Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi
Figures
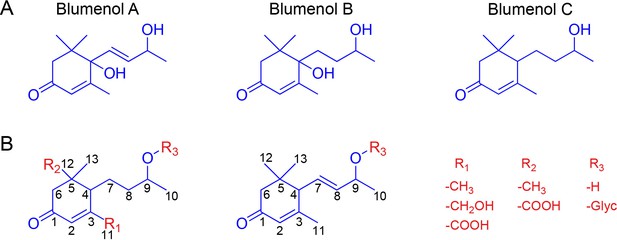
Blumenol core structures and exemplary modifications.
(A) Structure of blumenol A, blumenol B and blumenol C. (B) Exemplary blumenol C derivatives. Glyc, glycoside.
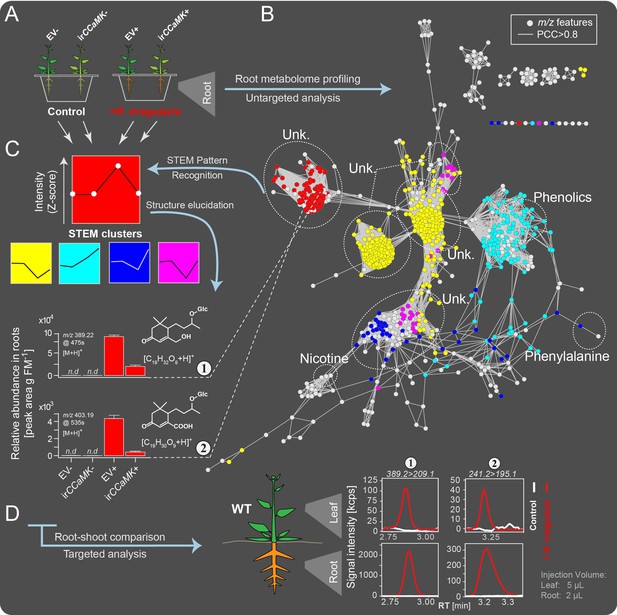
Combined targeted and untargeted metabolomics identified blumenol derivatives as AMF-indicative in-planta metabolic fingerprints in the roots and leaves of Nicotiana attenuata plants.
(A) Experimental set-up. EV and irCCaMK plants were co-cultured and inoculated with or without R. irregularis. Six weeks after inoculation (wpi), root samples were harvested for metabolite profiling. (B) Covariance network visualizing m/z features from UHPLC-qTOF-MS untargeted analysis (n = 8). Known compounds, including nicotine, phenylalanine and various phenolics, and unknown metabolites (Unk.) are enclosed in dashed ellipses. (C) Normalized Z-scores of m/z features were clustered using STEM Clustering; 5 of 8 significant clusters are shown in different colors and mapped onto the covariance network. The intensity variation (mean +SE) of 2 selected features (Compounds 1 and 2) are shown in bar plots (n.d., not detected). (D) Representative chromatograms of Compounds 1 and 2 in roots and leaves of plants with and without AMF inoculation, as analyzed by targeted UHPLC-triple quadrupole-MS metabolomics.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.37093.007
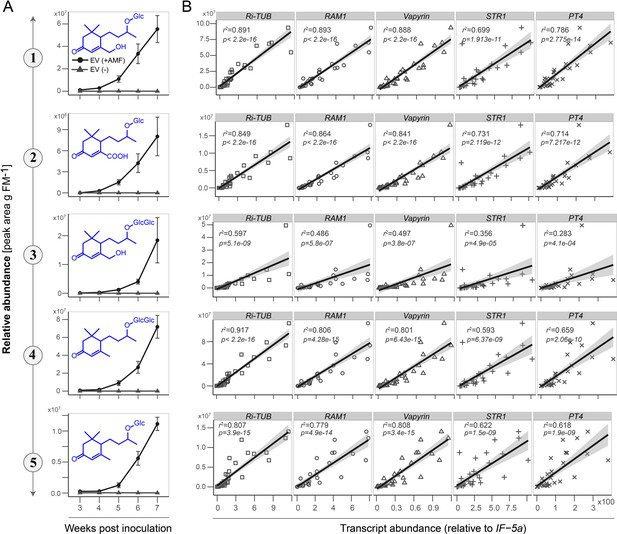
Abundance of root blumenol derivatives correlates positively with root AMF colonization.
(A) Time lapse accumulations (3–7 weeks post inoculation, n ≥ 3 for each time point) of Compounds 1, 2, 3, 4 and 5 in roots of plants with (EV+, black lines with circles) and without (EV-, grey lines with triangle) AMF inoculation. The experiment was conducted with empty vector (EV) transformed plants. Data are means ±SE. (B) Abundance of Compounds 1, 2, 3, 4 and 5 relative to the transcript abundance of the R. irregularis specific housekeeping gene, Ri-tub (GenBank: EXX64097.1), as well as to the plant-derived marker genes RAM1, Vapyrin, STR1 and PT4 (Gene ID and transcripts abundance are listed in Data Set 1). The transcript abundance was quantified by q-PCR, relative to NaIF-5a (NCBI Reference Sequence: XP_019246749.1). The correlations among blumenol derivatives and the transcript abundances of marker genes were analyzed by linear regression (lm) models.
-
Figure 2—figure supplement 1—source data 1
Source data for Source data for Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.37093.006
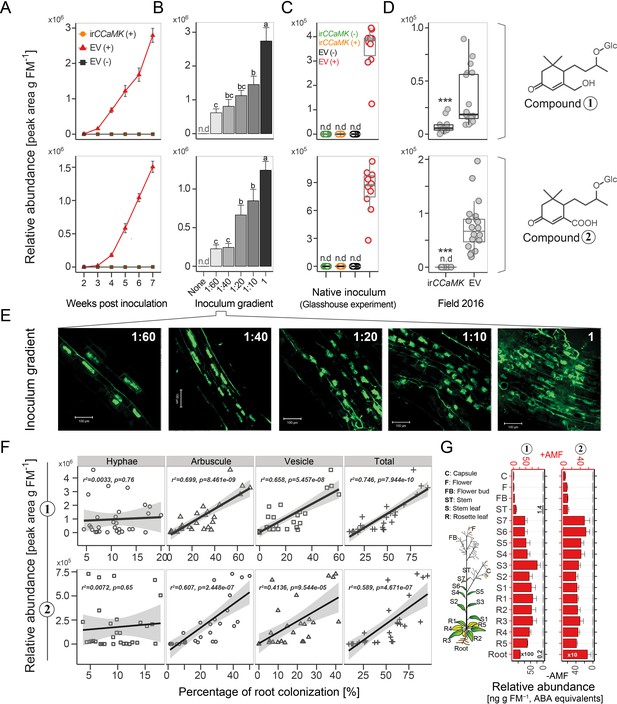
Compounds 1 and 2 are leaf markers of root AMF colonization in N. attenuata.
(A) Time lapse accumulations of Compounds 1 and 2 in leaves of EV plants with (EV+, red) or without (EV-, black) AMF-inoculation and of irCCaMK plants with AMF-inoculation (irCCaMK+, orange, covered by black) (means ± SE, n ≥ 5). (B) Leaf abundances of Compounds 1 and 2 (five wpi) of plants inoculated with different inoculum concentrations (means + SE, n ≥ 4); different letters indicate significant differences (p<0.05, one-way ANOVA followed by Fisher’s LSD). (C) Compounds 1 and 2 in leaf samples of EV and irCCaMK plants inoculated with (+) or without (-) AMF inoculum isolated from the plant’s native habitat (six wpi); different letters indicate significant differences (p<0.05, one-way ANOVA followed by Tukey’s HSD, n = 10). (D) Field experiment (Great Basin Desert, Utah, USA): Compounds 1 and 2 in leaf samples of EV (n = 20) and irCCaMK (n = 19) plants sampled eight weeks after planting. (Student’s t-test: ***p<0.001). (E) Representative images of WGA-488 stained roots of plants shown in B) (bar = 100 μm). (F) Leaf Compounds 1 and 2 relative to the percentage of root colonization by hyphae, arbuscules, vesicles and total root length colonization of the same plants (linear regression model). (G) Compounds 1 and 2 in 17 different tissues of plants with (+AMF, n = 3, red bars) or without (-AMF, n = 1, black bars) AMF-inoculation harvested at six wpi.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.37093.015
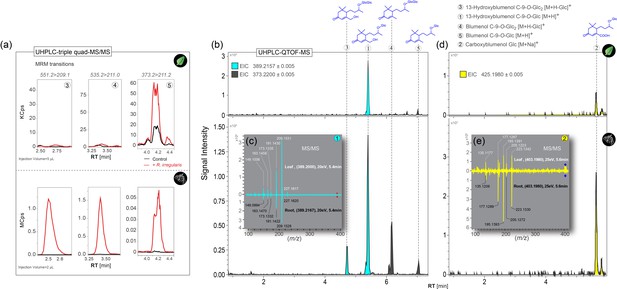
AMF-induced accumulation of blumenol derivatives in roots and leaves of N. attenuata.
(A) Representative chromatograms of targeted tandem MS-based analyses of Compounds 3, 4 and 5 in roots (bottom panel) and leaves (top panel) of N. attenuata plants after inoculation with R. irregularis (+R. irregularis, red line, 6wpi) and in untreated control plants (Control, black line). Experiments were conducted with wild type (WT) plants. The respective precursor-to-product ion transitions are indicated at the top. (B), (D) Representative chromatograms of a high resolution MS-based analysis of Compounds 1, 3, 4 and 5 (B), as well as Compound 2 (D) in roots (bottom panel) and leaves (top panel) of N. attenuata plants after inoculation with R. irregularis. Extracted ion chromatograms (EIC) are labeled by colors and settings listed at the top. (C), (E) Comparison of fragmentation patterns of Compounds 1 (C) and 2 (E) in both tissues by high resolution tandem MS.
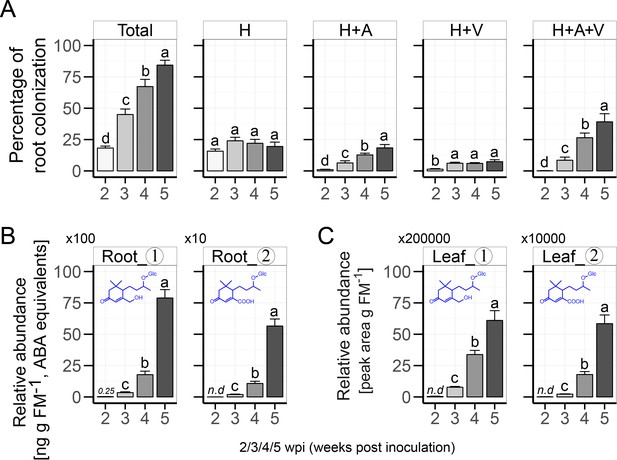
Time course analysis of the root colonization by AMF and the corresponding accumulation of Compounds 1 and 2 in roots and leaves of N. attenuata.
(A) Root colonization in EV plants at different time points after inoculation with R. irregularis (2/3/4/5 wpi). H: hyphae; A: arbuscules; V: vesicles; Total: total root length colonization (means +SE; n = 8). (B, C) Abundances of Compounds 1 and 2 in roots (B) and leaves (C) of plants at different time points after inoculation with R. irregularis (2/3/4/5 wpi; means +SE; n ≥ 5). Different letters indicate significant differences (p<0.05, one-way ANOVA followed by Tukey’s HSD). n.d., not detected.
-
Figure 3—figure supplement 2—source data 1
Source data for Source data for Figure 3—figure supplement 2.
- https://doi.org/10.7554/eLife.37093.011
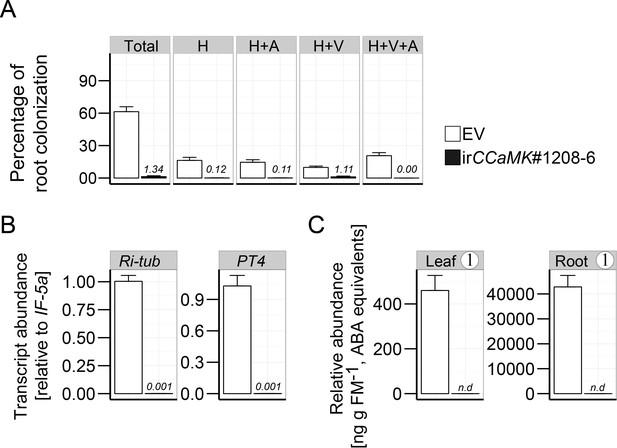
Root AMF colonization and abundance of Compound 1 in a second independently transformed irCCaMK line.
(A) Root colonization analysis in EV and irCCaMK (A-09-1208-6) plants. H: hyphae; A: arbuscules; V: vesicles; Total: total root length colonization (means +SE; EV, n = 9; irCCaMK; n = 7). (B) Transcript abundance of AMF marker genes in roots of EV and irCCaMK plants inoculated with R. irregularis (six wpi; means +SE; n = 6). (C) Compound 1 levels in roots and leaves of EV and irCCaMK plants inoculated with R. irregularis (six wpi; means +SE; n = 8).
-
Figure 3—figure supplement 3—source data 1
Source data for Source data for Figure 3—figure supplement 3.
- https://doi.org/10.7554/eLife.37093.013
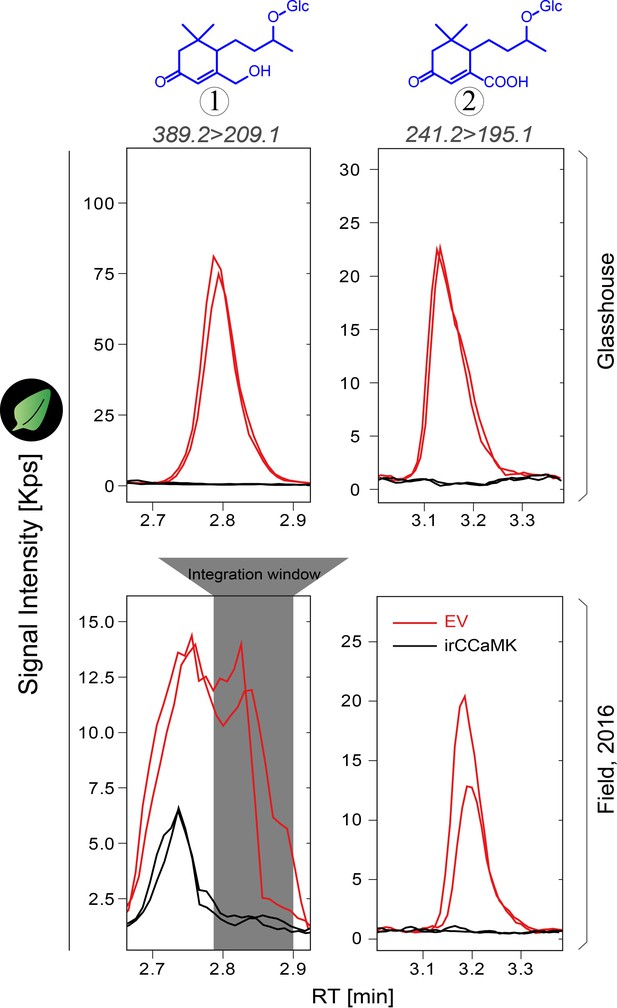
Signals from Compound 1 are partially disturbed in field samples, but not for Compound 2.
Leaf samples were harvested from glasshouse-(top panel) and field-grown, Utah, 2016 (bottom panel) plants for analysis. Representative chromatograms of two samples of each genotype, EV (red) and irCCaMK (black), are shown. Grey area indicates the peak integration window used for the quantification of Compound 1.
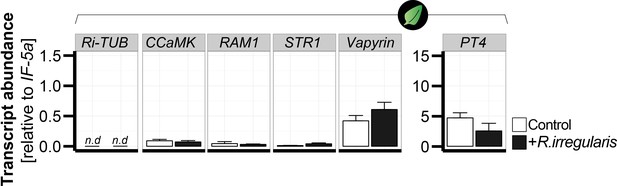
Transcript abundance of classical arbuscular mycorrhizal symbiosis-marker genes do not respond in leaves of mycorrhizal and control N. attenuata plants.
The transcript abundance (relative to NaIF-5a) of classical root marker genes was analyzed in leaves of N. attenuata plants in the presence (+R. irregularis, black bars) and absence (control, white bars) of root colonization with R. irregularis. The marker genes include the R. irregularis specific housekeeping gene, Ri-tub, as well as the plant-derived marker genes CCaMK, Vapyrin, PT4, STR1 and RAM1. Leaf samples were harvested six wpi and analyzed by qPCR. Data represent means +SE (n ≥ 3), n.d., not detected.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.37093.017
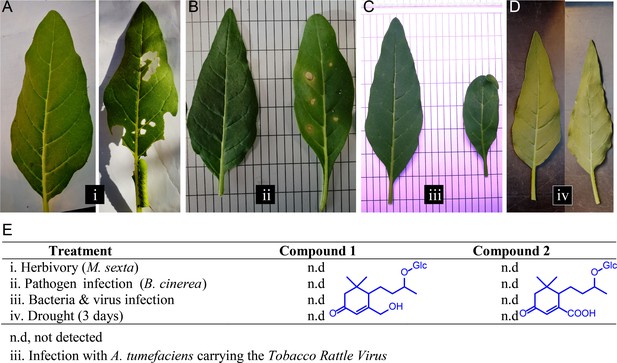
Different biotic and abiotic stresses do not elicit accumulations of Compounds 1 and 2 in leaves.
(A–D) Representative leaves of N. attenuata plants subjected to different stresses (right leaf), as well as the untreated controls (left leaf): (A) Manduca sexta feeding for 10 days; (B) Botrytis cinerea infection for five days. (C) Infection for two weeks with Agrobacterium tumefaciens carrying the Tobacco Rattle Virus; (D) Dehydration for three days. For each treatment, four biological replicates were used. (E) Accumulation of Compounds 1 and 2 in treated samples from (A–D). n.d., not detected.
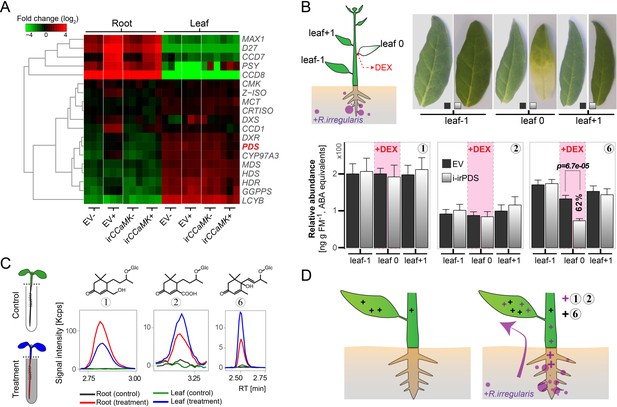
AMF-indicative Compounds 1 and 2 in shoots of mycorrhizal plants originate from the roots.
(A) Hierarchical clustering analysis of transcript abundance from RNA-seq of methylerythritol 4-phosphate (MEP) and (apo)carotenoid biosynthetic genes (for details see Figure 6—figure supplement 1A). (B) Compounds 1, 2 (AMF-specific) and 6 (not AMF-specific) in AMF-inoculated i-irPDS and EV plants. On each plant, a single stem leaf (leaf 0) was elicited with 100 μM DEX-containing paste for three weeks; treated and adjacent, untreated control leaves (leaf −1 and leaf +1) were harvested. Representative leaves are shown (bleaching indicates PDS silencing); (means +SE, n ≥ 6). The same leaf positions in i-irPDS and EV plants were compared by Student’s t-tests. (C) Contents of Compounds 1, 2 and 6 in the roots and shoots of seedlings whose roots were dipped for 1 d into an aqueous solution with (treatment) or without (control) AMF-indicative blumenols. (D) Model of blumenol distribution in plants with (right panel) and without (left panel) AMF colonization. The model illustrates constitutive blumenols (e.g., Compound 6 in N. attenuata) and AMF-indicative ones (e.g., Compounds 1 and 2 in N. attenuata) and their inferred transport.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.37093.024
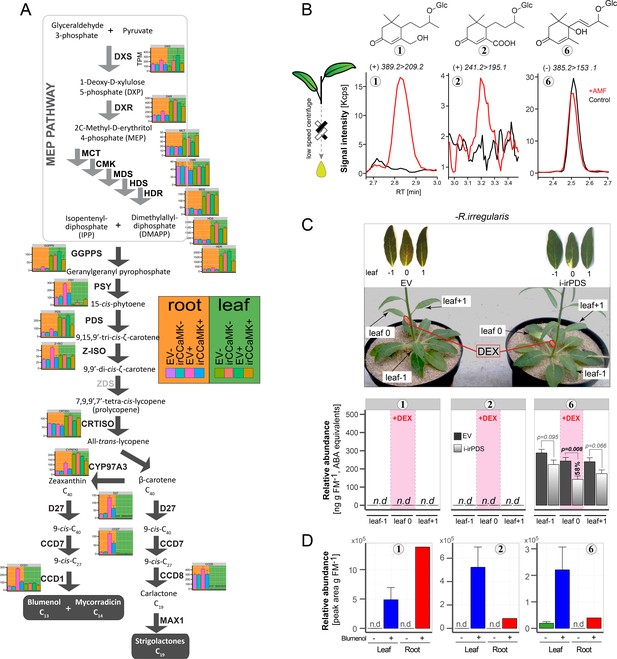
Foliar levels of Compounds 1 and 2 are derived from roots.
(A) Transcript abundance of MEP and apocarotenoid pathway biosynthetic genes (based on homologies to tomato, Arabidopsis and tobacco). Plant materials from the same experimental set-up as in Figure 2A were used for sequencing. Data are means +SE (n = 3) generated by RNA-seq and the abundance of each transcript is expressed in TPM (Transcripts per kilobase of exon model per million mapped reads). Transcripts were analyzed in roots (left panel, orange background) and leaf tissues (right panel, green background) of EV and irCCaMK plants with (EV+ and irCCaMK+ respectively) and without (EV- and irCCaMK- respectively) inoculations with R. irregularis. Gene abbreviations; CRTISO: carotenoid isomerase; GGPPs: geranylgeranyl diphosphate synthase; PSY: phytoene synthase; PDS: phytoene desaturase; ZDS: ζ-carotene de-saturase; Z-ISO: ζ- carotene isomerase; CCD: carotenoid cleavage dioxygenase; MAX1: cytochrome P450-type monooxygenase CYP711A1; DXS: 1-deoxy-D-xylulose 5-phosphate synthase; DXR: 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK: 4-(cytidine 5′-diphospho)−2-C-methyl-D-erythritol kinase; MDS: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS: 4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; D27: carotenoid isomerase. (B) Representative chromatograms from a targeted tandem MS-based analysis of Compounds 1, 2 and 6 in stem sap fluid of N. attenuata plants after R. irregularis inoculation (+AMF, red line, 6wpi) and of untreated control plants (Control, black line). The respective precursor-to-product ion transitions are indicated at top. (C) Accumulations of Compounds 1, 2 and 6 in non AMF-inoculated plants after local silencing of the carotenoid biosynthesis in the DEX-treated leaf. The experiment was performed with plants harboring a transformation construct mediating the chemically-inducible silencing of the phytoene desaturase (i-irPDS), as well as with empty vector (EV) plants. On each plant, a single stem leaf (leaf 0) was treated with a 100 μM dexamethasone (DEX) containing lanolin paste for 3 weeks. The adjacent, untreated leaves (leaf −1 and leaf +1) were harvested as controls. Representative leaves are shown (bleaching indicates functional PDS silencing). Data are means +SE (n ≥ 6). For statistical analysis, the samples from the same leaf positions in i-irPDS and EV plants were compared by Student’s t test. (D) Contents of Compounds 1, 2 and 6 in the roots (red bars) and shoots (blue bars) of seedlings whose roots were dipped into an aqueous solution with or without addition of the respective blumenols. Seedlings were incubated for 1d before analysis. Data are means +SE (shoot, n = 3; root, n = 1). The data originate from the same experiment presented in Figure 6C.
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.37093.021
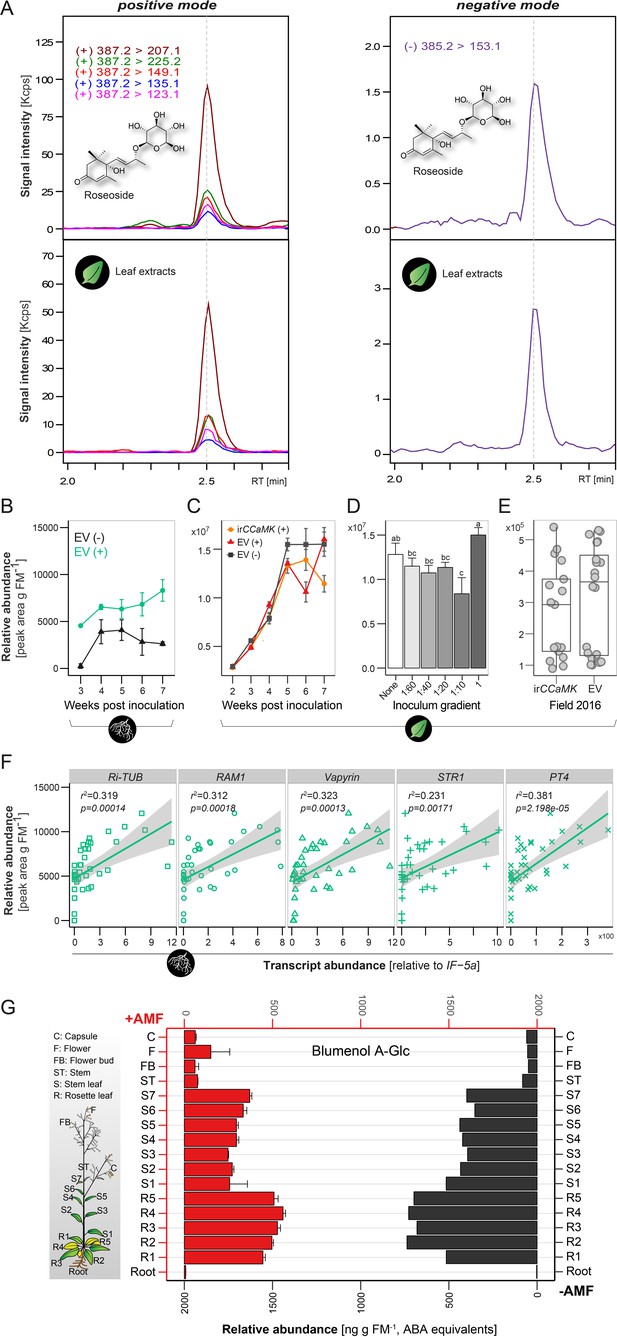
Compound 6 is constitutively produced in shoots of N. attenuata and not indicative of AMF associations.
(A) Representative chromatograms from a targeted tandem MS-based analysis of Compound 6 in leaves of N. attenuata (bottom panel) and as comparison, a blumenol A-9-O-glucoside (roseoside) standard (top panel). The precursor-to-product ion transitions are indicated. (B) Time lapse accumulations of Compound 6 in roots of EV plants with (EV+, green line) or without (EV-, black line) AMF-inoculation. Data represent means ±SE (n ≥ 3). (C) Time lapse accumulations of Compound 6 in leaves of EV plants with (EV+, red line) or without (EV-, black line) AMF-inoculation and of irCCaMK plants with AMF-inoculation (irCCaMK+, orange line). Data represent means ±SE (n ≥ 5). (D) Comparison of the abundances of Compound 6 in leaves of plants inoculated with different inoculum concentrations, samples were harvested at 5 weeks-post-inoculation (wpi). Data are means +SE (n ≥ 4). Different letters indicate significant differences (p<0.05, one-way ANOVA followed by Fisher’s LSD). (E) Field experiment (Great Basin Desert, Utah, USA): leaf samples of EV (n = 20) and irCCaMK (n = 19) plants were sampled 8 weeks after planting and amounts of Compound 6 were analyzed. For statistical analysis, Student’s t test was applied. (F) Abundance of Compound 6 relative to the transcript abundance of the R. irregularis specific housekeeping gene, Ri-tub (GenBank: EXX64097.1), as well as to the plant derived marker genes RAM1, Vapyrin, STR1 and PT4.The transcript abundance was quantified by q-PCR, relative to NaIF-5a (NCBI Reference Sequence: XP_019246749.1). The correlation between Compound 6 and transcript abundance of marker genes was analyzed by linear regression (lm) models. (G) Distribution of Compound 6 in different plant tissues, as indicated, of plants with (+AMF, n = 3, red bars) or without (-AMF, n = 1, black bars) AMF-inoculation. Samples were harvested at six wpi.
-
Figure 6—figure supplement 2—source data 1
Source data for Figure 6—figure supplement 2.
- https://doi.org/10.7554/eLife.37093.023
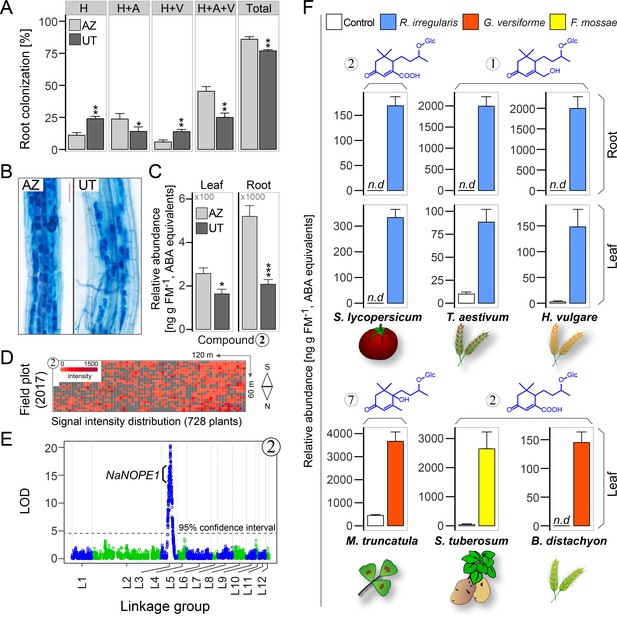
AMF-indicative changes in blumenols in aerial plant parts are valuable research tools providing accurate assessments of functional AMF associations in high-throughput screenings of multiple plant and AMF species.
(A) Root colonization analysis in two N. attenuata accessions (UT/AZ). H: hyphae; A: arbuscules; V: vesicles; Total: total root length colonization (n = 4; Student’s t-test, *p<0.05, **p<0.01, ***p<0.001). (B) Representative images of trypan blue stained roots (six wpi; bar = 100 μm). (C) Compound 2 in roots and leaves of UT and AZ plants with and without AMF-inoculation (means +SE, n = 8). (D) Heatmap of the normalized abundance of foliar Compound 2 in plants of a UT-AZ RIL population (728 plants) planted across a 7,200 m2 field plot. (E) QTL mapping analysis of the data from D. The QTL on linkage group three contains NaNOPE1, an AMF-associated gene, in addition to others. LOD, logarithm of the odds ratio. (F) Blumenol contents of different crop and model plants with and without AMF inoculation (S. lycopersicum (n = 6), T. aestivum (n = 10), H. vulgare (n = 5): eight wpi; M. truncatula (n = 3): seven wpi; S. tuberosum (n = 5): six wpi; B. distachyon (n = 4): five wpi). Different plant and AMF species were used as indicated (means +SE; n.d., not detected).
-
Figure 7—source data 1
Source data for Figure 7.
- https://doi.org/10.7554/eLife.37093.030
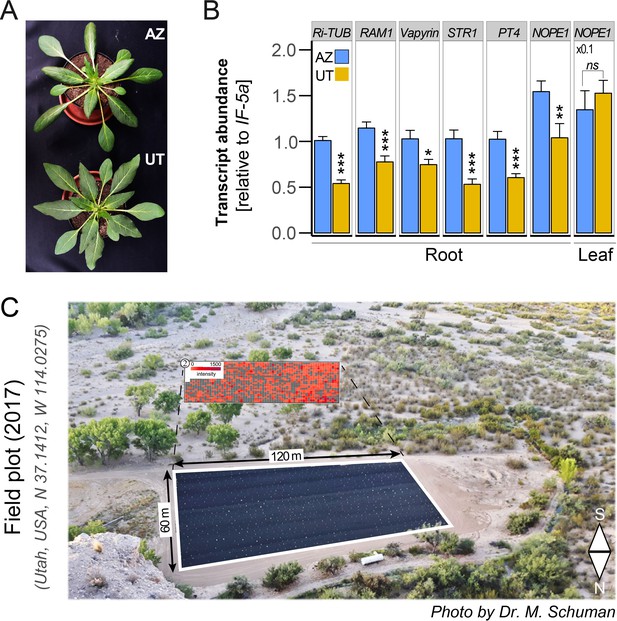
Phenotypes of UT and AZ accessions and field plot planting design.
(A) Representative N. attenuata plants of the UT and AZ accessions in the rosette stages of growth (12 days after potting). (B) Transcripts of marker genes in roots responding to AMF colonization in UT and AZ after six wpi inoculated with R. irregularis were quantified by qPCR in the same samples as in Figure 4A–C (n = 8); Student’s t test *p<0.05, **p<0.01, ***p<0.001. (C) Field plot of 728 sampled individual plants in Utah, USA, 2017.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.37093.027
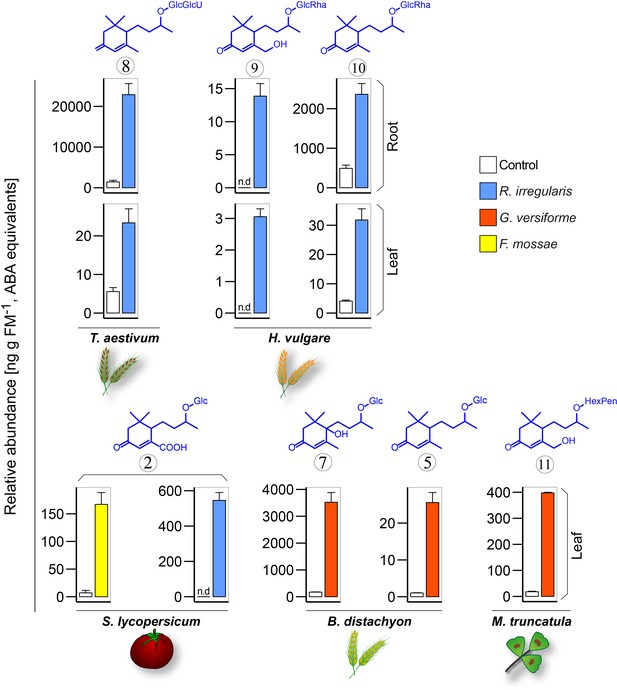
AMF-indicative changes in blumenols in aerial plant part are valuable research tools providing accurate assessments of functional AMF associations of multiple plant and AMF species (continued from Figure 7F).
Blumenol contents of different crop plants with and without AMF inoculation (T. aestivum: eight wpi, n = 5; H. vulgare: eight wpi, n = 10; S. lycopersicum with F. mossae: 6 wpi, n = 5; S. lycopersicum with R. irregularis: 11 wpi, n = 6; B. distachyon: five wpi, n = 4; M. truncatula: seven wpi, n = 3). Different plant and AMF species were used, as indicated; means +SE, n.d., not detected.
-
Figure 7—figure supplement 2—source data 1
Source Code File for QTL analysis.
- https://doi.org/10.7554/eLife.37093.029
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (N. attenuata) | A-09-1212-1 | Groten et al. (2015), DOI: 10.1111/pce.12561 | Stably silenced in CCaMK via RNAi | |
| Genetic reagent (N. attenuata) | A-09-1208-6 | Groten et al. (2015), DOI: 10.1111/pce.12561 | Stably silenced in CCaMK via RNAi | |
| Genetic reagent (N. attenuata) | A-11-92−4 × A-11-325-4 | Schäfer et al. (2013), DOI: 10.1111/tpj.12301 | Chemically-inducible silenced in PDS via RNAi | |
| Genetic reagent (N. attenuata) | A-04-266-3 | Bubner et al. (2006), DOI: 10.1007/s00299-005-0111-4 | Empty vector control | |
| Biological sample (N. attenuata) | AZ-UT RIL | Zhou et al. (2017), DOI: 10.1016/j.cub.2017.03.017 | Biparental QTL mapping population |
1H and 13C NMR data for compounds 1–4 and 7.
https://doi.org/10.7554/eLife.37093.031| No. | Compound 1 | Compound 2 | Compound 3 | Compound 4 | Compound 7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos. | δH | mult., J [Hz] | δC | δH | mult., J [Hz] | δC | δH | mult., J [Hz] | δC | δH | mult., J [Hz] | δC | δH | mult., J [Hz] | δC |
| 1 | - | - | 202.3 | - | - | 203.1 | - | - | 202.2 | - | - | 202.3 | - | - | 200.9 |
| 2 | 6.06 | dd, 1.8/1.8 | 121.3 | 6.40 | s | 128.5 | 6.06 | s br | 121.3 | 5.81 | s br | 125.2 | 5.82 | s | 126.4 |
| 3 | - | - | 172.4 | - | - | 172.6 | - | - | 172.2 | - | - | 169.8 | - | - | 171.5 |
| 4 | 1.92 | dd, 5.2/5.2 | 47.8 | 2.64 | m | 46.3 | 1.92 | dd, 5.2/5.2 | 48.0 | 1.97 | dd, 5.0/5.0 | 52.4 | - | - | 79.2 |
| 5 | - | - | 37.2 | - | - | 36.9 | - | - | 37.2 | - | - | 37.2 | - | - | 42.9 |
| 6 | 2.59 2.02 | d, 17.5 d, 17.5 | 48.5 | 2.03 2.60 | d, 17.4 d, 17.4 | 48.0 | 2.59 2.02 | d, 17.6 d, 17.6 | 47.7 | 2.49 1.98 | d, 17.3 d, 17.3 | 48.0 | 2.13 2.16 | d, 18.0 d, 18.0 | 50.8 |
| 7 | 1.66 1.82 | m m | 26.8 | 1.62 1.88 | m m | 27.6 | 1.66 1.82 | m m | 27.1 | 1.69 1.80 | m m | 26.5 | 1.83 2.07 | m m | 34.6 |
| 8 | 1.63 | m | 37.1 | 1.60 | m | 36.0 | 1.63 | m | 37.2 | 1.68 1.61 | m m | 37.4 | 1.49 1.78 | m m | 32.7 |
| 9 | 3.82 | dd, 6.2/ 11.7 | 77.2 | 3.80 | m | 77.7 | 3.83 | dd, 6.3/ 11.7 | 77.7 | 3.83 | dd, 6.3/ 11.6 | 77.7 | 3.80 | ddd, 6.3/ 11.8/11.8 | 78.2 |
| 10 | 1.24 | d, 6.2 | 21.6 | 1.21 | d, 6.1 | 21.9 | 1.24 | d, 6.3 | 21.9 | 1.25 | d, 6.3 | 21.9 | 1.24 | d, 6.3 | 21.9 |
| 11 | 4.32 4.16 | dd, 17.8/1.8 dd, 17.8/1.8 | 64.9 | - | - | 160.1 | 4.33 4.16 | m/m | 64.9 | 2.05 | d, 1.2 | 24.9 | 2.04 | d, 1.0 | 21.7 |
| 12 | 1.02 | s | 28.4 | 1.01 | s | 28.4 | 1.02 | s | 28.6 | 1.02 | s | 28.7 | 1.01 | s | 24.3 |
| 13 | 1.12 | s | 27.5 | 1.12 | s | 27.5 | 1.11 | s | 27.5 | 1.10 | s | 27.4 | 1.09 | s | 23.7 |
| 1' | 4.31 | d, 7.9 | 103.8 | 4.30 | d, 7.9 | 104.1 | 4.32 | d, 7.9 | 103.8 | 4.32 | d, 7.8 | 104.0 | 4.31 | d, 7.9 | 104.0 |
| 2' | 3.15 | dd, 7.9/9.0 | 75.0 | 3.13 | dd, 7.9/8.9 | 75.1 | 3.15 | dd, 7.9/ 9.0 | 75.1 | 3.16 | dd, 7.8/9.0 | 75.2 | 3.14 | dd, 7.9/8.9 | 75.1 |
| 3' | 3.33 | dd, 9.0/9.0 | 77.9 | 3.35 | dd, 8.9/8.9 | 78.0 | 3.33 | m | 77.8 | 3.34 | dd, 9.0/9.0 | 78.0 | 3.33 | dd, 8.9/8.9 | 77.8 |
| 4' | 3.27 | dd, 9.0/9.0 | 71.4 | 3.27 | dd, 8.9/8.9 | 71.5 | 3.33 | m | 71.4 | 3.33 | dd, 9.0/9.0 | 71.5 | 3.27 | dd, 8.9/8.9 | 71.4 |
| 5' | 3.25 | m | 77.6 | 3.25 | m | 77.6 | 3.45 | m | 77.0 | 3.44 | m | 76.9 | 3.24 | m | 77.7 |
| 6' | 3.85 3.65 | dd, 11.8/2.2 dd, 11.8/5.5 | 62.5 | 3.84 3.66 | dd, 2.0/12.3 dd, 5.0/12.3 | 62.6 | 4.11 3.78 | dd, 11.7/2.0 dd, 11.7/5.7 | 69.6 | 4.11 3.79 | dd, 11.6/1.6 dd, 11.6/5.9 | 69.7 | 3.85 3.65 | d, 11.7 dd, 4.5/11.7 | 62.3 |
| 1'' | 4.40 | d, 7.9 | 104.6 | 4.40 | d, 7.8 | 104.8 | |||||||||
| 2'' | 3.21 | dd, 7.9/9.0 | 74.9 | 3.21 | dd, 7.8/9.0 | 75.0 | |||||||||
| 3'' | 3.34 | dd, 9.0/9.0 | 77.9 | 3.34 | dd, 9.0/9.0 | 77.9 | |||||||||
| 4'' | 3.28 | dd, 9.0/9.0 | 71.4 | 3.28 | dd, 9.0/9.0 | 71.5 | |||||||||
| 5'' | 3.26 | m | 77.9 | 3.26 | ddd, 9.0/5.4/1.8 | 78.0 | |||||||||
| 6'' | 3.87 3.66 | dd, 11.9/2.0 dd, 11.9/5.2 | 62.5 | 3.86 3.66 | dd, 11.6/1.8 dd, 11.6/5.4 | 62.7 | |||||||||
-
s, singlet; s br, broad singlet; d, doublet; dd, doublet of doublet; m, multiplet
MRM-settings used for targeted blumenol analysis.
https://doi.org/10.7554/eLife.37093.032| Nr. | Compound name | RT | Q1 [m/z]*, † | Q3 [m/z] ‡, § (CE [V]) |
|---|---|---|---|---|
| 1 | 11-hydroxyblumenol C-Glc¶, ** | 2.82 | +389.22 | 227.16 (-2.5), 209.15 (-7.5), 191.14 (-12.5), 163.10 (-15), 149.10 (-17.5) |
| 2 | 11-carboxyblumenol C-Glcf¶, ** | 3.22 | +403.22 | 241.16 (-2.5), 223.15 (-7.5), 177.10 (-15), 195.14 (-12.5) |
| +241.16 # | 223.15 (-5), 177.10 (-15), 195.14 (-10) | |||
| 3 | 11-hydroxyblumenol C-Glc-Glc ¶, ** | 2.5 | +551.27 | 389.22 (-2.5), 227.16 (-7.5), 209.15 (-10), 191.14 (-15), 149.10 (-20) |
| 4 | Blumenol C – Glc-Glc ¶, ** | 3.47 | +535.27 | 373.22 (-2.5), 211.00 (-10), 193.10 (-17.5), 135.00 (-22.5), 109.00 (-22.5) |
| 5 | Blumenol C - Glc ¶, †† | 4.18 | +373.22 | 211.20 (-6), 193.16 (-9), 175.10 (-15), 135.12 (-16), 109.10 (-20) |
| 6 | Blumenol A - Glc¶, †† | 2.51 | - 385.20 | 153.10 (14) |
| +387.20 | 225.15 (-5), 207.14 (-8), 149.10 (-18), 135.12 (-16), 123.08 (-23) | |||
| 7 | Blumenol B - Glc¶, ** | 2.5 | +389.22 | 227.16 (-5), 209.15 (-7.5), 191.14 (-12.5), 153.10 (-17.5), 149.10 (-17.5) |
| 8 | Blumenol C – Glc-GlcU¶, ‡‡ | 3.25 | +549.27 | 373.22 (-2.5), 211.00 (-10), 193.10 (-17.5), 135.00 (-22.5), 109.00 (-22.5) |
| and 3.38 | ||||
| 9 | 11-hydroxylumenol C – Glc-Rha‡‡ | 2.8 | +535.27 | 389.22 (-2.5), 227.16 (-7.5), 209.15 (-10), 191.14 (-15), 149.10 (-20) |
| 10 | Blumenol C – Glc-Rha¶, ‡‡ | 4.1 | +519.27 | 373.22 (-2.5), 211.00 (-10), 193.10 (-17.5), 135.00 (-22.5), 109.00 (-22.5) |
| 11 | Hydroxyblumenol C-Hex-Pen‡‡ | 2.5 | +521.27 | 389.22 (-2.5), 227.16 (-7.5), 209.15 (-10), 191.14 (-15), 149.10 (-20) |
| D6-ABA†† | 4.5 | - 269.17 | 159.00 (10) |
-
RT: retention time
CE: collision energy
-
Glc: glucose
GlcU: glucuronic acid
-
Rha: rhamnose
Hex: hexose
-
Pen: pentose
*Resolution: 0.7
-
†[M + H]+ or [M-H]- if not stated differently
‡Resolution: 2
-
§ Quantifiers are depicted in bold
# [M + H-Glc]+
-
¶ Verified by high resolution MS
**Verified by NMR
-
††Optimized with commercial available standards
‡‡Transitions predicted based on structural similar compounds and literature information
MRM-settings for the analysis of selected blumenols in N. attenuata.
https://doi.org/10.7554/eLife.37093.033| Nr. | Compound name | RT | Q1 [m/z]*, † | Q3 [m/z]‡, § (CE [V]) |
|---|---|---|---|---|
| 1 | 11-hydroxyblumenol C-Glc¶, ** | 2.82 | +389.22 | 227.16 (-2.5), 209.15 (-7.5), 191.14 (-12.5), 163.10 (-15), 149.10 (-17.5) |
| 2 | 11-carboxyblumenol C-Glc¶, ** | 3.22 | +403.22 | 241.16 (-2.5), 223.15 (-7.5), 177.10 (-15), 195.14 (-12.5) |
| +241.16# | 223.15 (-5), 177.10 (-15), 195.14 (-10) | |||
| 6 | Blumenol A - Glc ¶, †† | 2.51 | - 385.20 | 153.10 (14) |
| +387.20 | 225.15 (-5), 207.14 (-8), 149.10 (-18), 135.12 (-16), 123.08 (-23) | |||
| D6-ABA†† | 4.0 | - 269.17 | 159.00 (10) |
-
RT: retention time
CE: collision energy
-
Glc: glucose
Hex: hexose
-
Pen: pentose
*Resolution: 0.7
-
†[M + H]+ or [M-H]- if not stated differently
‡Resolution: 2
-
§Quantifiers are depicted in bold
#[M + H-Glc]+
-
¶Verified by high resolution MS
**Verified by NMR
-
††Optimized with commercial available standards
Sequences of primers used for qPCR-based analysis of AMF-colonization rates.
https://doi.org/10.7554/eLife.37093.034| Gene | Forward primer | Reversed primer |
|---|---|---|
| NaIF-5a | GTCGGACGAAGAACACCATT | CACATCACAGTTGTGGGAGG |
| NaRAM1 | ACGGGGTCTATCGCTCCTT | GTGCACCAGTTGTAAGCCAC |
| NaVapyrin | GGTCCCAAGTGATTGGTTCAC | GACCTTCAAAGTCAACTGAGTCAA |
| NaSTR1 | TCAGGCTTCCACCTTCAATATCT | GACTCTCCGACGTTCTCCC |
| NaPT4 | GGGGCTCGTTTCAATGATTA | AACACGATCCGCCAAACAT |
| NaCCaMK | TTGGAGCTTTGTTCTGGTGGT | ATACTTGCCCCGTGTAGCG |
| NaNOPE1 | ACTTGATGCCATGTTTCAGAGC | TCCAATTCGCGATAAGCTGGT |
| Ri-TUB | TGTCCAACCGGTTTTAAAGT | AAAGCACGTTTGGCGTACAT |
Additional files
-
Source code 1
Source code for QTL analysis.
- https://doi.org/10.7554/eLife.37093.035
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37093.036





