Stem cell topography splits growth and homeostatic functions in the fish gill
Figures
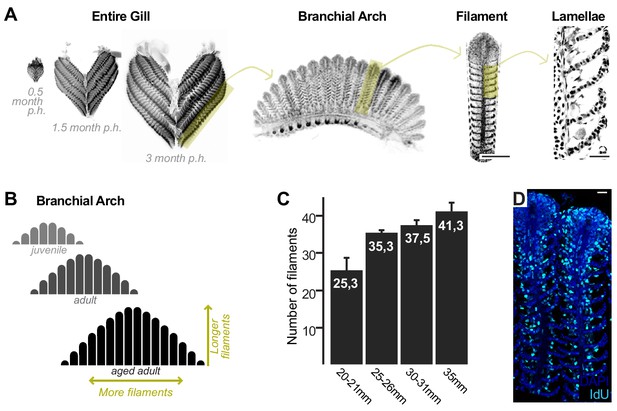
Growth and homeostasis in the medaka gill.
(A) Enucleated entire gills of medaka at different post-embryonic times show that organ size increases during post-embryonic growth (left). A gill contains four pairs of branchial arches (middle left) that display numerous filaments (middle right). Filaments are composed of lamella (right), where gas exchange occurs. (B) Scheme depicting that branchial arches grow by increasing the number of filaments, and filaments grow by increasing its length. (C) The number of filaments per branchial arch is higher in bigger fish - x axis represents fish length, and y axis the number of filaments in the second right branchial arch. (D) IdU incorporation in the adult gill reflects proliferating cells all along the longitudinal axis of a filament. Scale bars are 100 µm in (A) filament, and 20 µm in (A) lamella and (D).
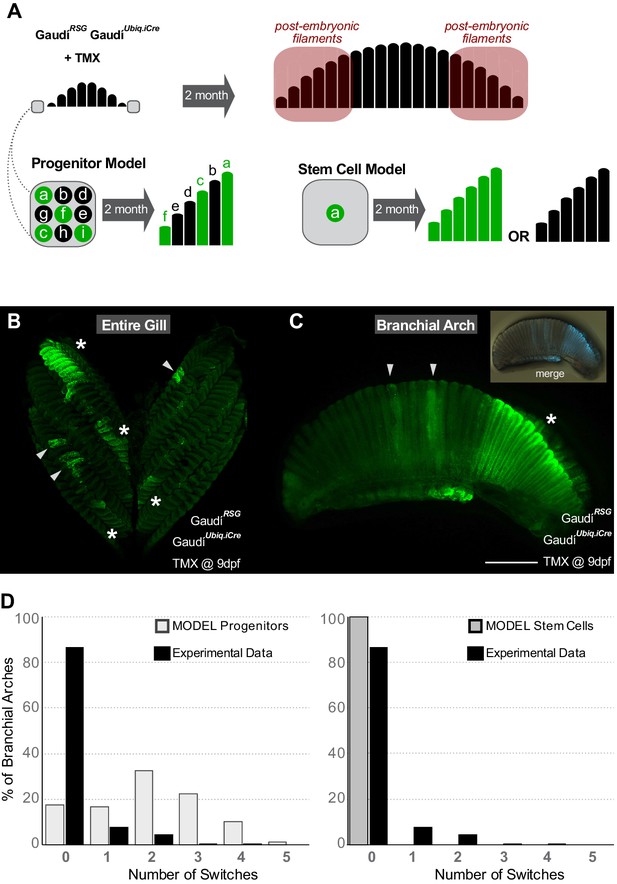
Gill stem cells located at the periphery of branchial arches generate more filaments life-long.
(A) Scheme showing the expected outcome assuming a progenitor (left bottom) or a stem cell (right bottom) model. Please note that the schemes considered as ‘labelled’ any filament containing EGFP+ cells all along their longitudinal axis. (B) Entire gill from a double transgenic GaudíUbiq.iCre GaudíRSG fish 2 month after induction with TMX. (C) Branchial arch from a double transgenic GaudíUbiq.iCre GaudíRSG fish 2 months after induction with TMX. Arrowheads in B and C indicate recombined embryonic filaments located at the centre of branchial arches, and asterisks indicate stretches of peripheral filaments with the same recombination status. Note that the present resolution does not allow revealing different recombination patterns in each filament. (D) Graphs showing the distribution of switches in stretches of the six most peripheral filaments. The graphs show a comparison of the experimental data (black) to the expected distribution according to a progenitor model (light gray, left) and to a stem cell model (gray, right). Scale bar is 500 µm in (C).
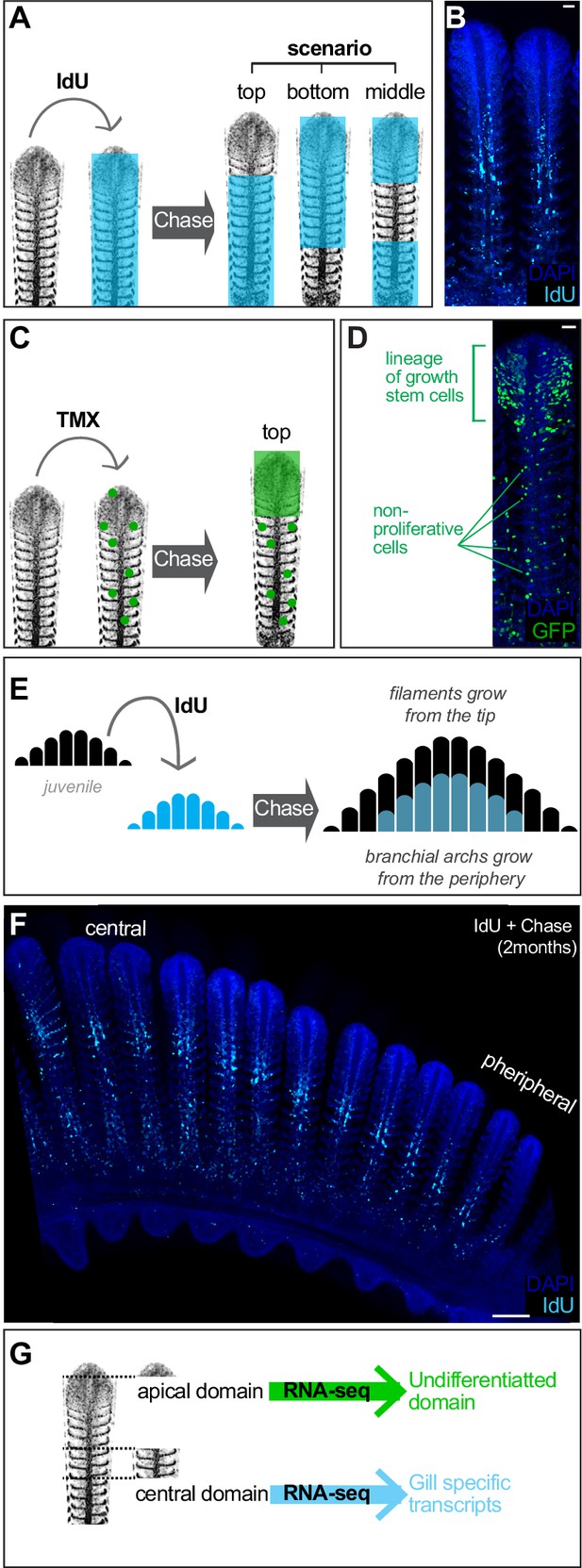
Filament growth stem cells are located at the apical tip.
(A) Scheme showing the expected outcome of IdU pulse and chase experiments depending on the location of growth stem cells. (B) IdU pulse and chase experiment shows the apical region devoted of signal, indicating these cells were generated after the IdU pulse. (C) Scheme showing the expected outcome of a filament in which growth stem cells were labelled. (D) A filament from a double transgenic GaudíUbiq.iCre GaudíRSG fish one month after induction with TMX shows an expanding clone in the apical region, indicating a high proliferative activity compared to clones located at other coordinates along the longitudinal axis. (E, F). Scheme (E) and data (F) showing an IdU pulse and chase experiment on branchial arches. The apical part of each filament and the more peripheral filaments are devoted of signal revealing the stereotypic growth of branchial arches. (G) Scheme showing the manual sorting of apical and middle regions for total RNA preparation. RNA-seq revealed differentiated cells in the middle region of filaments and undifferentiated cells at the apical domain. Scale Bars are 20 μm in (B) and (D), and 100 μm in (F).
-
Figure 3—source data 1
Transcriptome of apical and medial domains in a gill filament.
- https://doi.org/10.7554/eLife.43747.006
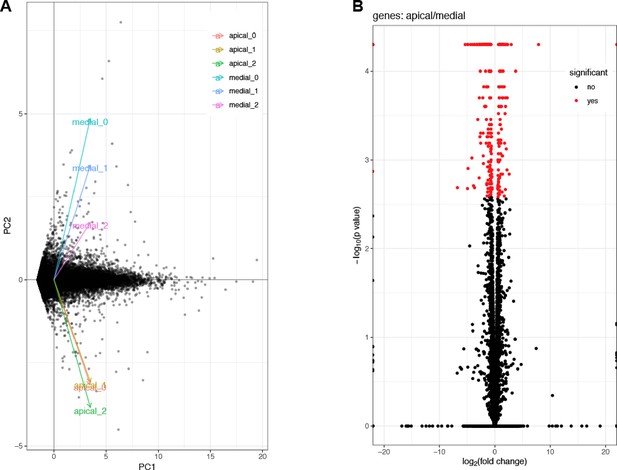
Technical analysis of the RNA-seq data.
(A) Principal Component Analysis (PCA) plot of the three biological replicates of transcripts from the apical and the medial part of the medaka gill filaments. (B) Volcano-plot representation of differential expression analysis of the genes identified from the RNA-seq libraries of the apical and the medial part of the gill filaments. Red points mark the genes with change in expression between the two conditions with a p-value<0.05.
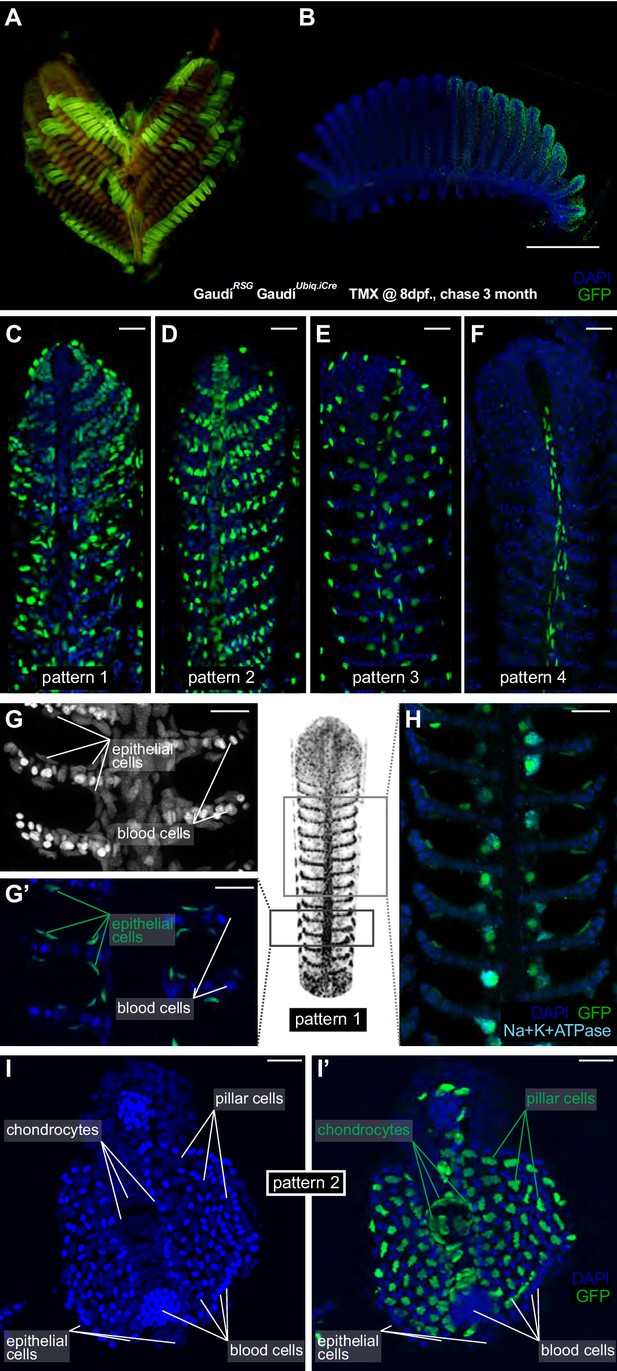
Filament growth stem cells are fate restricted.
(A–B) A gill (A) and a branchial arch (B) from a double transgenic GaudíUbiq.iCre GaudíRSG fish two month after induction with TMX. (C–F) Confocal images from filaments in A, B, stained for EGFP and DAPI to reveal the cellular composition of different clones. Four different recombination patterns were identified. (G, G’) A detailed view of pattern 1(C) shows recombined epithelial cells covering each lamella. (H) Co-staining with an anti-Na+K+ATP-ase antibody confirms that MRC cells are clonal to other epithelial cells in the filament. (I, I’) Cross-section of a filament that displays pattern 2 (D). DAPI staining allows identifying blood cells (strong signal, small round nuclei), pillar cells (weaker signal, star-shaped nuclei), and chondrocytes (elongated nuclei at the central core of the filament) (I). The lineage tracker EGFP reveals that chondrocytes and pillar cells are clonal along a filament (I’). Scale Bars are 500 µm in (B), 20 µm in (C–H) and (I).
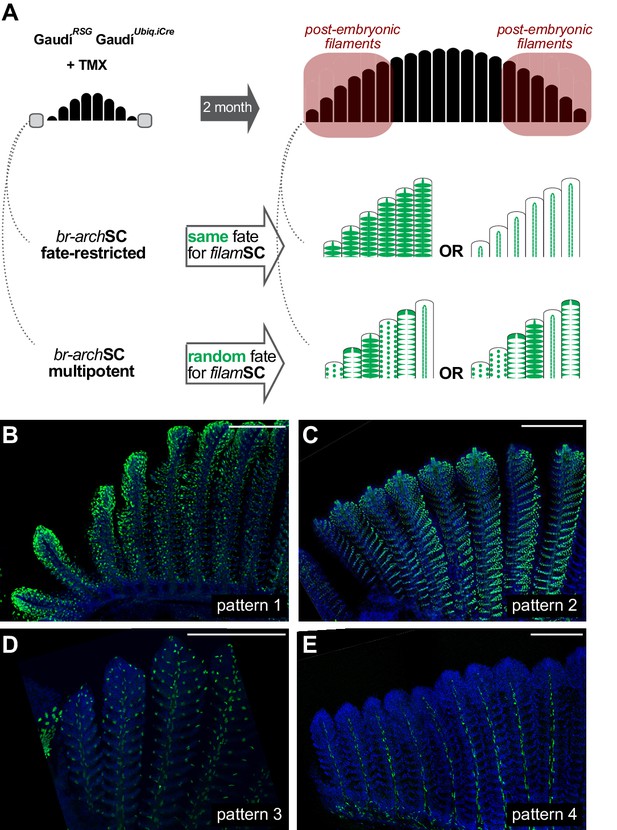
Branchial arch stem cells are fate restricted.
(A) Scheme showing the expected outcome assuming that br-archSCs are fate restricted (middle) or multi-potent (bottom). The recombination pattern of consecutive filaments would be identical if generated by fate restricted br-archSCs, and non-identical if derived from a multipotent br-archSC. (B–E) Confocal images show an identical recombination pattern in consecutive peripheral filaments for pattern 1 (B), pattern 2 (C), pattern 3 (D) and pattern 4 (E). Scale Bars are 200 μm in (B).
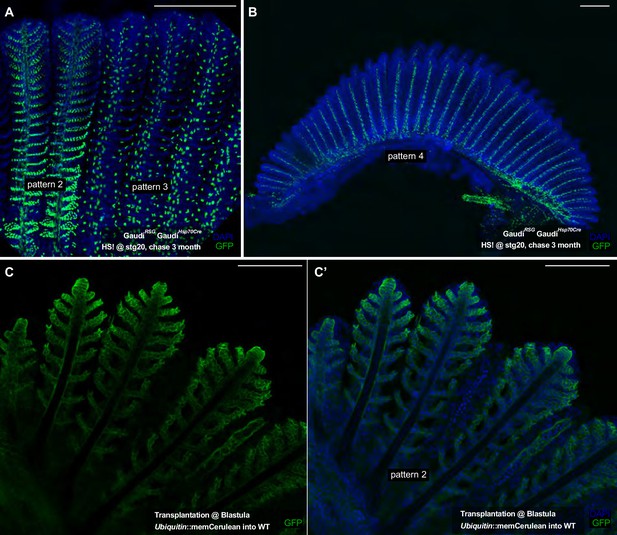
Early embryonic recombination and transplantations at blastula stage indicate fate-restricted stem cells in the fish gill.
(A) Consecutive filaments from a double transgenic GaudíHsp70A.Cre GaudíRSG fish 3 months after induction (heat-shock at stage 20). Filaments display recombination in specific patterns (pattern 2 in two filaments at the left, pattern 3 in three filaments at the right). (B) A Branchial arch from a double transgenic GaudíHsp70A.Cre GaudíRSG fish 3 months after induction (heat-shock at stage 20). All filaments are labelled in pattern 4 indicating that the earliest founder cell of that lineage was labelled. (C) Consecutive filaments of a chimeric fish generated by transplantations at blastula stage (Host: unlabelled WT, Donor: GaudiBBW - driving ubiquitously a membrane-tagged Cerulean fluorescent protein). Filaments display the recombination Pattern 2 - compare to Figure 4D and mind differences in nuclear vs membrane label. Scale Bars are 200 μm in (A, B), and 100 μm in (C).
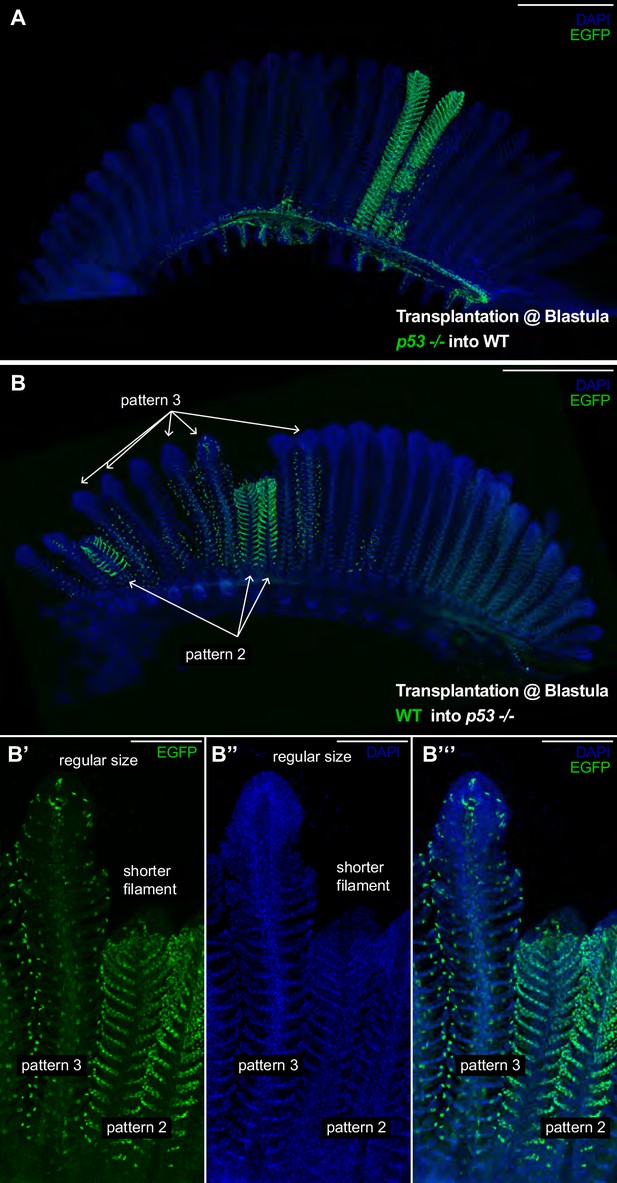
p53 Coordinates growth stem cells in a lineage-specific manner.
(A) Branchial arch of a p53-/- -to-WT chimera, where p53-/- mutant cells are labelled in green. Composite filaments display the proper length as compared to non-labelled neighbour filaments. (B) Branchial arch of a WT-to-p53-/- chimera, where WT cells are labelled in green. Composite filaments are shorter than their neighbours only when pattern 2 comes from the donor - Note the short size of the right filament in B’-B’’ compared to the filament at the left. Scale Bars are 500 μm in (A, B) and 100 μm in (B’–B’’’).
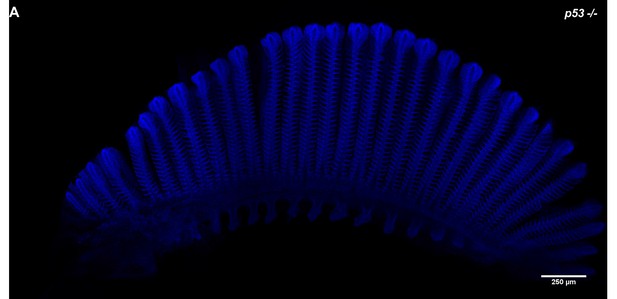
Branchial Arch of p53E241X mutant.
(A) DAPI staining on a branchial arch extracted from a p53 homozygous mutant showing the un-distinguishable organisation, length and size of filaments and lamellae than wild-type samples.
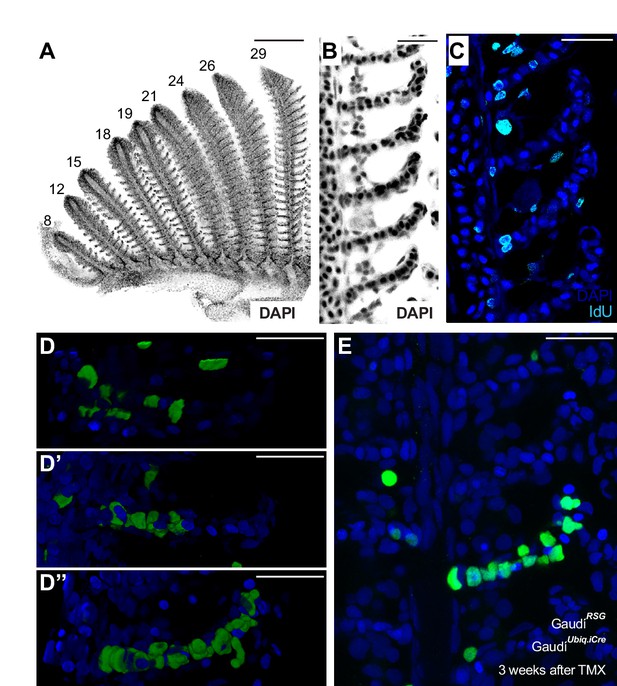
Homeostatic stem cells locate to the base of each lamella.
(A) DAPI image of peripheral filaments indicating the increasing number of lamellae per filament. (B) DAPI image of consecutive lamellae along a filament reveals that lamellae do not increase their size. (C) IdU pulse reveals proliferative cells at the base of the lamellae. (D–E) EGFP cells indicating clonal progression of clones in double transgenic GaudíUbiq.iCre GaudíRSG fish 1 month after induction with TMX during adulthood. Clones of pillar cells progress from the base to the distal part of a lamellae (D’’, E). Scale Bars are 200 μm in (A), and 20 μm in (B–E).
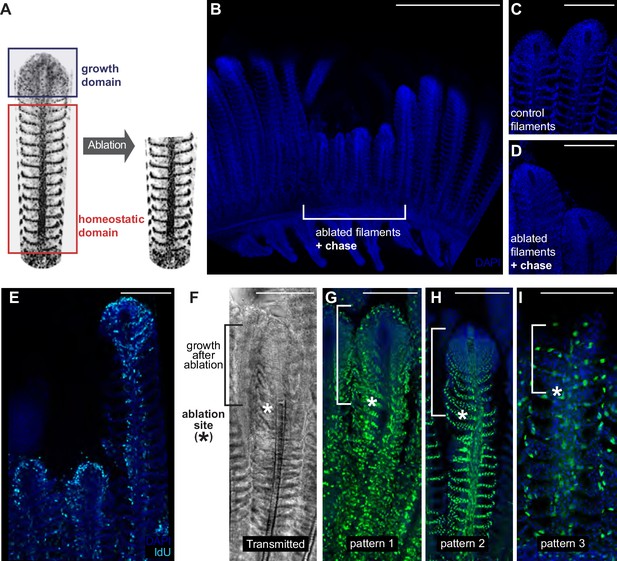
The homeostatic domain sustains growth after filament ablation.
(A) Scheme of the ablation procedure. The growth domain and the upper part of the homeostatic domain are mechanically ablated. (B) DAPI image of control filaments shows an intact growth domain at the top. (C) DAPI image of injured filaments after a chase of 1 month shows a regenerated growth domain. (D) During the duration of the experiment, ablated filaments were unable to reach the length of their neighbour, non-ablated filaments. (E) IdU-positive cells are detected at the growth domain of both intact and ablated treatment. (F–I) Transmitted (F) and fluorescent (G–I) images of gill filaments from recombined GaudíUbiq.iCre GaudíRSG fish, 3 weeks post-ablation. The upper fraction containing the growth domain was generated after the ablation (highlighted in F). The recombination pattern observed in the regenerated zone is identical to the recombination pattern that the filaments had before ablation, indicating that cells maintain their fate during the regenerative response. Scale Bars are 500 µm in (A) and 100 µm in (C–I).
Videos
3D reconstruction of a pattern 3-labelled filament.
A middle section of filament in an adult GaudíUbiq.iCre GaudíRSG fish that was induced for recombination at late embryonic stages. The filament shows the lineage of a growth stem cell that labels pattern 3.
3D reconstruction of a pattern 4-labelled filament.
A middle section of filament in an adult GaudíUbiq.iCre GaudíRSG fish that was induced for recombination at late embryonic stages. The filament shows the lineage of a growth stem cell that labels pattern 4.
3D reconstruction of a pattern 2-labelled filament.
A middle section of filament in an adult GaudíUbiq.iCre GaudíRSG fish that was induced for recombination at late embryonic stages. The filament shows the lineage of a growth stem cell that labels pattern 2.
3D reconstruction of a pattern 2-labelled lamella.
A middle section of filament in an adult GaudíUbiq.iCre GaudíRSG fish that was induced for recombination at late embryonic stages. The filament shows the lineage of a homeostatic stem cell that labels pattern 2 only in one lamella.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Oryzias latipes) | Cab | wild type stock, southern population | ||
| Genetic reagent (O. latipes) | p53 E241X mutant | Taniguchi et al., 2006 | ||
| Genetic reagent (O. latipes) | GaudíUbiq.iCre | Centanin et al., 2014 | ||
| Genetic reagent (O. latipes) | GaudíHsp70.A | Centanin et al., 2014 | ||
| Genetic reagent (O. latipes) | GaudíRSG | Centanin et al., 2014 | ||
| Genetic reagent (O. latipes) | GaudiLoxPOUT | Centanin et al., 2014 | ||
| Genetic reagent (O. latipes) | GaudiBBW | Centanin et al., 2014 | ||
| Antibody | a-EGFP (Rabbit IgG polyclonal) | Invitrogen (now Thermo Fischer) | CAB4211; RRID: AB_10709851 | Dilution 1:750 |
| Antibody | a-EGFP (Chicken IgY polyclonal) | life technologies | A10262; RRID: AB_2534023 | Dilution 1:750 |
| Antibody | a-Na + K + ATP-ase (Rabbit monoclonal) | Abcam | ab76020, EP1845Y | Dilution 1:200 |
| Antibody | a-BrdU/IdU (Mouse IgG monoclonal) | Becton Dickinson | 347580 | Dilution 1:50 |
| Antibody | Alexa 488 Goat a-Rabbit | Invitrogen (now Thermo Fischer) | A-11034 | Dilution 1:500 |
| Antibody | Alexa 488 Donkey a-Chicken | Invitrogen (now Thermo Fischer) | 703-545-155 | Dilution 1:500 |
| Antibody | Alexa 647 Goat a-Rabbit | Life Technologies | A-21245 | Dilution 1:500 |
| Antibody | Alexa 647 Goat a-Rabbit | Life Technologies | A-21245 | Dilution 1:500 |
| Antibody | Cy5 Donkey a-Mouse | Jackson | 715-175-151 | Dilution 1:500 |
| Chemical compound, drug | tamoxifen | Sigma | T5648 | |
| Chemical compound, drug | tricaine | Sigma-Aldrich | A5040-25G | |
| Chemical compound, drug | BrdU | Sigma-Aldrich | B5002 | final concentration of 0,4 g/l |
| Chemical compound, drug | IdU | Sigma-Aldrich | I7125 | final concentration of 0,4 g/l |
| Chemical compound, drug | Trizol | |||
| Software, algorithm | Ensemble | Public | ||
| Other | DAPI | Roth | final concentration of 5 ug/l | |
| Software, algorithm | DAVID 6.8 | https://david.ncifcrf.gov/home.jsp |
Additional files
-
Supplementary file 1
Simulation of recombined pattern in branchial arches assuming progenitor cells.
The table represents simulated values of recombination (0 = unlabelled; 1 = labelled) in five gills, according to the ‘progenitor’ model described in M and M. The labelling efficiency was calculated from real data from 5 GaudíUbiq.iCre GaudíRSG recombined gills.
- https://doi.org/10.7554/eLife.43747.018
-
Supplementary file 2
Simulation of recombined pattern in branchial arches assuming stem cells.
The table represents simulated values of recombination (0 = unlabelled; 1 = labelled) in five gills, according to the ‘stem cell’ model described in M and M. The labelling efficiency was calculated from real data from 5 GaudíUbiq.iCre GaudíRSG recombined gills.
- https://doi.org/10.7554/eLife.43747.019
-
Supplementary file 3
Experimental data of recombined pattern in adult branchial arches.
The table includes values for recombination (0 = unlabelled; 1 = labelled) in five gills enucleated from adult GaudíUbiq.iCre GaudíRSG fish that were induced for recombination at late embryonic stages.
- https://doi.org/10.7554/eLife.43747.020
-
Supplementary file 4
Objective function representation comparing data to progenitor and stem cell models.
The table displays the values for an objective function, comparing the recombination pattern obtained in 22 GaudíUbiq.iCre GaudíRSG recombined gills with the predicted values for a stem cell or a progenitor model (See M and M). Lower values for the objective function represent a better fit between experimental and simulated data.
- https://doi.org/10.7554/eLife.43747.021
-
Supplementary file 5
Enriched transcripts in the apical and medial domains of gill filaments.
Representation of transcripts obtained from the apical and the medial domain of gill filaments, ordered according to their differential expression.
- https://doi.org/10.7554/eLife.43747.022
-
Supplementary file 6
Experimental data of recombined patterns in adult branchial arches with cellular resolution.
The table displays values of recombination (0 = unlabelled; 1 = Pattern 1; 2 = Pattern 2; 3 = Pattern 3; 4 = Pattern 4) in gills enucleated from adult GaudíUbiq.iCre GaudíRSG fish that were induced for recombination at late embryonic stages.
- https://doi.org/10.7554/eLife.43747.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43747.024





