Adaptation to constant light requires Fic-mediated AMPylation of BiP to protect against reversible photoreceptor degeneration
Figures
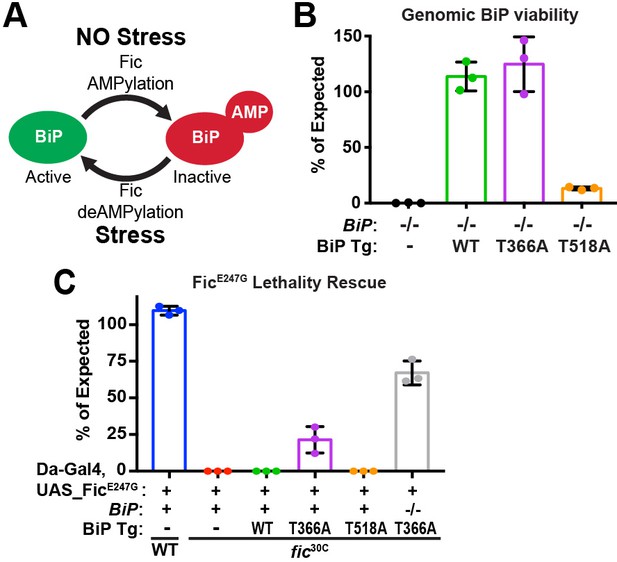
BiP is a target of Fic AMPylation and deAMPylation in vivo.
(A) BiP AMPylation during times of low ER stress reserves a portion of the chaperone to allow for a rapid, deAMPylation-driven, response to high ER stress (Casey et al., 2017; Preissler et al., 2017a). (B) Bar graphs show the percentage of null mutant BiPG0102/y males rescued by the indicated genomic BiPWT, BiPT366A or BiPT518A genomic transgene (Tg) relative to sibling controls. N = 3 biological replicas. At least 50 flies scored for each replica. Bar graphs show means ± Standard Deviation (SD). (C) Bar graphs show the percentage of viable flies of the indicated wild-type or fic30C genotypes expressing the overactive FicE247G under the ubiquitous Da-Gal4 driver relative to sibling controls. Among the indicated genomic BiP transgenes, only BiPT366A provides partial rescue of lethality in the BiP+/+ background and near complete rescue in a BiPG0102 null background. N = 3 biological replicas. At least 100 total flies scored for each replica. Bar graphs show means ± SD.
-
Figure 1—source data 1
Relates to Figure 1B and C.
Quantification of fly survival.
- https://doi.org/10.7554/eLife.38752.006
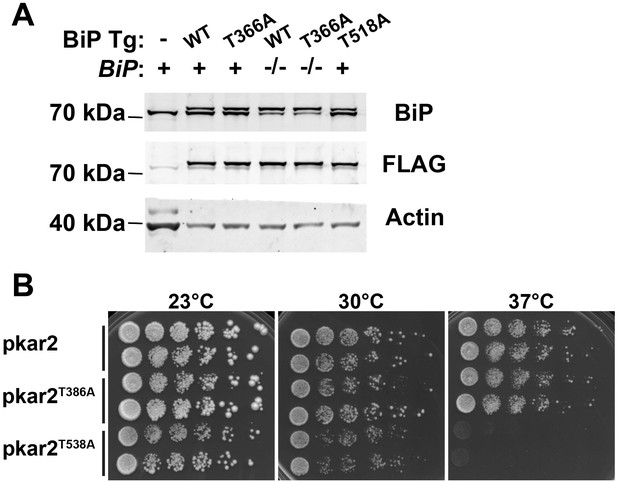
Expression of genomic BiP transgenes.
(A) Western blots for FLAG-tagged BiP transgenes and total BiP in whole head lysates in BiP wild type or homozygous mutant background as indicated. Actin (JLA-20) served as a loading control. (B) Kar2T538A mutants have temperature-sensitive growth defects. Yeast strains kar2Δ + pKar2, kar2Δ + pkar2T386A, and kar2Δ + pkar2T538A were grown at 25°C and five-fold serially diluted onto plates of rich media incubated at the indicated temperatures.
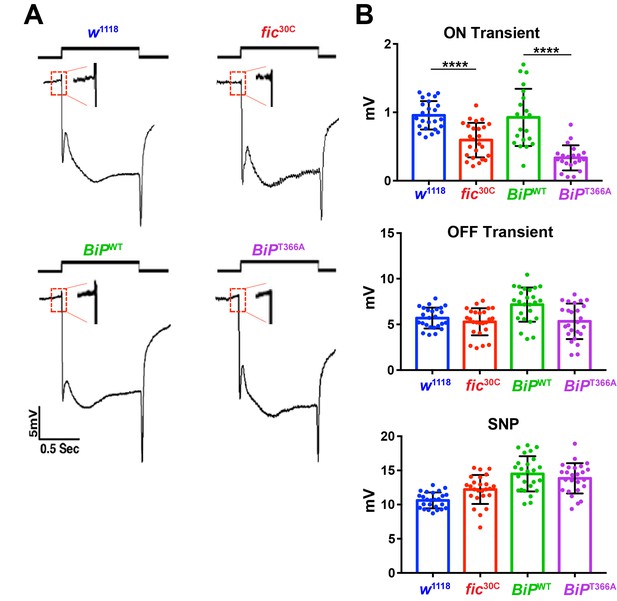
AMPylation-resistant BiPT366A phenocopies fic.
(A) ERGs of fic30C flies show reduced ON transients (arrows). Flies homozygous for a lethal BiPG0102 allele rescued by BiPWT transgene have normal vision but flies rescued with the mutant BiPT366A transgene display reduced ON transients. (B) Quantification of ERG traces. Bar graphs show means ± SD. ****p<0.0001; ***p<0.001; *p<0.05; n = 24 flies per genotype and condition.
-
Figure 1—figure supplement 2—source data 1
Relates to Figure 1—figure supplement 2B.
Quantification of ERG components (white-).
- https://doi.org/10.7554/eLife.38752.007
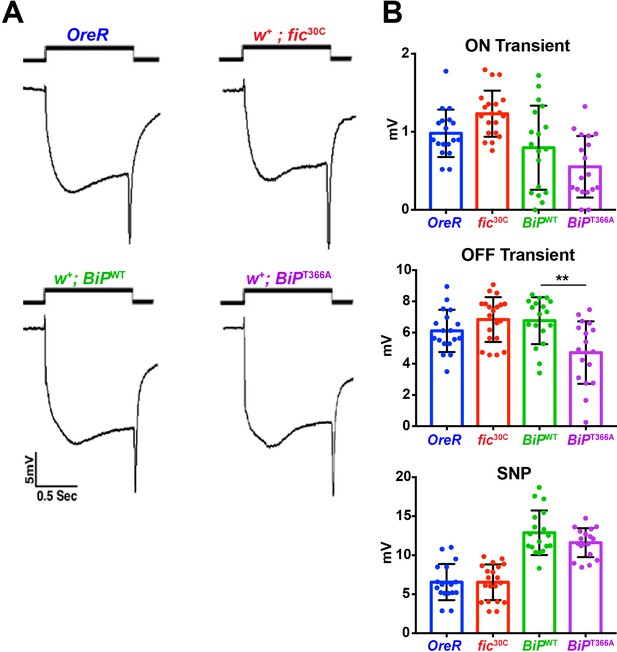
Red eye pigment suppresses ERG phenotypes of fic30C and BiPT366A mutants.
(A) ERGs of OreR and red-eyed fic30C flies as well as BiPWT and BiPT366A animals. (B) Quantification of ERG data. Bar graphs show means ± SD. **p<0.01, n = 18 flies per genotype and condition.
-
Figure 1—figure supplement 3—source data 2
Relates to Figure 1—figure supplement 3B.
Quantification of ERG components (white+).
- https://doi.org/10.7554/eLife.38752.008
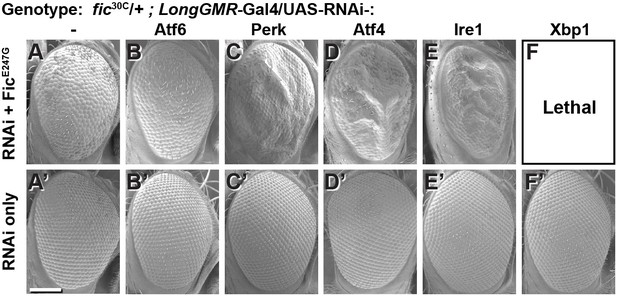
Genetic interactions between Fic and UPR genes.
Representative SEM images of heterozygote mutant fic30C/+ eyes expressing the indicated UAS-RNAi transgenes with (A–F) or without (A’–F’) UAS-FicE247G under longGMR-Gal4 control. See Figure 2—figure supplement 1 for quantification. Scale bar: 100 µM.
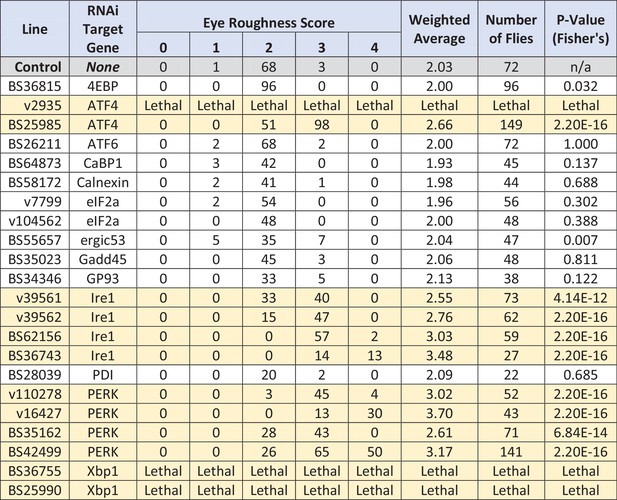
Genetic interactions between Fic and UPR genes.
UASScer-driven RNAi transgenes (either TRIP or VDRC lines) were used to silence candidate UPR and ER stress-related genes in a fic30C/+heterozygous background, with either LongGMR-Gal4, UAS-FicE247G or LongGMR-Gal4 only. Eye roughness was scored for individual flies and averaged for each cross. Table reports number of flies scored in each group (0 = no roughness, 2 = mildly rough (control flies), 4 = severely rough, 1 and 3 are intermediate phenotypes) and the weighted average of the eye roughness. Significance differences are highlighted in yellow, and p-values were determined using Fisher’s Exact Test for categorical data, comparing the effects of each gene knockdown with the control group (top line, fic30C/+; LongGMR-Gal4, UAS-FicE247G). Interactions were considered significant for any individual test if p<0.003 as determined using Bonferroni’s multiple comparison adjustment.
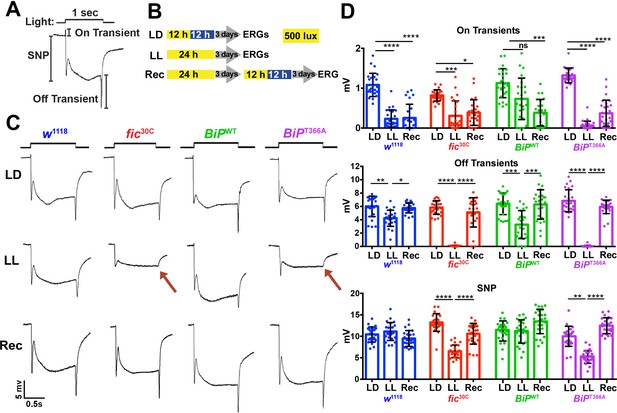
Fic-mediated AMPylation of BiP is required for photoreceptor maintenance.
(A) A representative ERG trace in response to a 1 s light pulse displaying the sustained negative potential (SNP), representing the depolarization within photoreceptor neurons, and the ON and OFF transients, reflecting post-synaptic activity of lamina neurons. (B) Representation of the different light treatments of flies before ERG recordings: 3 days of 12 hr light (500 lux) and 12 hr dark (LD), 3 days of continuous light (LL) or 3 days of continuous light followed by 3 days of LD (Rec). 1 s light pulses were performed at 4 s intervals. (C) Representative traces from w1118, fic30C, BiPWT and BiPT366A flies. Under LL, fic30C and BiPT366A mutants lose ON and OFF transients (red arrows) and have reduced SNPs. The changes are reversed after 3 days of recovery (Rec). (D) Quantification of key components of ERGs shown in panel C. Bar graphs show means ± SD. ****p<0.0001; ***p<0.001; **p<0.01; *p<0.05; n = 24 flies for each genotype/condition, pooled from three independent biological replicas.
-
Figure 3—source data 1
Relates to Figure 3D.
Quantification of ERG components with LD, LL, and Recovery.
- https://doi.org/10.7554/eLife.38752.015
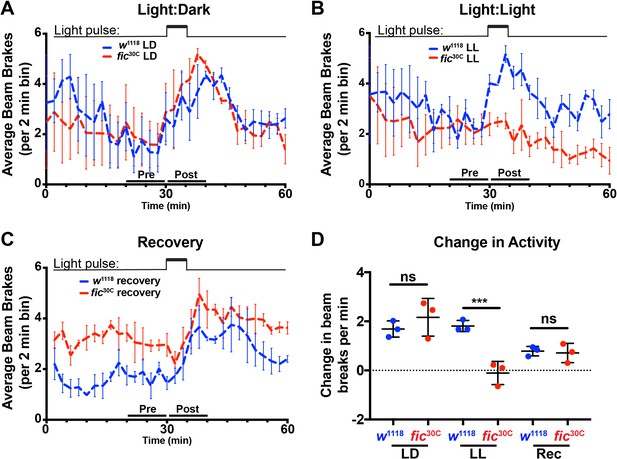
Light-induced defects in light-startle activity in fic30C mutants.
(A, B, and C) Actogram of w1118 or fic30C flies reared in LD for three days (A), LL for three days (B), or recovery condition (three days in LL then three days in LD) (C). Light pulse is indicated by upper bars. Data is averaged from three biological replicas, each containing 16 flies per genotype. Data were collected in two-minute bins. Error bars represent Standard Error. (D) Quantification of change in beam breaks per 2 min bin for the 10 min intervals before and after the onset of the light pulse in each experiment. Bar graphs show means ± SD. ***p<0.01, n = 3 experimental repeats with 16 flies per genotype and condition. Dead flies and those with a change in activity greater than three deviations from the median were excluded.
-
Figure 3—figure supplement 1—source data 1
Relates to Figure 3—figure supplement 1A, B, C and D.
Quantification of light-induced startle behavior- average activity and delta calculations.
- https://doi.org/10.7554/eLife.38752.016
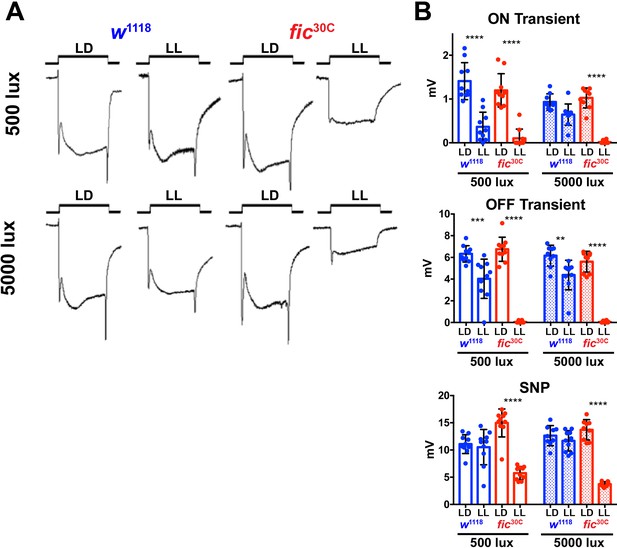
fic30C mutants are sensitive to constant light, regardless of total intensity.
(A) Representative ERGs of flies following 3 days of LL or LD with either 500 or 5000 lux light, showing fic30C null animals lose ON/OFF transients and have reduced SNPs with constant light, regardless of intensity, but under LD conditions, even at 5000 lux, have healthy ERG responses. (B) Quantification of ERG data. Bar graphs show means ± SD. ****p<0.0001; ***p<0.001; *p<0.05; n = 10 flies per genotype and condition.
-
Figure 3—figure supplement 2—source data 2
Relates to Figure 3—figure supplement 2B.
Quantification of ERG components with 500 and 5000 lux.
- https://doi.org/10.7554/eLife.38752.017
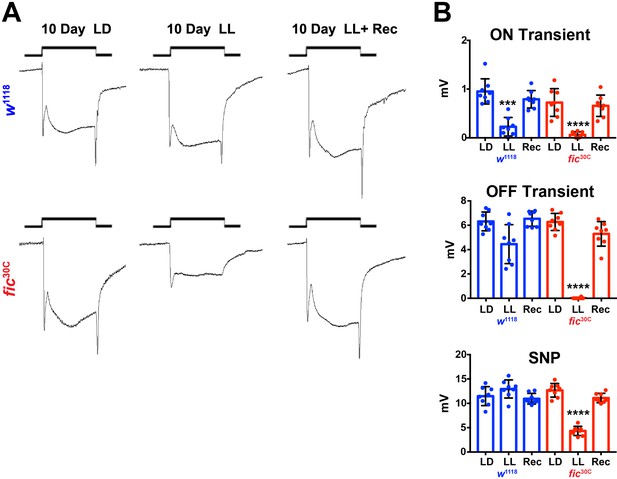
Fic30C mutants recover ERG properties in 72 hours after 10 days of LL.
(A) Representative ERGs of flies following 10 days of LL, 10 days LD (500 lux), and 3 days Recovery following 10 days. (B) Quantification of ERG data. Bar graphs show means ± SD. ****p<0.0001; ***p<0.001; *p<0.05; n = 8 flies per genotype and condition.
-
Figure 3—figure supplement 3—source data 3
Relates to Figure 3—figure supplement 3B.
Quantification of ERG components with 10 days of LL.
- https://doi.org/10.7554/eLife.38752.018
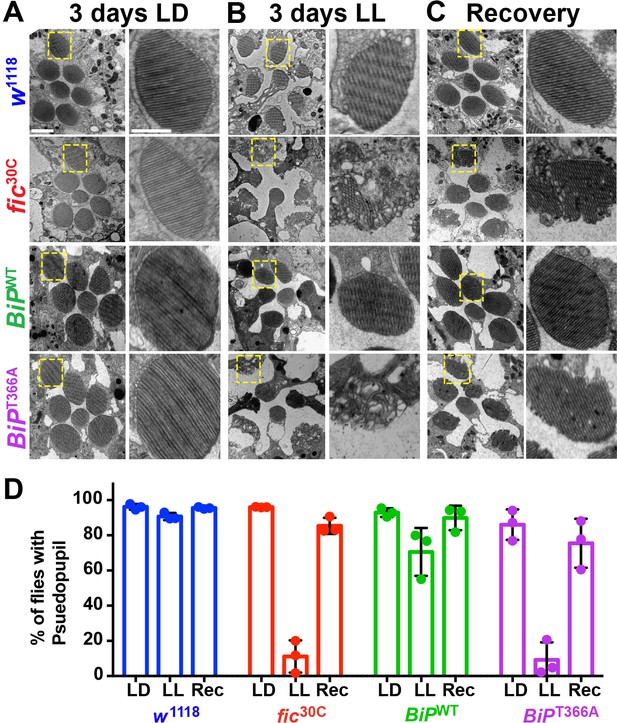
AMPylation of BiP is required for maintaining structural plasticity in the retina.
(A–C) Representative TEM images of retina thin sections from the indicated genotypes with either standard LD (A), the stress-inducing LL (B) or recovery treatment (C). Scale bars: 1 µM. Yellow boxes indicate rhabdomeres shown in high magnification images. High magnification scale bars: 0.5 µM. (D) Percentages of flies with intact deep pseudopupil following LD, LL and Rec. N = 3 independent biological replicas with approximately 50 flies scored per genotype per replica. Bar graphs show means ± SD.
-
Figure 4—source data 1
Relates to Figure 4D.
Scoring of flies for deep pseudopupil defects.
- https://doi.org/10.7554/eLife.38752.020
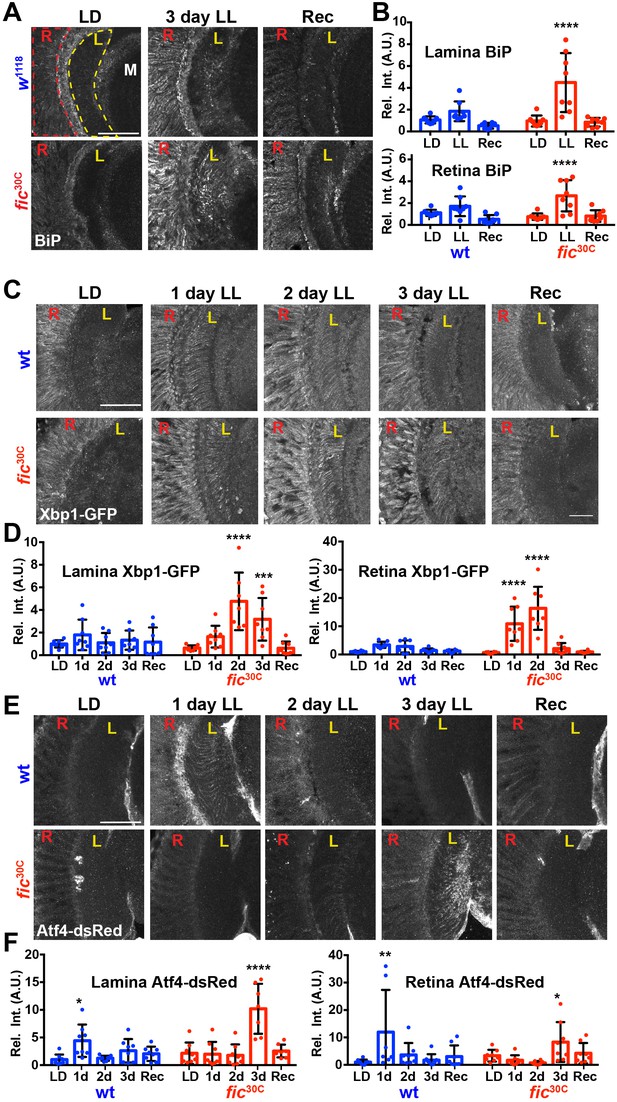
ER homeostasis is disturbed in fic mutants during prolonged light stimulation.
(A) Representative images of BiP immunohistochemistry in sections of w1118 and fic30C flies following 3 days LD, LL or Recovery treatments. (B) Quantification of BiP fluorescence intensity, normalized to wild-type LD controls, in the lamina neuropil and retina from two independent experiments. (C) Representative images of a Xbp1-GFP splicing reporter in either a Fic wild-type or the null fic30C background following LD, 1 day LL, 2 day LL, 3 day LL, and Recovery conditions. (D) Quantification of GFP fluorescence intensity, normalized to wild-type LD controls, in the lamina neuropil and retina from two independent experiments. (E) Representative images of an Atf4-dsRed reporter in either a wild-type or fic30C background following LD, 1 day LL, 2 day LL, 3 day LL, and Recovery conditions. (F) Quantification of Atf4-dsRed intensity, normalized to wild-type LD controls, in the lamina neuropil and retina from two independent experiments. For all experiments, n = 8 flies per genotype/condition, with exceptions of outliers falling three standard deviations outside the mean. Bar graphs show means ± SD. For all experiments, significance is indicated for treatment compared to the LD condition for the corresponding genotype. ****p<0.0001; ***p<0.001; **p<0.01; *p<0.05. All scale bars: 50 µM.
-
Figure 5—source data 1
Relates to Figures 5B, D and F.
Quantification of integrated intensity of BiP (5B), Xbp1-GFP (5D), and Atf4-DsRed (5F).
- https://doi.org/10.7554/eLife.38752.022
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | fic | NA | FLYB:FBgn0263278 | |
| Gene (D. melanogaster) | Hsc3-70 | NA | FLYB:FBgn0001218 | |
| Genetic reagent (D. melanogaster) | Da-Gal4 | Bloomington Drosophila Stock Center | BDSC:55851; FLYB:FBst0055851; RRID: BDSC_55851 | |
| Genetic reagent (D. melanogaster) | LongGMR-Gal4 | Bloomington Drosophila Stock Center | BDSC:8121; FLYB:FBst0008121; RRID:BDSC_8121 | |
| Genetic reagent (D. melanogaster) | W[1118] | Bloomington Drosophila Stock Center | BDSC:3605; FLYB:FBst0003605; RRID:BDSC_3605 | |
| Genetic reagent (D. melanogaster) | BiP[G0102]/FM7c | Bloomington Drosophila Stock Center | BDSC:11815; FLYB:FBal0098203; RRID:BDSC_11815 | |
| Genetic reagent (D. melanogaster) | UAS[Scer]-Xbp1- GFP.hg | Bloomington Drosophila Stock Center | BDSC:60731; FLYB:FBst0060731; RRID:BDSC_60731 | Sone et al. (2013) |
| Genetic reagent (D. melanogaster) | dsRed.crc(ATF4). 5'UTR.tub | DOI: 10.1371/journal.pone. 0126795; PMID:25978358 | FLYB: FBal0304834 | Gift from Don Ryoo, NYU. Tubulin promoter and ATF4 5'UTR drive DsRed expression (Flybase FBal0304834) |
| Genetic reagent (D. melanogaster) | genomic 3xFLAG- BiP[WT] | This paper | pAttb_gen3xFLAG-BiP[WT] inserted in AttP landing site at 89E11 | |
| Genetic reagent (D. melanogaster) | genomic 3xFLAG- BiP[T366A] | This paper | pAttb_gen3xFLAG-BiP[T366A] inserted in AttP landing site at 89E11 | |
| Genetic reagent (D. melanogaster) | genomic 3xFLAG- BiP[T518A] | This paper | pAttb_gen3xFLAG-BiP[T518A] inserted in AttP landing site at 89E11 | |
| Genetic reagent (D. melanogaster) | GMR-dsRNA[white] | This paper | pAttb_GMR-dsRNA[white] inserted in AttP landing site at 43A1 | |
| Genetic reagent (D. melanogaster) | fic[30C] | DOI: 10.1074/jbc.M117. 799296; PMID:29089387 | Casey et al. (2017) | |
| Genetic reagent (D. melanogaster) | UAS[Scer]-V5- Fic[E247G] | DOI: 10.1074/jbc.M117. 799296; PMID:29089387 | Casey et al. (2017) | |
| Genetic reagent (D. melanogaster) | UPR and ER protein RNAi lines screened are contained in Supplemental Table 1 | |||
| Genetic reagent (Saccharomyces cerevisiae) | KAR2::KAN | GE Healthcare Life Sciences | SGD:S000003571 | gene replacement generated using PCR-based gene deletion strategy yielding start- to stop-codon deletion |
| Strain, strain background (Saccharomyces cerevisiae) | BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | DOI: 10.1002/(SICI)1097- 0061(19980130)14:2 < 115:: AID-YEA204 > 3.0.CO;2–2; PMID: 9483801 | GenBank: JRIS00000000.1 | |
| Antibody | anti-Hsc70-3 (BiP) (Guinea Pig polyclonal) | DOI: 10.1038/sj.emboj. 7601477; PMID:17170705 | FLYB: FBgn0001218; RRID: AB_2569409 | Gift from Don Ryoo, NYU (1:2000 IHC, 1:8000 WB) |
| Antibody | anti-RFP (Rabbit polyclonal) | Rockland | Rockland:600-401-379; RRID:AB_2209751 | (1:1000 IHC) |
| Antibody | anti-GFP (Chicken polyclonal) | ThermoFisher Scientific | ThermoFisher Scientific:A10262; RRID: AB_2534023 | (1:1000 IHC) |
| Antibody | anti-Flag (mouse monoclonal) | Sigma | Sigma:F-3165; RRID:AB_259529 | (1:2000 WB) |
| Antibody | anti-Actin (mouse monoclonal) | Developmental Studies Hybridoma Bank | DSHB:JLA20; RRID: AB_528068 | 1:2000 (WB) |
| Antibody | Alexa 488- or 568- secondaries | Molecular Probes | (1:1000 IHC) | |
| Antibody | LICOR 800 or 700- secondaries | LICOR Biosciences | (1:20,000 WB) | |
| Recombinant DNA reagent | pAttb_gen3xFLAG- BiP[WT] | This paper | PCR in multiple steps from genomic DNA {sequence location = X: 11,801,696..11,807,117 [-]}. Cloned into modified pAttb vector | |
| Recombinant DNA reagent | pAttb_gen3xFLAG- BiP[T366A] | This paper | Progenitors: pAttb_gen3xFLAG- BiP[WT]. Mutated sequence synthesized with Geneblock (IDT) | |
| Recombinant DNA reagent | pAttb_gen3xFLAG- BiP[T518A] | This paper | Progenitors: pAttb_gen3xFLAG- BiP[WT]. Mutated sequence synthesized with Geneblock (IDT) | |
| Recombinant DNA reagent | pAttb_GMR- dsRNA[white] | This paper | Progenitors: pUASt_dsRNA[white] (gift from Dean Smith, UT Southwestern, PMID: 11804566). GMR sequence: Geneblock (IDT) | |
| Recombinant DNA reagent | pKAR2:LEU2 | This paper | Cloned from amplification of endogenous KAR2 with primers (see below) | |
| Recombinant DNA reagent | pKAR2[T386A]: LEU2 | This paper | Progenitor: pKAR2:LEU2. Site directed mutagenesis used to make mutation | |
| Recombinant DNA reagent | pKAR2[T538A]: LEU2 | This paper | Progenitor: pKAR2:LEU2. Site directed mutagenesis used to make mutation | |
| Sequence-based reagent | 5’-GCATCCGCGGATACT CTCGTACCCTGCCGC-3’ | This paper | Cloning for pKAR2:LEU2 | |
| Sequence-based reagent | 5’-ATGCGAGCTCCGTAT ATACTCAGTATAATC-3’ | This paper | Cloning for pKAR2:LEU2 | |
| Sequence-based reagent | 5’-GGTTGGTGGTTCTG CTAGAATTCCAAAGGT CCAACAATTGTTAGAA TCATACTTTGATGG-3’ | This paper | Mutagenesis primer for pKAR2[T386A]:LEU2 | |
| Sequence-based reagent | 5’-ACCTTTGGAATTCT AGCAGAACCACCAAC CAAAACGATATCATCA ACATCCTTCTTTTCC-3’. | This paper | Mutagenesis primer for pKAR2[T386A]:LEU2 | |
| Sequence-based reagent | 5’-AGATAAGGGAGCTGG TAAATCCGAATCTATCAC CATCACTAACG-3’ | This paper | Mutagenesis primer for pKAR2[T538A]:LEU2 | |
| Sequence-based reagent | 5’-GGATTTACCAGCTCC CTTATCTGTGGCAGACA CCTTCAGAATACC-3’. | This paper | Mutagenesis primer for pKAR2[T538A]:LEU2 | |
| Chemical compound, drug | VECTASHIELD Antifade Mounting Medium with DAPI | Vector Laboratories | Vector Laboratories: H-1200 | |
| Software, algorithm | Adobe Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38752.023





