Schnyder corneal dystrophy-associated UBIAD1 inhibits ER-associated degradation of HMG CoA reductase in mice
Figures
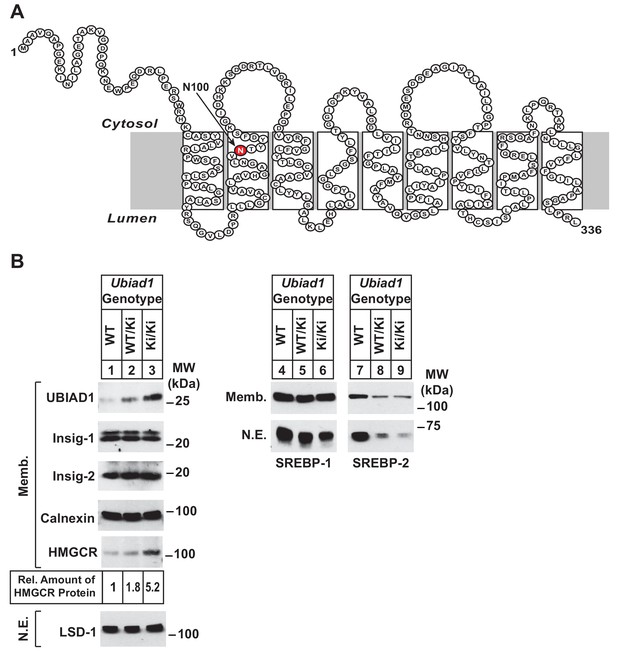
Accumulation of HMGCR protein in livers of Ubiad1Ki/Ki mice with mixed C57BL/6 × 129 genetic background.
(A) Amino acid sequence and predicted topology of mouse UBIAD1 protein. Asparagine-100 (N100), which corresponds to the most frequently mutated amino acid residue in SCD, is enlarged, shaded in red and indicated by an arrow. (B) Male WT, Ubiad1WT/Ki, and Ubiad1Ki/Ki littermates (8–9 weeks of age, eight mice/group) were fed an ad libitum chow diet prior to sacrifice. Livers of the mice were harvested and subjected to subcellular fractionation as described in ‘Materials and methods.’ Aliquots of resulting membrane (Memb.) and nuclear extract (N.E.) fractions (80–160 µg of total protein/lane) for each group were pooled and subjected to SDS-PAGE, followed by immunoblot analysis using antibodies against endogenous HMGCR, SREBP-1, SREBP-2, UBIAD1, Insig-1, Insig-2, calnexin, and LSD-1. Although shown in a separate panel, LSD-1 serves as a loading control for the nuclear SREBP immunoblots. The amount of hepatic HMGCR protein in Ubiad1Ki/Ki mice was determined by quantifying the band corresponding to HMGCR using ImageJ software.
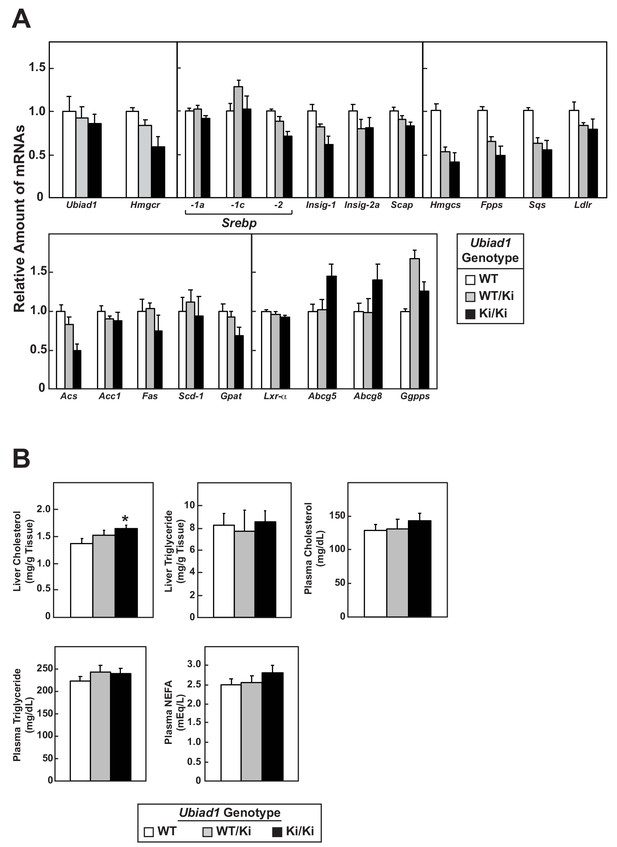
Relative amounts of hepatic mRNAs encoding components of the Scap-SREBP pathway and lipid analysis in WT and Ubiad1Ki/Ki mice.
(A) Total RNA isolated from livers of mice used in Figure 1B (8 mice/group) was separately isolated. Equal amounts of RNA from the individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; cyclophilin mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in WT mice, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from eight mice. (B) The amount of cholesterol, triglycerides, and non-esterified fatty acids (NEFA) in livers and plasma from WT or Ubiad1 knockin mice used in Figure 1B was determined by a colorimetric assay as described in ‘Materials and methods.’ Error bars, S.E. The p value was calculated using Student’s t test: *, p ≤ 0.05. Hmgcs, HMG coenzyme A synthase; Fpps, farnesyl pyrophosphate synthase; Sqs, squalene synthase; Acs, acetyl coenzyme A synthetase; Acc1, acetyl coenzyme A carboxylase-1; Fas, fatty acid synthase; Scd-1, stearoyl coenzyme A desaturase-1; Gpat, glycerol-3-phosphate acyltransferase; Abcg5 and Abcg8, ATP-binding cassette subfamily G member 5 and 8, respectively; Ggpps, geranylgeranyl pyrophosphate synthase.
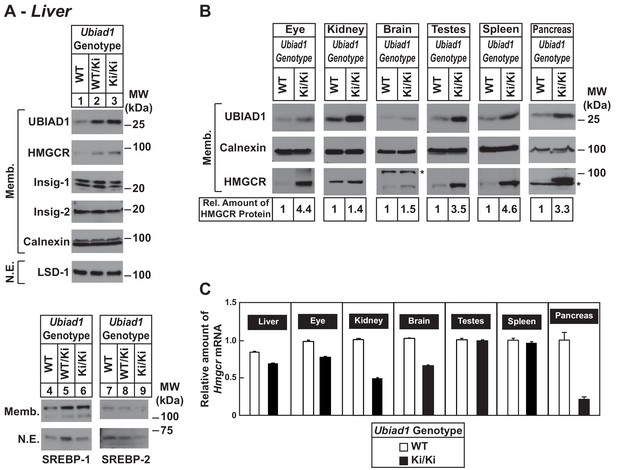
Accumulation of HMGCR protein in tissues of WT and Ubiad1Ki/Ki mice with C57BL/6 genetic background.
(A and B) Eight to nine-week old male WT, Ubiad1WT/Ki, and Ubiad1Ki/Ki littermates (six mice/group) were fed an ad libitum chow diet prior to study. Aliquots of membrane (Memb.) and nuclear extract (N.E.) fractions from homogenized livers, enucleated eyes, kidneys, brains, testes, and spleens (23–50 µg of total protein/lane) were analyzed by immunoblot using antibodies against the indicated proteins. The asterisk indicates a non-specific cross-reactive band observed in the anti-HMGCR immunoblot from brain and pancreas. Although shown in separate panels, LSD-1 serves as a loading control for the nuclear SREBP-1 and SREBP-2 immunoblots. In (B), the amount of HMGCR protein in the indicated tissues from Ubiad1Ki/Ki mice was determined by quantifying the band corresponding to HMGCR using Image J software. (C) For mRNA analysis, equal amounts of RNA from the indicated tissue of individual mice were subjected to quantitative real-time RT-PCR using primers against the Hmgcr mRNA and cyclophilin mRNA as an invariant control. Error bars, S.E.
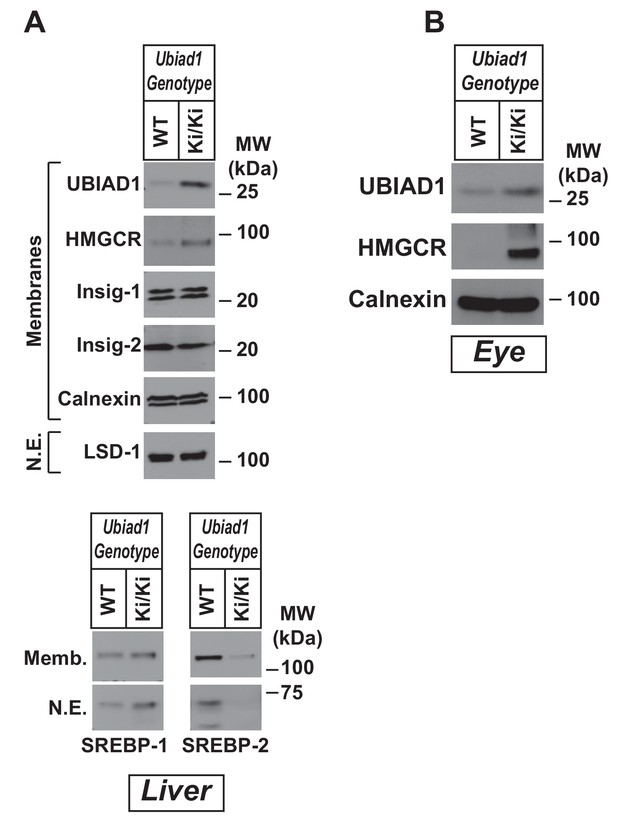
Accumulation of HMGCR protein in eyes and livers of WT and Ubiad1Ki/Ki mice.
Female WT, Ubiad1WT/Ki, and Ubiad1Ki/Ki littermates of animals used in Figure 2 (six mice/group, 8–9 weeks of age) were fed an ad libitum chow diet prior to study. Aliquots of membrane (Memb.) and nuclear extract (N.E.) fractions from homogenized livers (A) and enucleated eyes (B) (50–80 µg of total protein/lane) were analyzed by immunoblot using antibodies against the indicated proteins. Although shown in separate panels, LSD-1 serves as a loading control for the nuclear SREBP-1 and SREBP-2 immunoblots in (A).
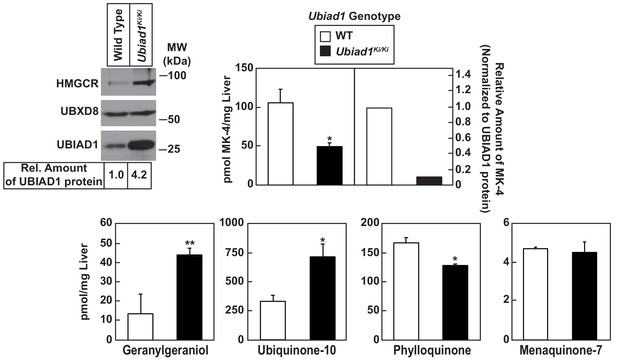
Analysis of nonsterol isoprenoids in WT and Ubiad1Ki/Ki mice.
Male mice (10–12 weeks of age, five mice/group) were fed ad libitum a chow diet prior to study. Livers were collected for subcellular fractionation and immunoblot analysis of resulting membrane fractions (80 µg total protein/lane) using antibodies against the indicated proteins or to determine the amount of menaquinone-4 (MK-4), geranylgeraniol, ubiquinone-10, phylloquinone, and menaquinone-7 (MK-7) by LC-MS/MS as described in ‘Materials and methods.’ The relative amount of hepatic MK-4 in Ubiad1Ki/Ki mice was determined by normalizing the amount of the vitamin K2 subtype to the amount of UBIAD1 protein, which was quantified using ImageJ software. Error bars, S.E. The p value was calculated using Student’s t test: *, p < 0.05; **, p < 0.01.
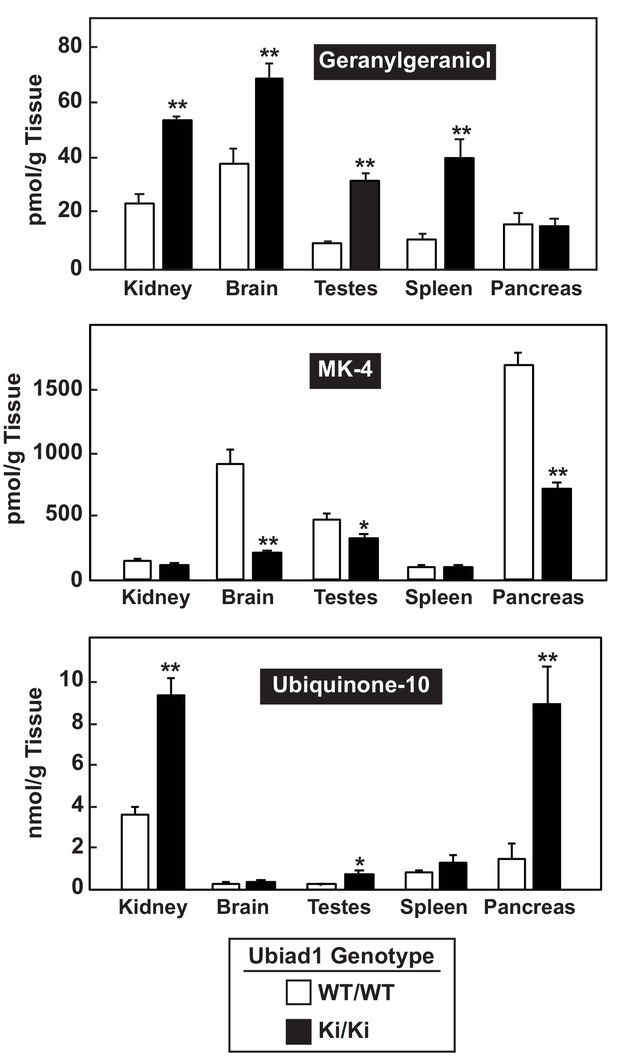
Analysis of nonsterol isoprenoids in various tissues of WT and Ubiad1Ki/Ki mice.
The indicated tissues from mice used in Figure 3 were collected and the amount of menaquinone-4 (MK-4), geranylgeraniol, and ubiquinone-10 by LC-MS/MS as described in ‘Material and methods.’ Error bars, S.E. The p value was calculated using Student’s t test: *, p < 0.05; **, p < 0.01.
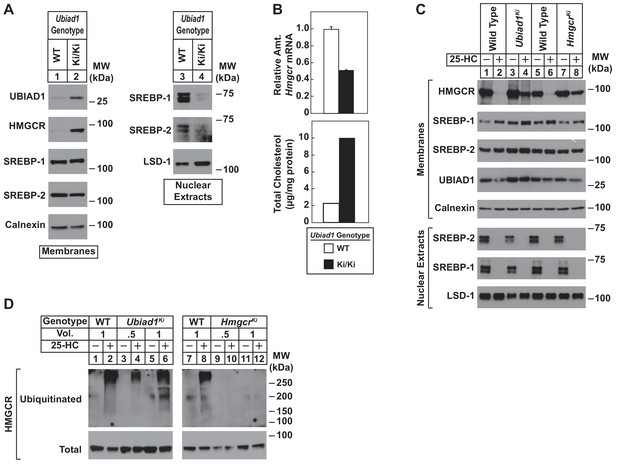
Sterol-mediated regulation of HMGCR in mouse embryonic fibroblasts (MEFs) from WT and Ubiad1Ki/Ki mice.
MEFs from WT and Ubiad1Ki/Ki mice were set up for experiments on day 0 at 2 × 105 cells per 10 cm dish in MEF medium supplemented with 10% fetal calf serum (FCS). (A) On day 3, cells were harvested for subcellular fractionation. Aliquots of resulting membrane and nuclear extract fractions (35–50 µg total protein/lane) were subjected to SDS-PAGE, followed by immunoblot analysis using antibodies against the indicated proteins. (B) On day 3, cells were harvested for measurement of Hmgcr mRNA levels by quantitative RT-PCR and total cholesterol levels using a colorimetric assay as described in ‘Materials and methods.’ (C and D) On day 2, cells were depleted of isoprenoids through incubation for 16 hr at 37°C in MEF medium containing 10% lipoprotein-deficient serum, 10 µM sodium compactin, and 50 µM sodium mevalonate. The cells were subsequently treated with 1 µg/ml 25-HC as indicated; in (D), the cells also received 10 µM MG-132. (C) After 4 hr at 37°C, cells were harvested for preparation of membrane and nuclear extract fractions (35–50 µg total protein/lane) that were analyzed by immunoblot with antibodies against the indicated protein. (D) Following incubation for 1 hr at 37°C, cells were harvested, lysed in detergent-containing buffer, and immunoprecipitated with 30 µg polyclonal anti-HMGCR antibodies. Immunoprecipitated material was subjected to SDS-PAGE and immunoblot analysis with IgG-A9 (against HMGCR) and IgG-P4D1 (against ubiquitin).
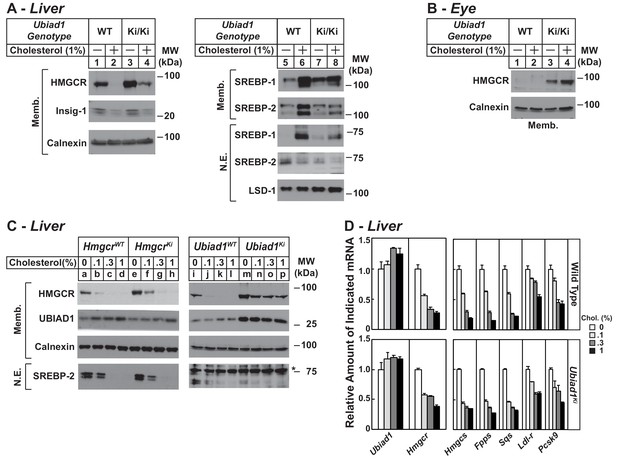
Regulation of HMGCR in livers of cholesterol-fed WT, Ubiad1Ki/Ki, and HmgcrKi/Ki mice.
Male mice (12–13 weeks of age, five mice/group) were fed an ad libitum chow diet supplemented with the indicated amount of cholesterol for 5 days. Aliquots of membrane (Memb.) and nuclear extract (N.E.) fractions from homogenized livers (A and C) or enucleated eyes (B) (70 µg protein/lane) were analyzed by immunoblot analysis with antibodies against the indicated proteins as described in the legend to Figure 1. The asterisk denotes a nonspecific band observed in the nuclear SREBP-2 immunoblot. (D) For mRNA analysis, equal amounts of RNA from livers of mice were subjected to quantitative real-time RT-PCR using primers against the indicated mRNAs and cyclophilin mRNA as an invariant control. Error bars, S.E. Pcsk9, proprotein convertase subtilisin/kexin type 9.
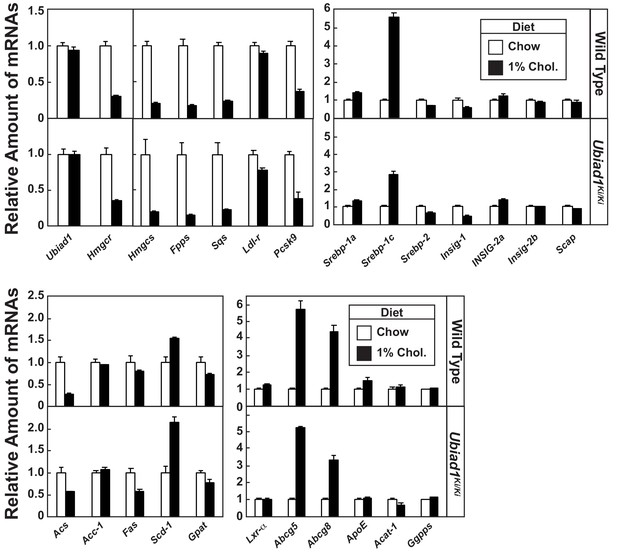
Effect of dietary cholesterol on expression of mRNAs encoding components of the Scap-SREBP pathway in livers of WT and Ubiad1 knock-in mice.
Total RNA from livers of mice used in Figure 5A (5 mice/group) was separately isolated. Equal amounts of RNA from the individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; cyclophilin mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in WT mice fed a chow diet, which was arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from five mice. ApoE, apolipoprotein E; Acat-1, acyl-coenyzme A:cholesterol acyltransferase-1.
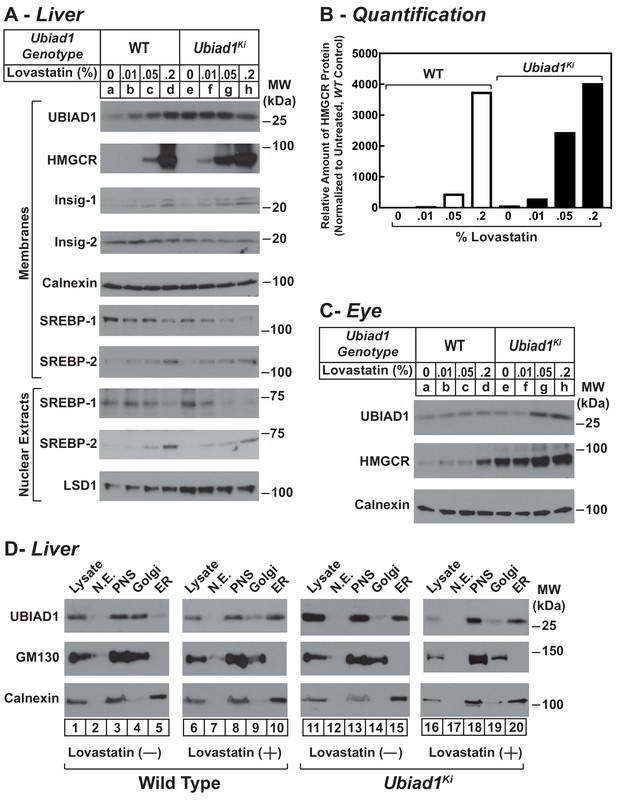
Statin-mediated regulation of HMGCR and UBIAD1 in WT and Ubiad1Ki/Ki mice.
Male mice (6–8 weeks of age, five mice/group) were fed an ad libitum chow diet supplemented with the indicated amount (A and C) or 0.2% (D) lovastatin for 5 days. (A and C) Aliquots of membrane and nuclear extract fractions from homogenized livers (A) or enucleated eyes (C) (70 µg protein/lane) were analyzed by immunoblot analysis with antibodies against the indicated proteins. In (B), the amount of HMGCR protein in livers of Ubiad1Ki/Ki mice shown in (A) was determined by quantifying the band corresponding to HMGCR using Image J software and normalizing to the amount of the protein in untreated WT controls. (D) Post nuclear supernatants (PNS) obtained from liver homogenates were fractionated on a discontinuous sucrose gradient (7.5–45%) that yielded a light membrane fraction enriched in Golgi and a heavy membrane fraction enriched in ER. Aliquots of the homogenates (lysate), nuclear extracts (N.E.), PNS, Golgi-enriched membranes, and ER-enriched membranes were subjected to SDS-PAGE, followed by immunoblot analysis with antibodies against the indicated proteins.
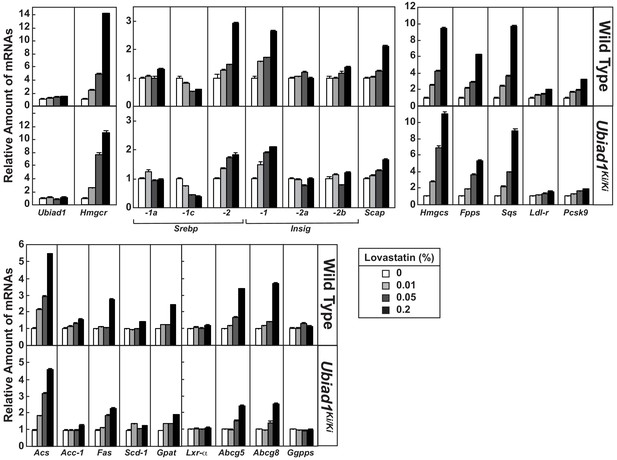
Effect of lovastatin on expression of mRNAs encoding components of the Scap-SREBP pathway in livers of WT and Ubiad1 knock-in mice.
Total RNA from livers of mice used in Figure 6A (5 mice/group) was separately isolated. Equal amounts of RNA from the individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; cyclophilin mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in WT mice fed a chow diet, which was arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from five mice.
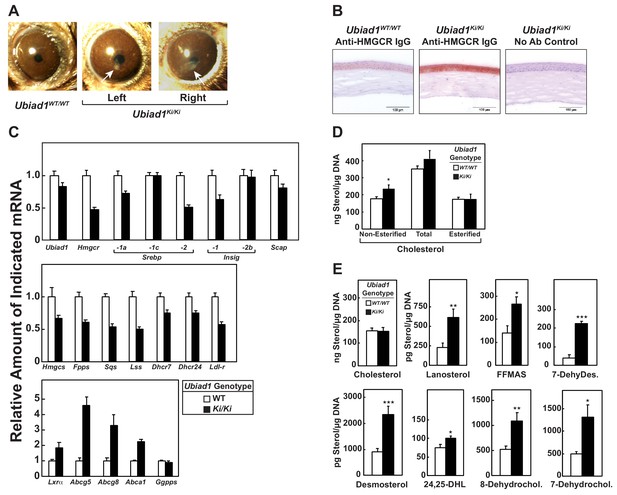
Ubiad1Ki/Ki mice exhibit signs of corneal opacification upon aging.
(A) Male and female mice (15 WT, 24 Ubiad1Ki/Ki, 50 weeks of age) consuming an ad libitum chow diet were analyzed by stereomicroscopic examination. Corneal opacification is indicated by white arrows. (B–E) Mice analyzed in (A) were sacrificed, corneas were then harvested and analyzed by immunohistochemical staining with anti-HMGCR polyclonal antibodies (B), quantitative RT-PCR (C), and LC-MS/MS (D and E) as described in the legend to Figure 1 and ‘Materials and methods.’ Error bars, S.E. The p value was calculated using Student’s t test: *, p < 0.05; **, p < 0.01; ***, p 0.005. Dhcr7, 7-dehydrocholesterol reductase; Dhcr24, 24-dehydrocholesterol reductase; 7-DehyDes., 7-dehydrodesmosterol; 8-Dehydrochol., 8-dehydrocholesterol; 7-Dehydrochol., 7-dehydrocholesterol.
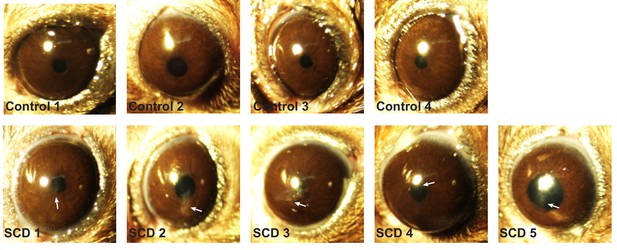
Ubiad1Ki/Ki mice exhibit signs of corneal opacification upon aging.
Female mice (15 WT, 24 Ubiad1Ki/Ki, 50 weeks of age) consuming an ad libitum chow diet were analyzed by stereomicroscopic examinations as described in Figure 7. Corneal opacification is indicated by white arrows.
Tables
Comparison of wild type (WT) and ubiad1ki/ki mice.
Male WT and Ubiad1Ki/Ki littermates (8–9 weeks of age, eight mice/group) were fed an ad libitum chow diet prior to study. WT mice were littermates of Ubiad1Ki/Ki mice. Each value represents the mean ±S.E. of 8 values. The p value was calculated using Student’s t test: *, p≤0.05.
| Parameter | WT | Ubiad1Ki/Ki |
|---|---|---|
| Body Weight (g) | 19.8 ± 0.4 | 20.1 ± 0.6 |
| Liver Weight (g) | 1.0 ± 0.05 | 0.9 ± 0.03 |
| Plasma Triglycerides (mg/dL) | 123.6 ± 31.2 | 94.5 ± 5.7 |
| Plasma Cholesterol (mg/dL) | 100.4 ± 8.4 | 90.3 ± 9.0 |
| Plasma Nonesterified Fatty Acids (mEq/L) | 1.3 ± 0.2 | 1.1 ± 0.03 |
| Liver Triglycerides (mg/g) | 9.61 ± 1.8 | 16.3 ± 5.0 |
| Liver Cholesterol (mg/g) | 1.17 ± 0.06 | 1.65 ± 0.24* |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Mouse/Ubiad1Ki/Ki (UBIAD1 (N100S)):C57BL/6J | This paper | N/A | Heterozygous knockin mice harboring mutations in the endogenous Ubiad1 gene that change Asparagine-100 to a Serine residue |
| Genetic reagent (M. musculus) | Mouse/Ubiad1Ki/Ki (UBIAD1 (N100S)): C57BL/6 | This paper | Homozygous knockin mice harboring mutations in the endogenous Ubiad1 gene that change Asparagine-100 to a Serine residue | |
| Genetic reagent (M. musculus) | Mouse/HmgcrKi/Ki (HMGCR K89R/ K248R):C57BL/6 | PMID: 27129778 | N/A | |
| Cell line | Mouse Embryonic Fibroblast- Ubiad1WT/WT | This paper | N/A | Mouse embryonic fibroblasts from wild type C57BL/ 6 mice |
| Cell line | Mouse Embryonic Fibroblast- Ubiad1Ki/Ki | This paper | N/A | Mouse embryonic fibroblasts from Ubiad1Ki/Ki C57BL/6 mice |
| Cell line | Mouse Embryonic Fibroblast- HmgcrWT/WT | This paper | N/A | Mouse embryonic fibroblasts from wild type C57BL/ 6 mice |
| Cell line | Mouse Embryonic Fibroblast- Hmgcr Ki/Ki | This paper | N/A | Mouse embryonic fibroblasts from HmgcrKi/Ki C57BL/6 mice |
| Antibody | Rabbit monoclonal anti-SREBP-1 | PMID: 28244871 | IgG-20B12 | |
| Antibody | Rabbit monoclonal anti-SREBP-2 | PMID: 25896350 | IgG-22D5 | |
| Antibody | Rabbit polyclonal anti-UBIAD1 | This paper | IgG-205 | Rabbit polyclonal antibody raised against amino acids 2–21 of mouse UBIAD1; used at 1–5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti- HMGCR | PMID: 27129778 | IgG-839c | used at 1–5 µg/ml for immunoblots |
| Antibody | Mouse monoclonal anti- HMGCR | PMID: 22143767 | IgG-A9 | used at 1–5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti- Insig-1 | PMID: 27129778 | anti-Insig-1 | used at 1:1000 dilution for immunoblots |
| Antibody | Rabbit polyclonal anti-Insig-2 | This paper | IgG-492 | Rabbit polyclonal antibody raised against a C-terminal peptide (CKVIPEKSHQE) of hamster Insig-2; used at 5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti-UBXD8 | PMID: 27129778 | IgG-819 | used at 1–5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti-Calnexin | Novus Biologicals | Cat#NB100-1965; RRID:AB_10002123 | used at 1–5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti-GM130 | Abcam | Cat#ab30637; RRID:AB_732675 | used at 1–5 µg/ml for immunoblots |
| Antibody | Rabbit polyclonal anti-LSD-1 | Cell Signaling Technology | Cat#2139; RRID:AB_2070135 | used at 1–5 µg/ml for immunoblots |
| Antibody | Mouse monoclonal anti-ubiquitin (IgG-P4D1) | Santa Cruz | Cat#SC8017;RRID:AB_628423 | used at 1–5 µg/ml for immunoblots |
| Recombinant DNA reagent | ||||
| Sequence-based reagent | Ubiad1Ki/Ki genotyping primers: Forward, GGAACACTTGGCTCTCATCT; Reverse, GGGAGCAGTGTTCATAATCC | This paper | N/A | Genotyping was determined by PCR analysis of genomic DNA prepared from tails of mice. |
| Sequence-based reagent | HmgcrKi/Ki genotyping primers: K89R- Forward, GTCCATGAACATGTTCACCG; Reverse, CAGCACGTCCTATTGGCAGA K248R – Forward, TCGGTGATGTTCCAGTCTTC; Reverse, GGTGGCAAACACCTTGTATC | PMID: 27129778 | N/A | |
| Sequence-based reagent (qRT-PCR) | UBIAD1 Forward, GACAGAACTTTGGTGGACAGAATTC; Reverse, CAGCCCAAGGTGTAGAGGAAGA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | SREBP-1a Forward, GGCCGAGATGTGCGAACT; Reverse, TTGTTGATGAGCTGGAGCATGT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | SREBP-1c Forward, GGAGCCATGGATTGCACATT; Reverse, GGCCCGGGAAGTCACTGT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | SREBP-2 Forward, GCGTTCTGGAGACCATGGA; Reverse, ACAAAGTTGCTCTGAAAACAAATCA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | HMGCR Forward, CTTGTGGAATGCCTTGTGATTG; Reverse, AGCCGAAGCAGCACATGAT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | Insig-1 Forward, TCACAGTGACTGAGCTTCAGCA; Reverse, TCATCTTCATCACACCCAGGAC | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | Insig-2a Forward, CCCTCAATGAATGTACTGAAGGATT; Reverse, TGTGAAGTGAAGCAGACCAATGT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | Insig-2b Forward, CCGGGCAGAGCTCAGGAT; Reverse, GAAGCAGACCAATGTTTCAATGG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | SCAP Forward, ATTTGCTCACCGTGGAGATGTT; Reverse, GAAGTCATCCAGGCCACTACTAATG | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | HMGCS Forward, GCCGTGAACTGGGTCGAA; Reverse, GCATATATAGCAATGTCTCCTGCAA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | FPPS Forward, ATGGAGATGGGCGAGTTCTTC; Reverse, CCGACCTTTCCCGTCACA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | SqS Forward, CCAACTCAATGGGTCTGTTCCT; Reverse, TGGCTTAGCAAAGTCTTCCAACT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | LDLR Forward, AGGCTGTGGGCTCCATAGG; Reverse, TGCGGTCCAGGGTCATCT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | PCSK9 Forward, CAGGCGGCCAGTGTCTATG; Reverse, GCTCCTTGATTTTGCATTCCA | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | ACS Forward, GCTGCCGACGGGATCAG; Reverse, TCCAGACACATTGAGCATGTCAT | Integrated DNA Technologies | N/A | |
| Sequence-based reagent (qRT-PCR) | ACC1 Forward, TGGACAGACTGATCGCAGAGAAAG; Reverse, TGGAGAGCCCCACACACA | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | FAS Forward, GCTGCGGAAACTTCAGGAAAT; Reverse, AGAGACGTGTCACTCCTGGACTT | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | SCD1 Forward, CCGGAGACCCCTTAGATCGA; Reverse, TAGCCTGTAAAAGATTTCTGCAAACC | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | GPAT Forward, CAACACCATCCCCGACATC; Reverse, GTGACCTTCGATTATGCGATCA | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | LXRα Forward, TCTGGAGACGTCACGGAGGTA; Reverse, CCCGGTTGTAACTGAAGTCCTT | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | ABCG5 Forward, TGGATCCAACACCTCTATGCTAAA; Reverse, GGCAGGTTTTCTCGATGAACTG | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | ABCG8 Forward, TGCCCACCTTCCACATGTC; Reverse, ATGAAGCCGGCAGTAAGGTAGA | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | GGPS Forward, CGTCTACTTCCTTGGACTGGAAA; Reverse, AGCTGGCGTGTGAAAAGCTT | Integrated DNA Technologies | N/A | |
| Sequence- based reagent (qRT-PCR) | Cyclophilin Forward, TGGAGAGCACCAAGACAGACA; Reverse, TGCCGGAGTCGACAATGAT | Integrated DNA Technologies | N/A | |
| Commercial assay or kit | TaqMan Reverse Transcription | Applied Biosystems | Cat#N8080234 | |
| Commercial assay or kit | Power SYBR Green PCR Master Mix | Applied Biosystems | Cat#4367659 | |
| Commercial assay or kit | Cholesterol/ Cholesterol Ester Assay Kit - Quantitation | Abcam | Cat#ab65359 | |
| Chemical compound, drug | Cholesterol | Bio-Serv; | Cat#5180; | |
| Chemical compound, drug | Sigma-Aldrich | Cat#C8667 | ||
| Chemical compound, drug | Coenzyme Q-10 | Cerilliant | Cat#V-060 | |
| Chemical compound, drug | Geranylgeraniol | Sigma-Aldrich | Cat#G3278 | |
| Chemical compound, drug | Geranylgeranyl pyrophosphate | Cayman Chemical Company | Cat#63330 | |
| Chemical compound, drug | Lovastatin | Abblis Chemicals LLC, Houston, TX | Cat#AB1004848 | |
| Chemical compound, drug | Menaquinone-4 | Sigma-Aldrich | Cat#809896 | |
| Chemical compound, drug | Cerilliant | Cat#V-031 | ||
| Chemical compound, drug | Menaquinone-7 | Cerilliant | Cat#V-044 | |
| Chemical compound, drug | Phylloquinone (Vitamin K1) | Cerilliant | Cat#V-030 | |
| Chemical compound, drug | 25-Hydroxycholesterol | Avanti Polar Lipids | Cat#700019P | |
| Software, algorithm | Image Studio v5.0 | LiCor Biosciences | ||
| Software, algorithm | Image J (Fiji) | NIH |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44396.016





