Wnt11 directs nephron progenitor polarity and motile behavior ultimately determining nephron endowment
Figures
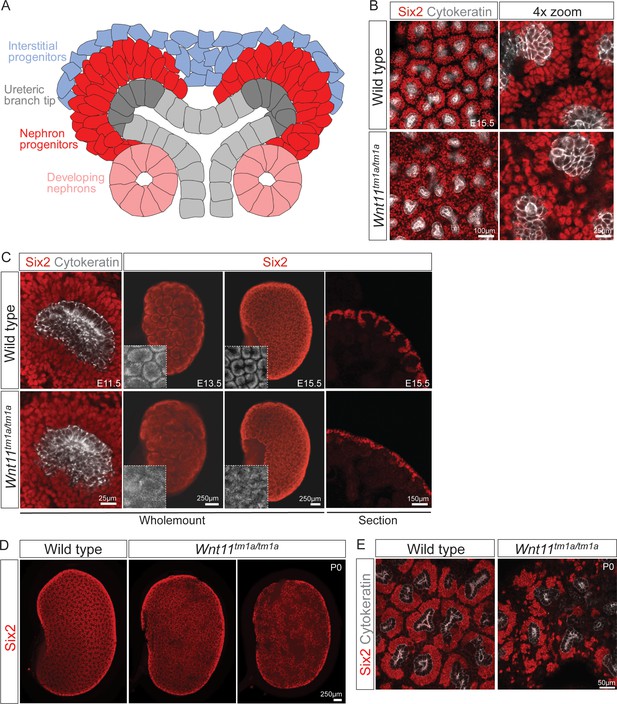
Wnt11 mutants have persistent loose, disorganized nephron progenitor niches that prematurely dropout.
(A) Schematic of the nephrogenic niche. Wnt11 is secreted by the ureteric branch tip cells. (B) Wholemount immunostained kidneys were analyzed at E15.5. Confocal images show that Wnt11tm1a/tm1a Six2 +nephron progenitors (red) are dispersed from the ureteric branch tips (grey) and rounded in appearance compared to wild type cells. A 4x zoom in the far-right panel highlights the differences in cellular distribution and morphology. (C) Wholemount immunostained wild type and Wnt11 mutant kidneys were analyzed at E11.5, E13.5, and E15.5. Wnt11tm1a/tm1a Six2 +nephron progenitors are more rounded and less organized than wild type counterparts beginning at E11.5 (view shows slice from a z-stack). The disorganized phenotype is clear in whole kidney views (maximum intensity projection) at E13.5 and E15.5. Insets highlight the disorganization in whole kidney views. The far-right panel shows a cryosection of E15.5 wild type and Wnt11 mutant kidneys immunostained for Six2, highlighting the dispersed phenotype is also evident in section. (D) Wholemount immunostains of P0 kidneys show the persistent disorganized phenotype and associated premature dropout of Six2 +nephron progenitor niches (red). (E) High resolution confocal views highlight the premature dropout of Six2 +nephron progenitor niches (red) around cytokeratin +ureteric epithelium (grey) in Wnt11tm1a/tm1a kidneys.
-
Figure 1—source data 1
Quantitation of P0 kidney metrics.
- https://doi.org/10.7554/eLife.40392.005

The Wnt11tm1a allele recapitulates expression of Wnt11.
(A) Schematic of Wnt11 protein. (B) Schematic of the Wnt11tm1a allele. The coding region is reflected by the colors which correlate with the protein structure depicted in A). (C) Wnt11tm1a/+ tissue sections through a whole E15.5 embryo were subjected to in situ hybridization for Wnt11 or β-galactosidase staining. The lacZ reporter in the Wnt11tm1a allele recapitulates expression of Wnt11. (D) Kidneys from E11.5, E15.5, and P2 animals were processed for in situ hybridization for Wnt11 or β-galactosidase staining. The Wnt11tm1a allele recapitulates expression of Wnt11 in the ureteric branch tips throughout development.
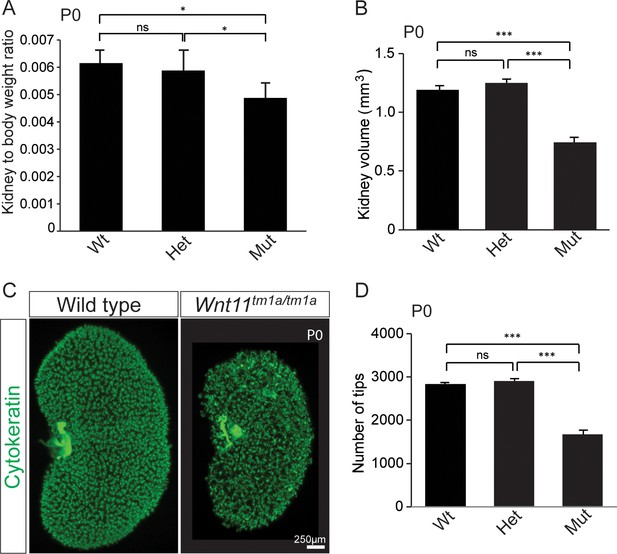
Kidney metrics at P0 reveal deficits in Wnt11 mutants.
(A) Quantitation of kidney to body weight ratio at P0 shows a significant reduction in Wnt11 mutants compared to either wild type or heterozygous animals. Error bars show standard deviation (SD) from 6 samples of each genotype. (B) Quantitation of kidney volume at P0 by three dimensional analyses show that Wnt11 mutant kidneys are smaller, error bars represent standard error mean (SEM) from 8 kidneys of each genotype. (C) Staining of the P0 ureteric tree with cytokeratin (green) reveals the small kidney structure and reduced branching present in Wnt11 mutants. (D) Quantitation of cytokeratin +ureteric branch tips at P0 by confocal analyses reveals a deficit in Wnt11 mutants, error bars are SEM from 8 kidneys of each genotype. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
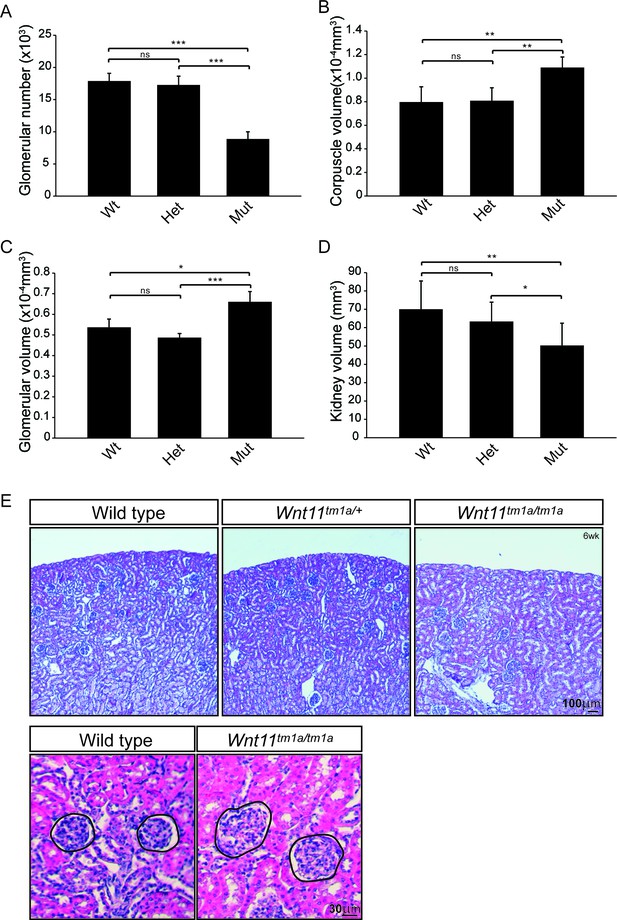
Nephron numbers are reduced in Wnt11tm1a/tm1a adults leading to compensatory hypertrophy.
(A) The physical disector/fractionator method was used to estimate glomerular number in 6 week old animals. A significant reduction is found in Wnt11 mutants. (B) Renal corpuscle volume is increased in Wnt11 mutant kidneys. (C) Glomerular volume is larger in Wnt11tm1a/tm1a kidneys. (D) Estimated kidney volume is reduced in Wnt11 mutants. (E) Histological sections stained with hematoxylin and eosin highlight that overall morphology is preserved, although glomerular/corpuscle volume is increased in the Wnt11 mutants. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. n = 6 for each genotype.
-
Figure 2—source data 1
Quantitation of adult kidney metrics.
- https://doi.org/10.7554/eLife.40392.007
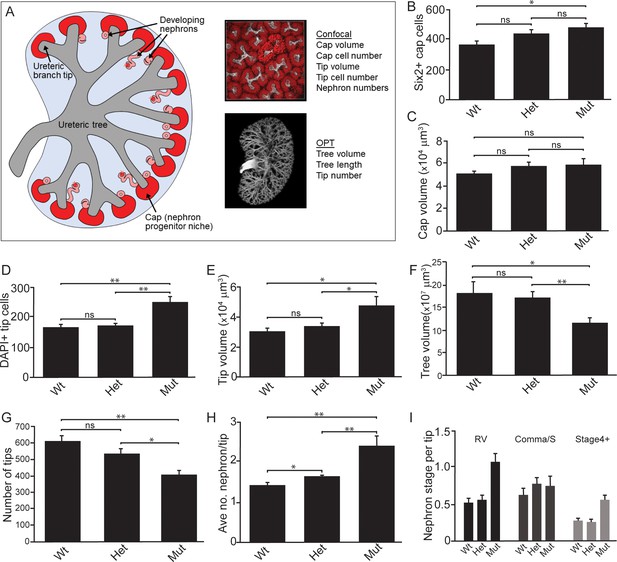
Quantitative analyses reveal significant alterations to niche metrics and accelerated nephrogenesis in Wnt11tm1a/tm1a kidneys.
(A) Schematic of the developing kidney highlighting the niche and structure metrics quantified after imaging by either confocal or optical projection tomography (OPT). E15.5 wholemount immunostains were performed with α-Six2 and α-pan cytokeratin antibodies and kidneys subsequently imaged by confocal microscopy for analyses in B-E, H, and I or OPT for analyses in F-G. (B) The number of Six2 +nephron progenitors per niche were quantified and reveal an increase in Wnt11 mutants. (C) The overall volume of each nephron progenitor niche (cap) is not significantly different between wild type and Wnt11 mutants. (D) The number of DAPI +cells were quantified in each cytokeratin +ureteric branch tip niche and were increased in Wnt11 mutants. (E) Ureteric branch tip volumes were measured and are larger in Wnt11 mutants, correlating with the increase in tip number. (F) Ureteric tree volume was quantified and is reduced in Wnt11tm1a/tm1a kidneys. (G) Quantitation of ureteric branch tip number reveals a significant decrease in Wnt11tm1a/tm1a kidneys. (H) The average number of nephrons per ureteric branch tip are increased in Wnt11 mutant kidneys. (J) Classification of the developing nephron structures associated with each tip highlight a bias in Wnt11 mutants towards renal vesicles (RV) and stage 4 + nephrons versus comma/s-shaped bodies. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. n = 6 kidneys of each genotype for confocal analyses. N = 8 of each genotype for OPT analyses.
-
Figure 3—source data 1
E15.5 confocal analyses.
- https://doi.org/10.7554/eLife.40392.010
-
Figure 3—source data 2
E15.5 OPT analyses.
- https://doi.org/10.7554/eLife.40392.011
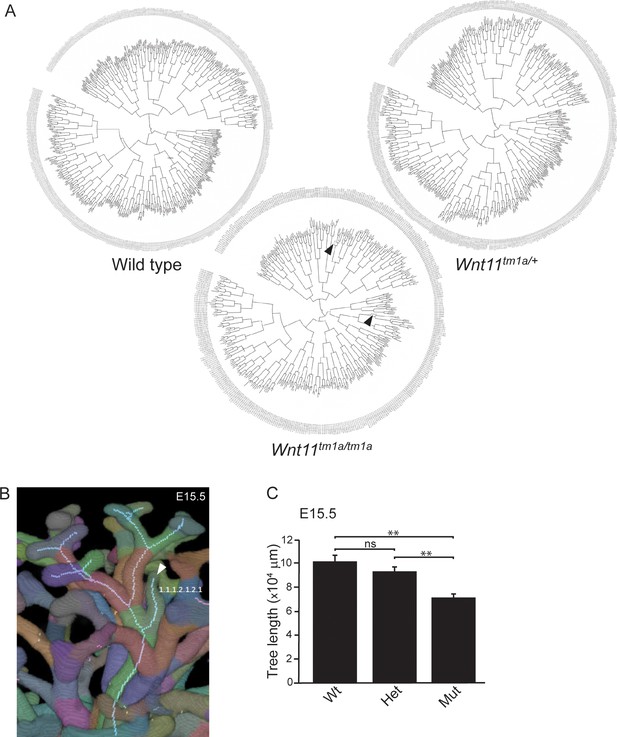
Branching patterns are similar among genotypes despite smaller ureteric trees and premature branch truncations in Wnt11 mutants.
(A) Clade diagrams of the branching patterns of a representative E15.5 kidney from wild type, Wnt11tm1a/+, and Wnt11tm1a/tm1a animals. Arrowheads point to tips that stopped branching prematurely. (B) Optical projection tomography (OPT) image rendered Tree Surveyor highlighting a prematurely truncated branch tip (arrowhead) in a Wnt11 mutant. (C) Ureteric tree length was measured with Tree Surveyor software and is decreased in mutant kidneys. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
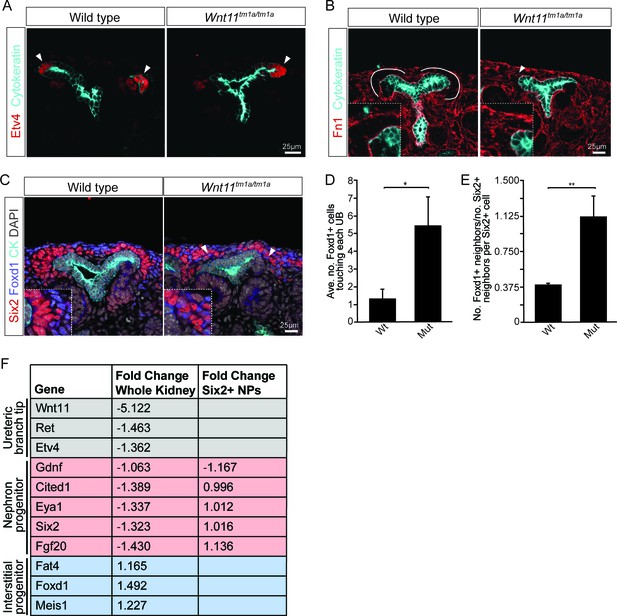
Nephron progenitors intermix with interstitial progenitors but no significant changes in gene expression are observed.
(A) E15.5 kidneys were sectioned and immunostained for the tip marker Etv4 (red) and cytokeratin (cyan). Distinct ureteric branch tip domains are still present in Wnt11 mutants as indicated (arrowheads). (B) E15.5 sections were stained for the matrix protein fibronectin (Fn1; red) and cytokeratin (cyan). Note the exclusion of fibronectin from the nephron progenitor niche in wild type animals (line marks boundary between the nephron progenitors and interstitial progenitors). In Wnt11tm1a/tm1a kidneys the fibronectin boundary is disrupted and staining observed in the nephron progenitor niche (arrowheads). Insets show zoomed view of the progenitor niche. (C) Immunolocalization of Foxd1 +interstitial progenitors (blue) in conjunction with Six2 +nephron progenitors (red) at E15.5 reveals mixing of the two cell populations in Wnt11 mutant kidneys (insets show zoomed view of cell mixing). Foxd1 +cells can infiltrate (arrowheads) the nephron progenitor niche and are found near the ureteric branch tips (cyan). (D) Quantitation of tissue sections immunostained for both Six2, Foxd1, and cytokeratin. There is an increase in the number of Foxd1 +cells touching the ureteric branch tips in Wnt11 mutants. 30 ureteric tip domains from n = 3 biological replicates were quantified. (E) Quantitation of Six2 cell neighbors. The number of Foxd1 +cells touching a Six2 +cell is divided by the number of Six2 +cells touching the same cell. In Wnt11 mutants a Six2 +cell is just as likely to have as many Foxd1 +neighbors as Six2 +neighbors. Three biological replicates were quantified, 10 Six2 +cells per sample. (F) Fold-changes associated with RNA-seq of either whole kidneys or Six2 +cells from wild type and Wnt11 mutant kidneys. The fold-change was calculated from the average of n = 6 for each genotype in whole kidney analysis and n = 3 for each genotype in the nephron progenitor analysis. Example genes which define each progenitor population (ureteric branch tip, nephron progenitor, and interstitial progenitor) are shown. No significant changes (>1.5 fold change) are observed. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
-
Figure 4—source data 1
Quantitation of nephron progenitor metrics.
- https://doi.org/10.7554/eLife.40392.014

Several Wnt receptors are expressed in the nephron progenitors and display weakly penetrant phenotypes upon deletion.
(A) RNA-seq from E16.5 Six2 +nephron progenitors reveals the expression of several Wnt pathway receptors and co-receptors. RPKM values are listed for each gene. Only genes with an RPKM >1 are shown. (B) β-galactosidase stains of Fzd2+/- and Fzd7+/- kidneys at E15.5. Alleles have a lacZ reporter inserted into the coding region. (C) Wholemount in situ hybridization for Fzd2 and Fzd7 on E15.5 urogenital systems show that they largely recapitulate the β-gal expression patterns. (D) Immunostaining for Six2 (red), β-galactosidase (green), and cytokeratin (blue) on E15.5 kidney sections from Fzd2 and Fzd7 heterozygous animals to show the expression domains of each receptor. Both are found within the nephron progenitors, but Fzd7 to a lesser extent. (E) Wholemount immunostains for Six2 (green) and cytokeratin (red) were performed on mutants and appropriate controls at E15.5 for each receptor listed (Wnt11 is included for comparison). For Ror2, the conditional line was crossed to the Six2TGCtg line to specifically remove Ror2 in the nephron progenitors (Ror2c/c;Six2TGCtg/+). No receptor mutant completely recapitulates the Wnt11 mutant phenotype. Insets show 2X magnification. (F) High resolution view shows the slightly disorganized Six2 +nephron progenitors (green) in E15.5 Ptk7-/- kidneys although cap boundaries are still visible and the phenotype only occurred in one of two mutants analyzed. (G) Wholemount immunostains for Six2 (green) on E15.5 kidneys from Fzd2-/- and Fzd2-/-;Fzd7+/- ±. Slightly disorganized caps were observed in one of three kidneys examined.
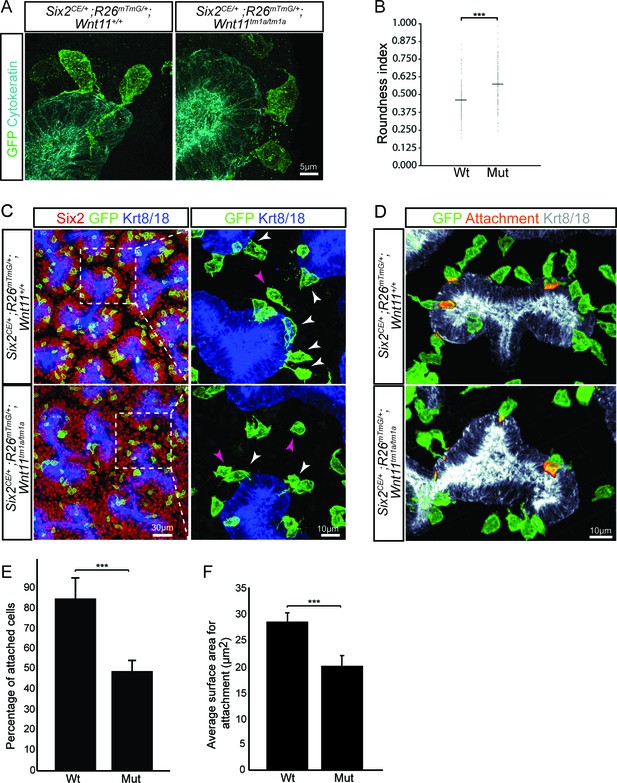
Wnt11 mutants show a significant reduction in membranous attachments of nephron progenitors to the ureteric branch tip.
(A) Sections from E15.5 Six2CE/+; R26mTmG/+; Wnt11+/+ and Six2CE/+; R26mTmG/+; Wnt11tm1a/tm1a kidneys (recombination induced 24 hr prior) were immunostained for GFP (green) and cytokeratin (cyan) to visualize the GFP +cells in relation to the ureteric branch tips. Nephron progenitors make long, membranous projections which attach to the ureteric bud. Less extensive projections make contact with the ureteric branch tip in Wnt11 mutants and appear rounder. (B) Quantitation of GFP +cell roundness from samples in A) (length/width = roundness index) reveals that Wnt11 mutant nephron progenitors are rounder. n = 100 cells for each genotype. (C) Wholemount immunostains were carried out on E15.5 Six2CE/+; R26mTmG/+; Wnt11+/+ and Six2CE/+; R26mTmG/+; Wnt11tm1a/tm1a kidneys (recombination induced 24 hr prior) for Six2 (red), GFP (green), and cytokeratin 8/18 (Krt8/18; blue). Confocal images show the dispersion of GFP +cells within the Six2 +nephron progenitor niche. Magnified views point to attached (white arrowhead) and detached (pink arrowhead) cells, with more detached cells present in Wnt11 mutants. (D) Kidneys similar to those from C) showing the overlay of GFP (green) and Krt8/18 (grey) signal as an area of attachment (orange). More extensive areas of attachment are observed in wild type kidneys. (E) Quantitation of attachments from samples similar to C) showing a significant reduction in the percentage of attached cells in Wnt11 mutants. n = 200–400 cells from each of 4 biological replicates were analyzed. (F) Quantitation of attachment area as shown in D). The average surface area of attachments is reduced in Wnt11 mutants. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
-
Figure 5—source data 1
Quantitation of nephron progenitor attachments.
- https://doi.org/10.7554/eLife.40392.018

Quantitation of cellular extensions reveals no significant differences between wild type and Wnt11 mutant nephron progenitors.
(A) Average extension lengths for attached nephron progenitor cells. (B) Average extension length for unattached nephron progenitors. (C) Angle of the extension relative to the ureteric bud for attached nephron progenitors. (D) Angle of extension relative to the ureteric bud for unattached nephron progenitors. An average of 20 cells from three replicates were analyzed. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
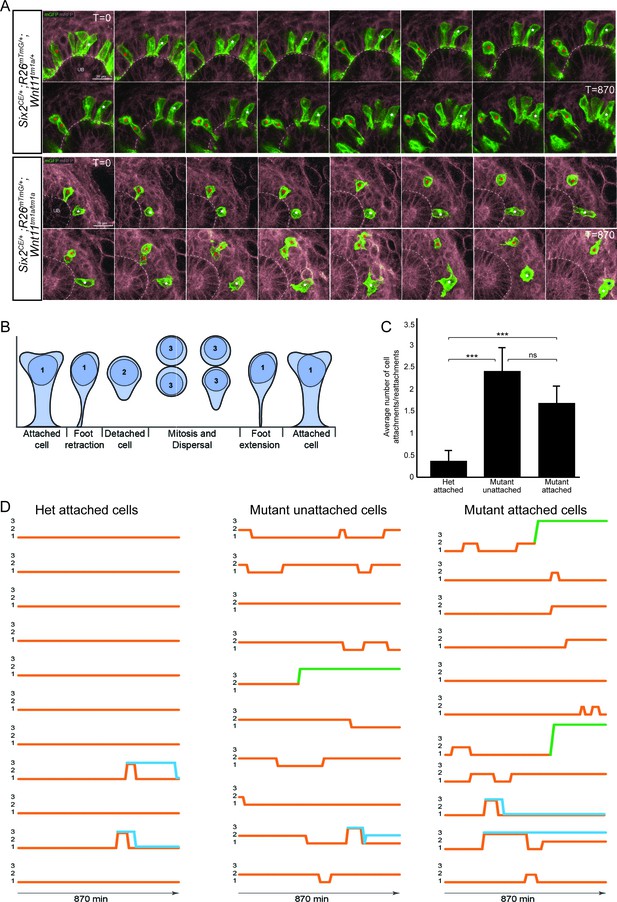
The nephron progenitors of Wnt11 mutants display dynamic attachments and reattachments to the ureteric bud.
(A) Still images from Six2CE/+; R26mTmG/+; Wnt11tm1a/+ and Six2CE/+; R26mTmG/+; Wnt11tm1a/tm1a kidney explant cultures live-imaged over 870 min (14.5 hr). GFP +cells are in green, the tdTomato +membrane of all other cells is in grey. Two cells are marked with an asterisk (red or white) in each genotype and followed across the time. The cells marked with red asterisk go through a cell division, the white cells do not. In the control kidneys, the white cell stays attached throughout. The red cell detaches briefly to divide and quickly reattaches. In Wnt11 mutants, both cells display dynamic attachments and reattachments. (B) Schematic of the classification scheme utilized to define cellular dynamics of the nephron progenitors. (C) Quantitation of the average number of attachments and reattachments of cells during live imaging. All het control cells generally begin attached and display few attachments/reattachments. Wnt11 mutant cells, whether they were attached or detached when imaging began, both display numerous attachments/reattachments. (D) Representative tracks of 10–11 individual cells classified as in B) over the course of live imaging. 11 control and 33 mutant cells were analyzed in total. Orange tracks highlights the transition between stages. Control cells stayed attached and only detached to divide. Mutant cells, whether initially attached or detached, show dynamic movements. Blue track = new cell from a division. Green = cell migrated out of the imaging field. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
-
Figure 6—source data 1
Analyses of nephron progenitor movements.
- https://doi.org/10.7554/eLife.40392.021
-
Figure 6—source code 1
Matlab script for analyses of movements.
- https://doi.org/10.7554/eLife.40392.022
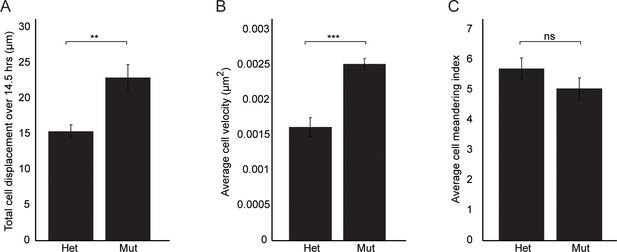
Wnt11 mutant nephron progenitors show greater displacement and velocities, but meander similarly to wild type cells.
(A) Total cell displacement was measured for Six2CE/+; R26mTmG/+; Wnt11tm1a/+ and Six2CE/+; R26mTmG/+; Wnt11tm1a/tm1a nephron progenitors imaged for 870 min (14.5 hr). GFP +cells were tracked and the total distance they moved recorded. Wnt11 mutant cells show greater displacement. (B) The average velocity of nephron progenitors was quantitated and Wnt11 mutant nephron progenitors display greater average speeds when moving. (C) The cell meandering index was calculated and Wnt11 mutants meander similarly to wild type cells. 87 control cells and 62 mutant cells were analyzed for all quantifications. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
Time lapse of Wnt11 mutant and control kidneys reveal differences in nephron progenitor dynamics.
Tamoxifen induced Six2CE/+; R26mTmG/+; Wnt11tm1a/+ and Six2CE/+; R26mTmG/+; Wnt11tm1a/tm1a kidneys were cultured on filters and live-imaged for 870 min (14.5 hr). Green cells are nephron progenitors that underwent recombination to express membrane-targeted GFP. Membrane targeted tdTomato is present in all other cells. Movies correlate to still frames in Figure 6.
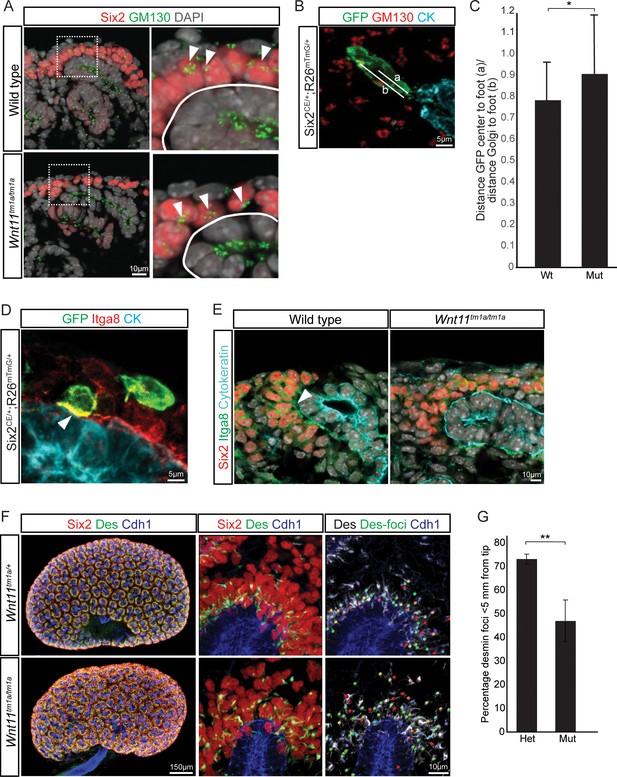
Nephron progenitor polarity is disrupted in Wnt11 mutants.
(A) E15.5 kidney sections were immunostained for Six2 (red), GM130 (Golgi; green) and DAPI (grey). The Golgi (white arrowheads) show a polarization to the distal end of nephron progenitors in wild type kidneys and this polarization is disrupted in Wnt11 mutants where they are localized closer to the ureteric branch tip (white outline). (B) Staining for GFP (green), GM130 (red), and cytokeratin (cyan) highlighting the normal polarization of Golgi within the nephron progenitor at E15.5. The letter ‘a’ marks the distance from the cell center to the foot of the cell (contact with tip) and ‘b’ marks the distance from the Golgi to the foot. (C) Ratio of the distances described in B) for wild type and mutant cells. Wild type cells show a smaller ratio indicating the Golgi lie farther from the ureteric tip than Wnt11 mutant cells, supporting their polarized nature and the loss in Wnt11 mutants. Three biological replicates and ~20 cells from each replicate were quantified. (D) E15.5 kidney sections immunostained for GFP (green), integrin a8 (Itga8; red) and cytokeratin (CK, cyan). Image shows the overlap of GFP +signal with Itga8 in cells close to the tip (white arrowhead), suggesting polarization of Itga8. (E) E15.5 kidney sections immunostained for Six2 (red), Itga8 (green), and cytokeratin (cyan). Arrowhead points to Itga8 polarization towards the ureteric branch tip in wild type kidneys, which is lost in Wnt11 mutants. (F) E15.5 wholemount immunostains for Six2 (red), desmin (Des; green), and cadherin 1 (Cdh1; blue) show the polarization of desmin foci (Des-foci) toward the ureteric branch tip which is disrupted in Wnt11 mutant kidneys. Since the desmin stain appears aster-like with wispy projections, the central focal point (foci) were identified for ease of distance quantitation. Foci were automatically located by Imaris imaging software as the most intense focal point of the desmin stain. (G) Quantitation of the percentage of desmin foci located greater than 5 μm from the ureteric tip in each genotype showing the greater dispersion from the tip in Wnt11 mutants. 385 foci from five control tips and 353 foci from four tips were analyzed. All error bars represent SEM. All significance values were determined by t-test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
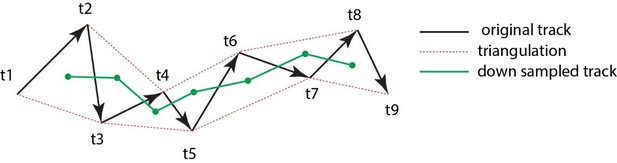
Triangulation to improve accuarcy of cell tracking.
Schematic showing the method of triangulaton used to improve the accuracy of tracking nephron progenitors.
Additional files
-
Supplementary file 1
RNA-seq from nephron progenitors.
RNA-seq of E15.5 GFP +FAC sorted nephron progenitors from wild type and Wnt11 mutants. Biological triplicates were performed for each genotype. Fold changes and RPKM are reported for each gene. Genes with RPKM <0 in wild type and mutant samples were removed for simplicity.
- https://doi.org/10.7554/eLife.40392.026
-
Supplementary file 2
RNA-seq from whole kidneys.
RNA-seq of E15.5 whole kidneys from wild type and Wnt11 mutant animals. Six biological replicates were performed for each genotype. Fold changes and RPKM are reported for each gene. Genes with RPKM <0 in wild type and mutant samples were removed for simplicity.
- https://doi.org/10.7554/eLife.40392.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40392.028





