of GCaMP fluorescence are shown. Paired Student’s t-test for normally distributed data, **p<0.01, ns: not significant (Materials and methods). NO-onset slightly suppressed the GCaMP6 signal in ASJ.
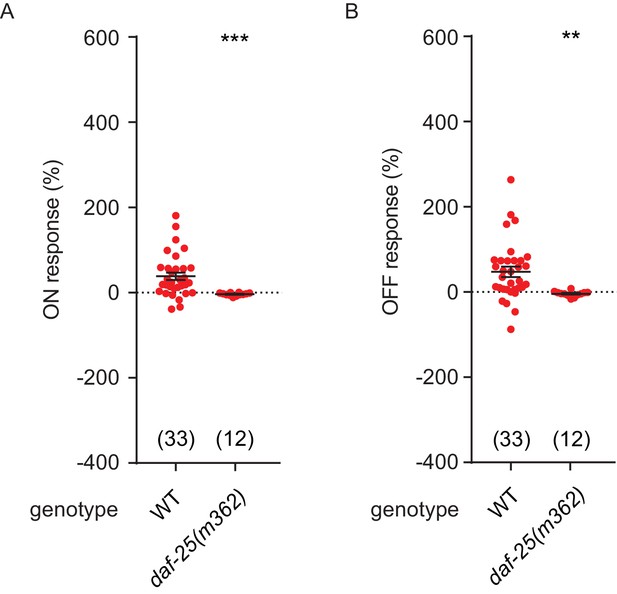
NO response amplitudes for data in Figure 6.
(A, B) NO evoked ON (A) and OFF (B) response amplitudes (scatter plots and mean ± SEM) are shown for recordings in Figure 6. ON and OFF response amplitudes are defined in the Methods. For statistical analysis, responses of all mutant genotypes in Figures 4–7 were compared with a common set of wild type controls using Kruskal–Wallis one-way analysis of variance. Significant differences from wild type are indicated as follows: **p<0.01; ***p<0.001. The number of animals analyzed is indicated for each genotype.
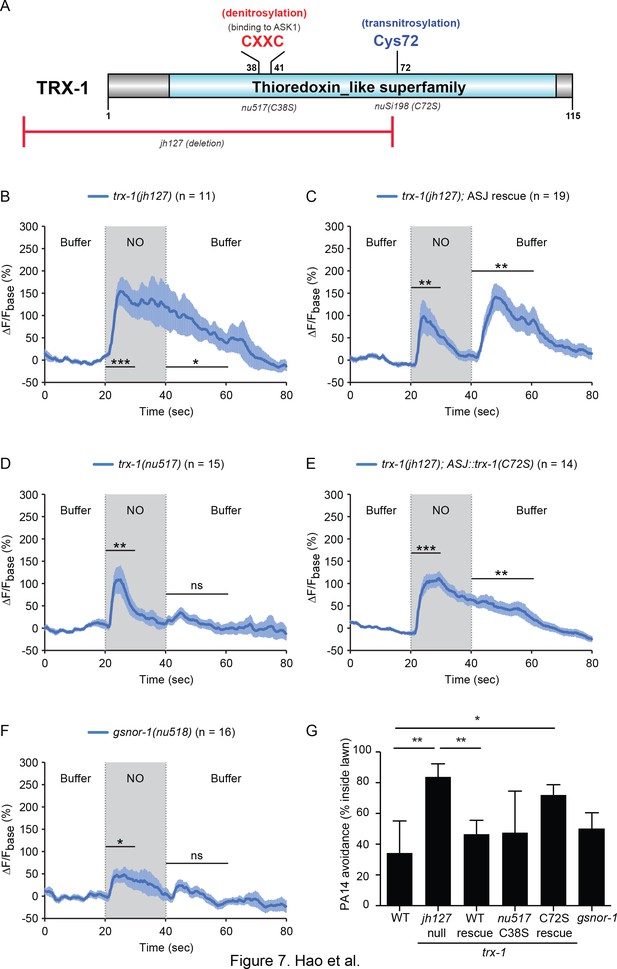
TRX-1/Thioredoxin regulates the ASJ response to NO.
(A) Domain organization of C. elegans thioredoxin TRX-1. Cysteines forming the de-nitrosylation active site (Cys38XXCys41) in TRX-1 are highlighted in red. Cys72, which is involved in trans-nitrosylation is highlighted in blue. The deleted region in jh127 as well as point mutations in nu517 (C38S) and nuSi198 (C72S) are indicated. (B, C) A trx-1(jh127) null mutant exhibited a prolonged ASJ response to the onset of NO simulation, which slowly returned to baseline following NO removal (B). This defect was rescued by a single copy transgene expressing wild-type trx-1 in ASJ (C). (D) A mutation in the active site for de-nitrosylation, trx-1(nu517 C38S) eliminated the ASJ response to NO removal. (E) In contrast to the full rescue by wild type TRX-1 (C), expressing a mutant TRX-1(C72S) single copy transgene failed to rescue the defective ASJ response to NO in trx-1(jh127) null mutants. (F) A mutation inactivating another de-nitrosylation enzyme (gsnor-1) disrupted the response of ASJ to the removal of NO stimulation, similar to the phenotype exhibited by the de-nitrosylation defective trx-1(nu517 C38S) mutant (D). Mean (solid line) and SEM (shaded area) of GCaMP fluorescence are shown. Wilcoxon signed rank test for data that were not normally distributed (for response to NO onset in C and for response to NO removal in B, (C); paired Student’s t-test for normally distributed data (for response to NO onset in B, D-F and for response to NO removal in D-F). ***p<0.001, **p<0.01, *p<0.05, ns: not significant (Materials and methods). (G) PA14 avoidance was analyzed in the indicated genotypes. PA14 avoidance was significantly reduced in trx-1 null mutants. This defect was rescued by a single copy transgene expressing wild type TRX-1 but was not rescued by a TRX-1(C72S) mutant transgene. No significant differences were observed for the other genotypes. For (G), *p<0.05, and **p<0.01 (one-way ANOVA, Tukey’s multiple comparisons test) (F = 5.50, p=0.003) followed by Tukey’s multiple comparisons test. Values represent means of four independent experiments, with ~40 animals analyzed in each replicate. Error bars indicate SEM.
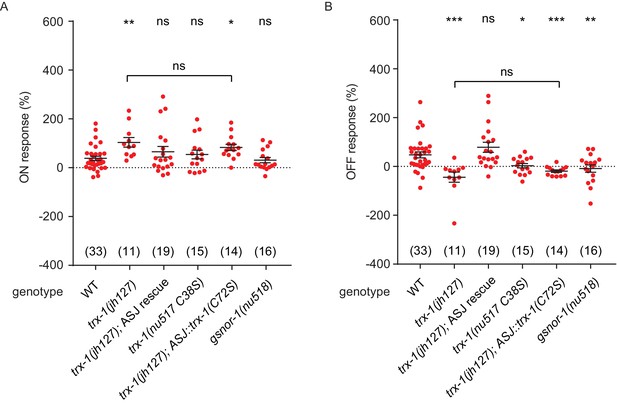
NO response amplitudes for data in Figure 7.
(A, B) NO evoked ON (A) and OFF (B) response amplitudes (scatter plots and mean ± SEM) are shown for recordings in Figure 7. ON and OFF response amplitudes are defined in the Methods. For statistical analysis, responses of all mutant genotypes in Figures 4–7 were compared with a common set of wild type controls using Kruskal–Wallis one-way analysis of variance. Significant differences from wild type are indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ns, not significant. The number of animals analyzed is indicated for each genotype.
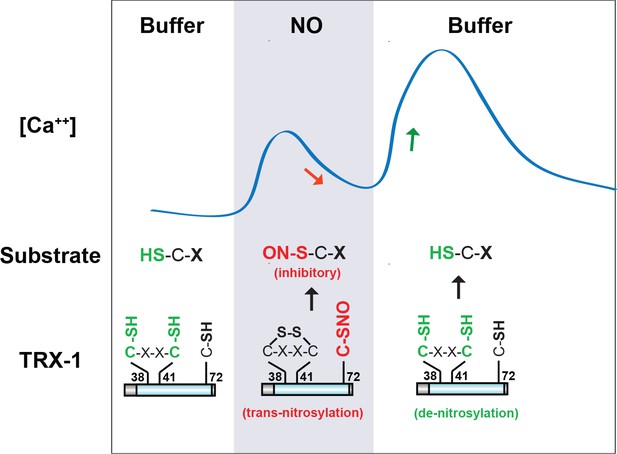
A model for TRX-1/Thioredoxin function in NO sensation.
Our data suggest that two enzymatic functions of TRX-1 endow ASJ neurons with a bi-phasic response to environmental NO. The trans-nitrosylation activity is proposed to be active during NO exposures. Trans-nitrosylation of an unidentified protein substrate is proposed to inhibit the ASJ ON response. The de-nitrosylation activity is proposed to be active following NO removal and is proposed to activate the ASJ OFF response (by reversing the inhibitory SNO-protein adducts formed during NO exposures). This model is described in greater detail in the Discussion section.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (C. elegans) | N2 Bristol | http://www.wormbase.org | N2 | N/A |
| Strain (C. elegans) | tax-4(p678) | http://www.wormbase.org | PR678 | N/A |
| Strain (C. elegans) | tax-2(p671) | http://www.wormbase.org | PR671 | N/A |
| Strain (C. elegans) | tax-4(p678);nuEx1965[podr-3::TAX-4] | This paper | KP9653 | 10ng/ul KP#3587 injected |
| Strain (C. elegans) | tax-4(p678);nuEx1966[pgcy-36::TAX-4] | This paper | KP9654 | 10ng/ul KP#3588 injected |
| Strain (C. elegans) | tax-4(p678);nuEx1967[ptrx-1::TAX-4] | This paper | KP9655 | 10ng/ul KP#3589 injected |
| Strain (C. elegans) | nuIs556;nuEx1968[ptrx-1::EGL-1] | This paper | KP9656 | 10ng/ul KP#3590 injected |
| Strain (C. elegans) | nuIs556[ptrx-1::GCaMP6.0s] | This paper | KP9672 | KP#3311 (25 ng/ul), UV integrated |
| Strain (C. elegans) | unc-13(s69);nuIs556 | This paper | KP9673 | N/A |
| Strain (C. elegans) | tax-4(p678);nuIs556 | This paper | KP9657 | N/A |
| Strain (C. elegans) | tax-4(p678);nuIs556;nuSi211[ptrx-1::TAX-4] | This paper | KP9658 | nuSi211 single copy miniMos |
| Strain (C. elegans) | daf-11(m47) | http://www.wormbase.org | DR47 | N/A |
| Strain (C. elegans) | daf-11(ks67) | http://www.wormbase.org | FK183 | N/A |
| Strain (C. elegans) | daf-11(m47);nuSi196 [pssu-1::DAF-11] | This paper | KP9659 | nuSi196 single copy miniMos |
| Strain (C. elegans) | daf-11(m47);nuIs556 | This paper | KP9660 | N/A |
| Strain (C. elegans) | daf-11(m47);nuIs556;nuSi196 | This paper | KP9661 | N/A |
| Strain (C. elegans) | gcy-27(ok3653) | http://www.wormbase.org | RB2622 | N/A |
| Strain (C. elegans) | gcy-27(ok3653);nuIs556 | This paper | KP9662 | N/A |
| Strain (C. elegans) | daf-11(nu629 ΔHNOBA);nuIs556 | This paper | KP9663 | nu629 isolated by CRISPR; Sequence in Methods |
| Strain (C. elegans) | osm-12(n1606) | http://www.wormbase.org | MT3645 | N/A |
| Strain (C. elegans) | bbs-9(gk471) | http://www.wormbase.org | VC1062 | N/A |
| Strain (C. elegans) | daf-25(m362) | http://www.wormbase.org | DR2386 | N/A |
| Strain (C. elegans) | daf-25(m362);nuEx1969[pssu-1::DAF-25] | This paper | KP9664 | 10 ng/ul KP#3593 injected |
| Strain (C. elegans) | daf-25(m362);nuIs556 | This paper | KP9665 | N/A |
| Strain (C. elegans) | trx-1(jh127) | http://www.wormbase.org | KJ412 | N/A |
| Strain (C. elegans) | trx-1(jh127);nuIs556 | This paper | KP9666 | N/A |
| Strain (C. elegans) | trx-1(jh127);nuSi197 [ptrx-1::TRX-1] | This paper | KP9674 | nuSi197 single copy miniMos |
| Strain (C. elegans) | trx-1(jh127);nuIs556;nuSi197 | This paper | KP9667 | N/A |
| Strain (C. elegans) | trx-1(nu517 C38S) | This paper | KP9675 | nu517 isolated by CRISPR; Sequence in Methods |
| Strain (C. elegans) | trx-1(nu517 C38S);nuIs556 | This paper | KP9668 | N/A |
| Strain (C. elegans) | trx-1(jh127);nuSi198 [ptrx-1::TRX-1(C72S)] | This paper | KP9676 | nuSi198 single copy miniMos |
| Strain (C. elegans) | trx-1(jh127);nuIs556;nuSi198 | This paper | KP9669 | N/A |
| Strain (C. elegans) | gsnor-1(nu518) | This paper | KP9670 | nu518 isolated by CRISPR; Sequence in Methods |
| Strain (C. elegans) | gsnor-1(nu518);nuIs556 | This paper | KP9671 | N/A |
| Strain (E. coli) | OP50 | (Brenner, 1974) | OP50 | N/A |
| Strain (P. aeruginosa) | PA14 | Fred Ausubel lab | PA14 | N/A |
| Strain (P. aeruginosa) | PA14 gacA | Fred Ausubel lab | PA14 gacA | N/A |
| Strain (P. aeruginosa) | PA14 nirS | Constantine Haidaris lab | PA14 nirS | (Van Alst et al., 2009) |
| Recombinant DNA reagent | pmyo-2::NLS-mCherry | Kaplan lab | KP#1480 | N/A |
| Recombinant DNA reagent | punc-122::mCherry | This paper | KP#2186 | 796bp unc-122 promoter |
| Recombinant DNA reagent | podr-3::TAX-4 | This paper | KP#3587 | 3kb odr-3 promoter; tax-4 cDNA from C. Bargmann |
| Recombinant DNA reagent | pgcy-36::TAX-4 | This paper | KP#3588 | gcy-36 promoter from C. Bargmann |
| Recombinant DNA reagent | ptrx-1::TAX-4 | This paper | KP#3589 | 1028bp trx-1 promoter |
| Recombinant DNA reagent | ptrx-1::EGL-1 | This paper | KP#3590 | 321bp egl-1 cDNA |
| Recombinant DNA reagent | ptrx-1::GCaMP6.0s | This paper | KP#3311 | GCaMP6.0s from Jihong Bai |
| Recombinant DNA reagent | ptrx-1::TAX-4 miniMos | This paper | KP#3591 | N/A |
| Recombinant DNA reagent | pssu-1::DAF-11 miniMos | This paper | KP#3592 | 543bp ssu-1 promoter and 3234bp daf-11 cDNA |
| Recombinant DNA reagent | pssu-1::DAF-25 | This paper | KP#3593 | 543 bp ssu-1 promoter; 1167bp daf-25 cDNA |
| Recombinant DNA reagent | ptrx-1::TRX-1 miniMos | This paper | KP#3594 | 1028bp trx-1 promoter; 723bp trx-1b genomic fragment |
| Recombinant DNA reagent | ptrx-1::TRX-1(C72S) miniMos | This paper | KP#3595 | created by Site-Directed Mutagenesis |
| Commercial assay or kit | QIAprep Spin Miniprep Kit | QIAGEN | 27106 | N/A |
| Commercial assay or kit | QIAquick Gel Extraction Kit | QIAGEN | 28706 | N/A |
| Commercial assay or kit | HiSpreed Plasmid Midi Kit | QIAGEN | 12643 | N/A |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | NEW ENGLAND BIOLABS INC | E0552S | N/A |
| Chemical compound, drug | DPTA NONOate | Cayman Chemical | 82110 | N/A |
| Chemical compound, drug | MAHMA NONOate | Sigma | M1555 | N/A |
| Chemical compound, drug | BDM (2,3-Butanedione monoxide) | Sigma | B0753 | N/A |
| Software, algorithm | Metamorph 7.1 | Molecular Devices | N/A | N/A |
| Software, algorithm | Fiji | https://fiji.sc/ | N/A | N/A |
| Software, algorithm | Prism 6 | https://www.graphpad.com/scientific-software/prism/ | N/A | N/A |
| Software, algorithm | IBM SPSS statistics 25 | https://www.ibm.com/products/spss-statistics | N/A | N/A |
| Other | olfactory microfluidic chips | (Chronis et al., 2007) | N/A | N/A |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36833.015





