Vps8 overexpression inhibits HOPS-dependent trafficking routes by outcompeting Vps41/Lt
Figures
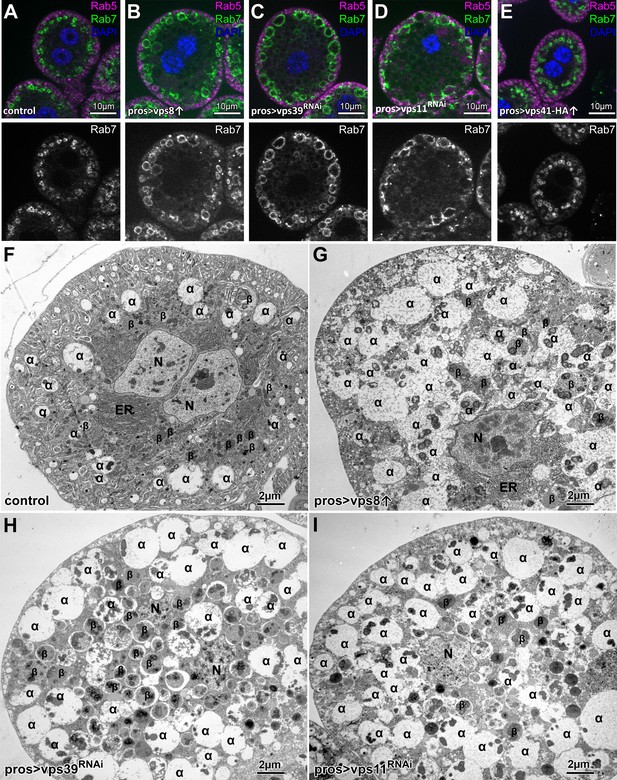
Overexpression of Vps8, but not Vps41 impairs endosome–lysosome fusion in Drosophila nephrocytes.
(A–E) Rab7+ late, but not Rab5+ early endosomes are enlarged in Vps8 overexpressing garland nephrocytes (B) similar to vps39 (C) or vps11 (D) RNAi cells. The size and location of endosomes are similar in control (A) and Vps41-9xHA overexpressing cells (E). (F–I) Ultrastructural analyses of nephrocytes reveal that compared to controls (F), late endosomes (also known as α-vacuoles – indicated by α in the panels) are enlarged and contain multiple dense cores in Vps8 overexpressing cells (G), similar to vps39 (H) or vps11 (I) RNAi cells. β: β-vacuoles/lysosomes, ER: endoplasmic reticulum, N: nucleus.
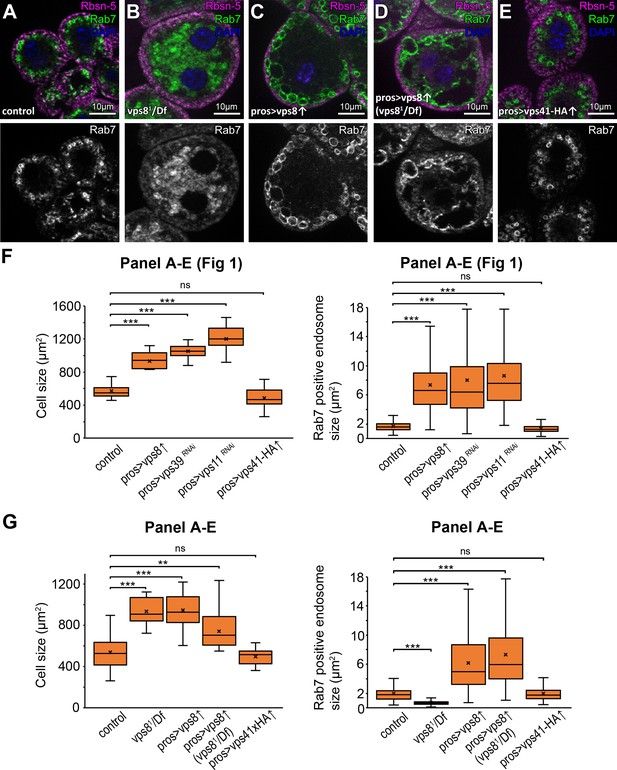
additional nephrocyte data and the quantification of fluorescent data.
(A–E) Rab7+ late, but not Rbsn-5+ early endosomes are enlarged in Vps8 overexpressing garland nephrocytes (C), which is observed both on a wild type (C) or vps8 mutant (D) genetic background compared to control (A) or vps8 mutant cells (B), respectively, meaning that the phenotype of Vps8 overexpression prevails over the mutant phenotype (cf. panels B-D). The size and distribution of endosomes are similar to controls in Vps41-9xHA overexpressing cells (E). (F–G): Statistical analysis of cell size and Rab7+ late endosome size data from panels A-E and Figure 1, panels A-E. The median and the average are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, **: p≤0.01, ***: p<0.001.
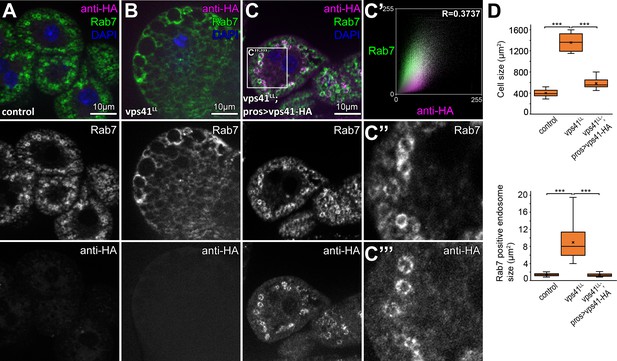
Overexpression of Vps41-9xHA rescues the vps41 mutant phenotype and the protein localizes to late endosomes.
Compared to controls (A), vps41 mutant nephrocytes are swollen and contain enlarged Rab7+ late endosomes (B). Overexpression of Vps41-9xHA in the nephrocytes of vps41 mutant animals rescues the mutant phenotype as both cell and late endosome size are normalized. Note that Vps41-9xHA colocalizes with endogenous Rab7 at the rim of endosomes (C). Averaged scatter plot (generated from 12 cells) shows the intensity correlation profile of Vps41-9xHA (labeled with anti-HA) with endogenous Rab7 (C’). Pearson correlation coefficient shown at the top right corner indicates substantial colocalization. (D) Quantification of data from A–C. The median and the average are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ***: p<0.001.
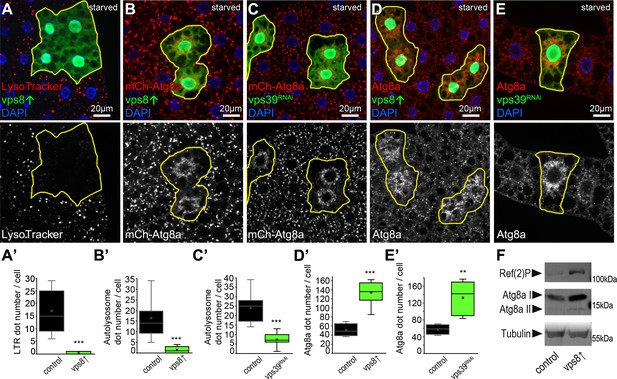
Overexpression of Vps8 inhibits autolysosome formation in starved fat cells, similar to HOPS (vps39) RNAi.
(A) Overexpression of Vps8 in GFP+ fat cells impairs LysoTracker Red dot formation compared with neighboring non-GFP control cells, similarly to HOPS loss-of-function (Takáts et al., 2014). (B and C) Both Vps8 overexpression (B) and vps39 RNAi (C) impairs the proper formation of 3xmCherry-Atg8a+ autophagic vesicles in GFP+ cells: these red dots are bigger and brighter in surrounding control cells and GFP+ cells contain smaller and fainter dots (likely autophagosomes) in both cases. (D and E) The number of endogenous Atg8a puncta (autophagosomes) increases in GFP+ Vps8 overexpressing (D) or vps39 RNAi (E) cells compared to GFP-negative control cells. (A’–E’) Quantification of data from panels A–E. The median and the average are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, and significant differences are indicated. (F) Western blot from well-fed adult lysates shows the obvious accumulation of Ref(2)P/p62 and both unlipidated (I) and autophagosome-associated, lipidated (II) forms of Atg8a in animals systemically overexpressing Vps8.
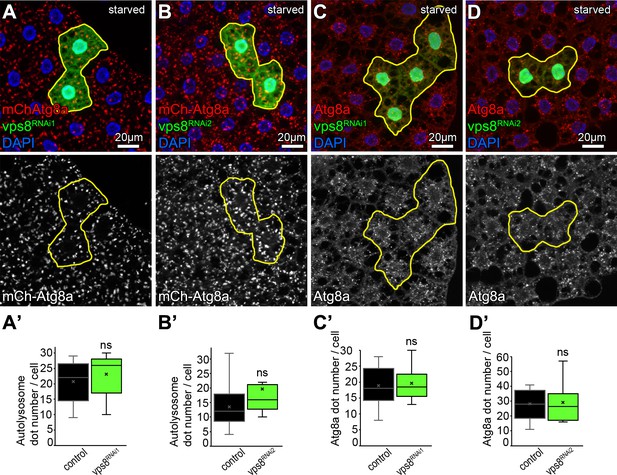
Vps8 (miniCORVET) is dispensable for autophagy in starved fat cells.
The size and distribution of both 3xmCherry-Atg8a autophagic vesicles (A,B) and endogenous Atg8a labeled autophagosomes (C,D) in GFP-marked vps8 RNAi cells are similar to those in GFP-negative control cells. (A’–D’) Quantification of data from panels A–D. The median and the average are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ns: not significant.
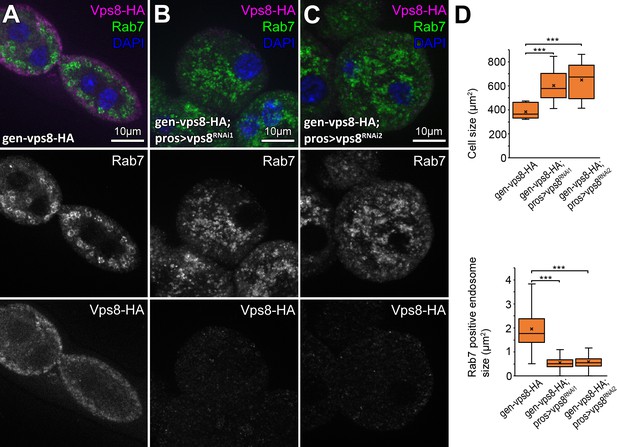
Validation of knockdown efficiencies for Vps8 RNAi lines used in this study.
Compared to gen-Vps8-9xHA controls (A), vps8 RNAi nephrocytes (B,C, note that these are also on a gen-Vps8-9xHA background) are swollen and filled with small Rab7+ late endosomes. As expected, the HA signal disappears in RNAi cells, indicating efficient knock-down. (D) Quantification of data from panels A–C. The median and the average are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ***: p<0.001.
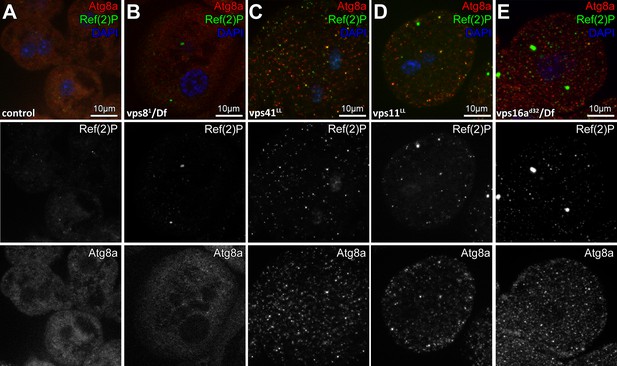
Analysis of basal autophagy in nephrocytes.
(A–E) Anti-Atg8a and anti-Ref(2)P/p62 stainings visualize autophagosomes and protein aggregates, respectively. Very few structures positive for either Atg8a or Ref(2)P are seen in control (A) and vps8 null mutant (B) nephrocytes, unlike vps41 (C), vps11 (D) and vps16a (E) mutants, which all accumulate both Atg8a+ autophagosomes and Ref(2)p+ protein aggregates.
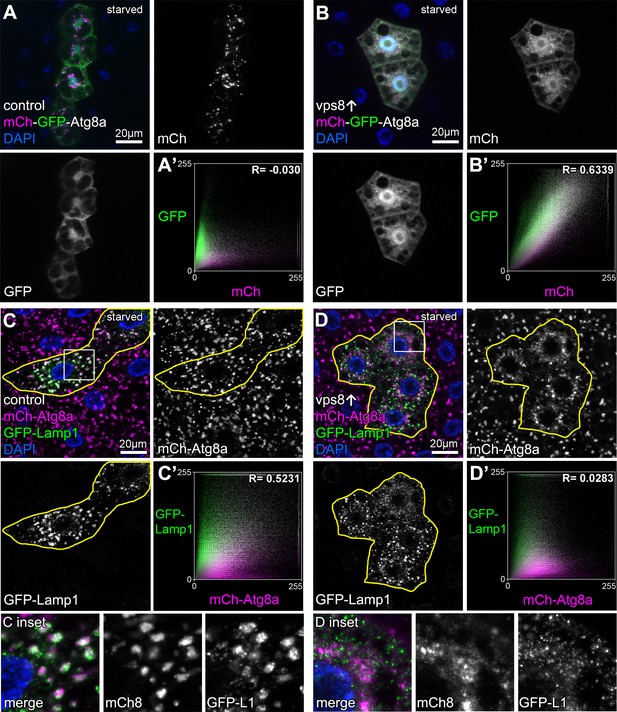
Overexpression of Vps8 inhibits autophagosome clearance in starved fat cells.
Tandem mCherry-GFP-Atg8a experiments demonstrates autophagic flux in starved control cells, as GFP is quenched in lysosomes while punctate mCherry signal is retained (A). In contrast, GFP also remains fluorescent and colocalizes with mCherry in Vps8 overexpressing cells (B). (A’ and B’) Averaged scatter plots (generated from 16 control and 13 UAS-Vps8 cells) show the intensity correlation profile of GFP with mCherry. Pearson correlation coefficients shown at the top of the panels indicate that in controls (A’) there is hardly any colocalization due to the autolysosomal quenching of GFP, whilst its signal persists in Vps8 overexpressing cells (B’), indicating the impairment of autophagic flux. (C and D) Large GFP-Lamp1 and 3xmCherry-Atg8a dots overlap in starved control cells (C) indicating proper autolysosome formation, while there is dramatically decreased overlap in Vps8 overexpressing cells (D). Note that insets show the boxed regions enlarged from C and D, respectively. (C’ and D’) Averaged scatter plots (generated from 16 control and 20 UAS-Vps8 cells) show the intensity correlation profile of GFP-Lamp1 with 3xmCherry-Atg8a. Pearson correlation coefficients shown at the top of the panels indicate that the two signals colocalize in controls (C’), unlike in Vps8 overexpressing cells (D’), pointing to the impairment of autophagosome-lysosome fusion.
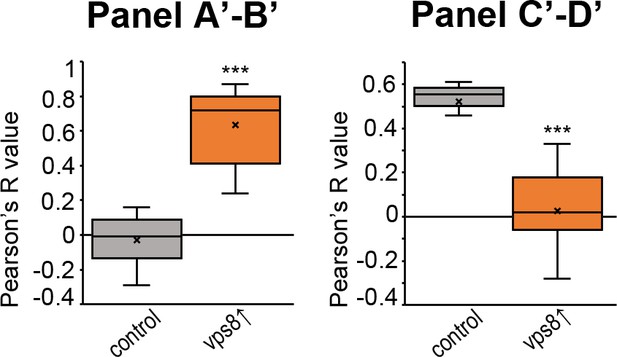
Quantification of data from Figure 3, panels A’-D’.
The median and the average of data are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ***: p<0.001. .
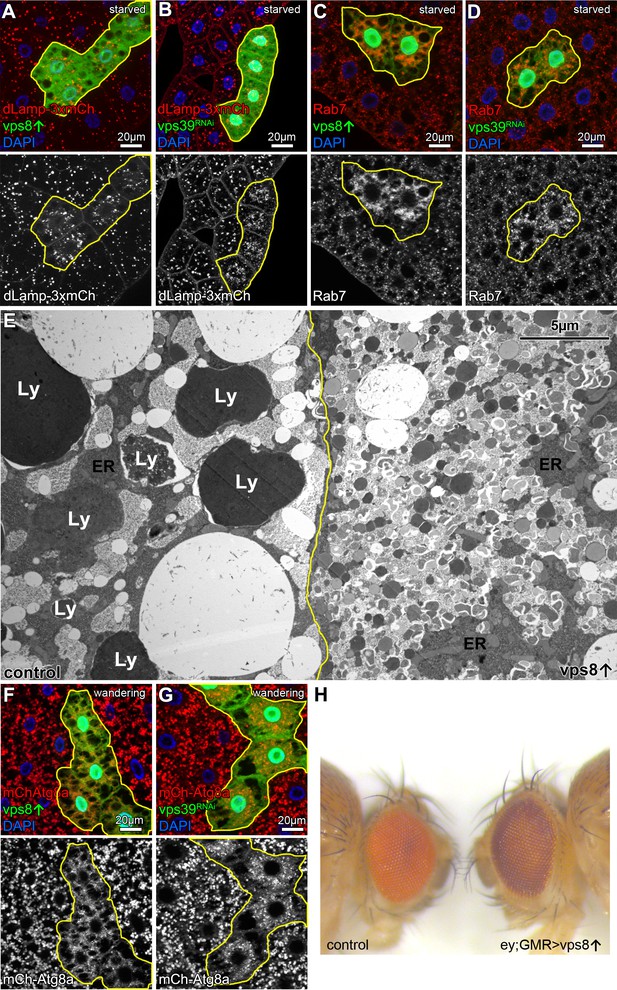
Overexpression of Vps8 impairs lysosome and LRO formation.
(A and B) The size of lysosomes (marked by dLamp-3xmCherry) decreases, but their number increases in GFP marked starved fat cells overexpressing Vps8 (A) or vps39 RNAi compared to neighboring GFP-negative control cells. (C and D) Starved, GFP+ fat cells that are overexpressing Vps8 (C) or vps39 RNAi (D) are full of small Rab7+ vesicles, while the surrounding control cells contain fewer Rab7+ dots. (E) Developmental autophagy proceeds normally in a control fat cell (left) of a wandering staged larva, indicated by the presence of large lysosomes containing electron-dense, protein-rich material (Ly). In contrast, the Vps8 overexpressing cell (right) is devoid of such structures and is filled with double-membrane autophagosomes and small lysosomes. ER: endoplasmic reticulum. (F and G) Large 3xmCherry-Atg8a+ autolysosomes are absent from GFP marked Vps8 overexpressing cells compared to neighboring control cells (F), similar to vps39 RNAi cells (G) in the fat tissue of wandering larvae. (H) The bright red color of the compound eye of a control fly (left) becomes darker upon eye specific overexpression of Vps8 (right). Note that Vps8 overexpression also eliminates the pseudopupil, which is seen as a dark spot in the middle of the control eye (left).
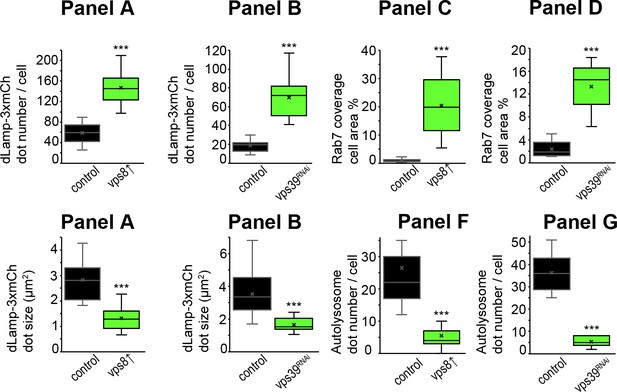
Quantification of data from Figure 4, panels A–D, F and G.
The median and the average of data are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, and significant differences are indicated in panels.
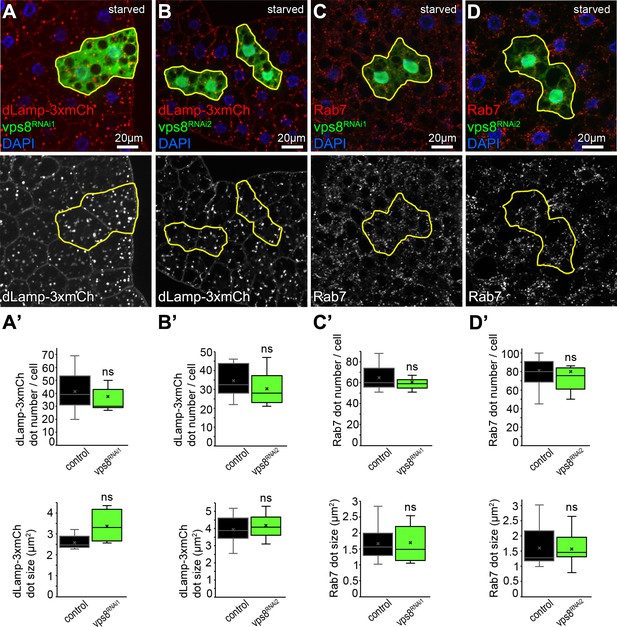
Vps8 (miniCORVET) is dispensable for lysosome formation in starved fat cells.
The patterns of dLamp-3xmCherry reporter (A,B) or endogenous Rab7 staining (C,D) in GFP marked vps8 RNAi starved fat cells are similar to surrounding control cells. (A’–D’) Quantification of data from panels A–D. The median and the average of data are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ns: not significant.
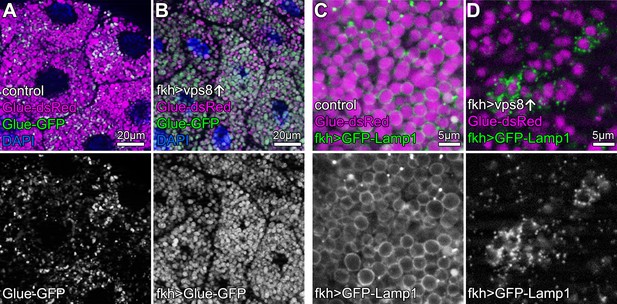
Overexpression of Vps8 impairs crinophagy.
(A and B) Crinophagy in salivary glands at puparium formation. Glue granule degradation proceeds normally in control cells (A) based on the lysosomal quenching of Glue-GFP, with Glue-dsRed+, Glue-GFP- structures representing crinosomes. (B) Glue-GFP signal persists upon salivary gland-specific overexpression of Vps8, indicating an impairment of crinophagic flux. (C,D) Secretory granule-lysosome fusion is inhibited upon Vps8 overexpression. 2 hr before pupariation, lysosomes (marked by Lamp1-GFP) fuse with secretory granules as seen by the formation of Lamp1-GFP ‘rings’ at the rim of Glue-dsRed vesicles in controls (C). In contrast, Lamp1-GFP lysosomes accumulate and no Lamp1-GFP rings are present in Vps8 overexpressing cells (D), indicating impaired crinosome formation due to the lack of secretory granule-lysosome fusion.
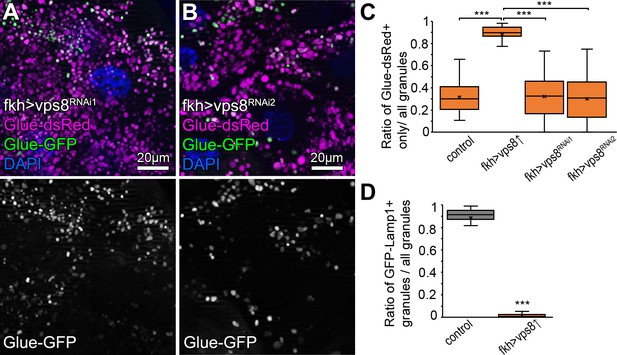
Vps8 (miniCORVET) is dispensable for crinophagy.
(A and B) Crinophagy proceeds normally in vps8 RNAi salivary glands, as expression of neither RNAi construct prevents the quenching of Glue-GFP. (C) Quantification of data from Figure 6, panels A, B and from panels A and B of this figure. The median and the average of data are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ***: p<0.001. (D) Quantification of data from Figure 6, panels C, D. The median and the average of data are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, ***: p<0.001.
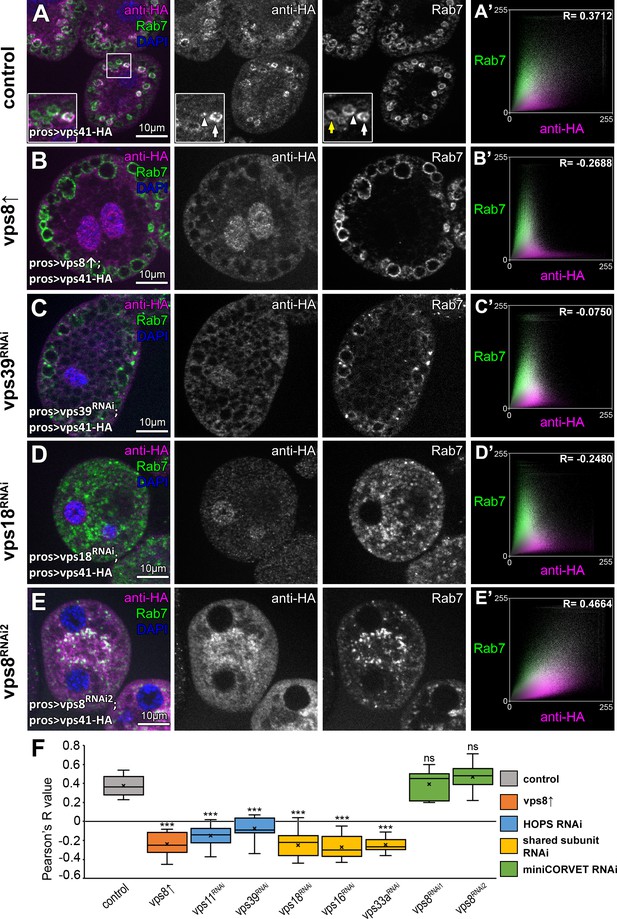
The late endosomal localization of Vps41 is lost upon Vps8 overexpression or loss of any other HOPS subunit.
All images show nephrocytes expressing Vps41-9xHA in different genetic backgrounds, stained with anti-HA (magenta) and anti-Rab7 (green). (A–E) Vps41-9xHA is recruited to a subset of Rab7+ endosomes (white arrow) in control nephrocytes (A). White arrowhead points to a patch of Vps41-9xHA on a Rab7+ endosome. Yellow arrow points to a Rab7 endosome with no Vps41-9xHA signal. Upon Vps8 overexpression (B) or HOPS-specific vps39 RNAi (C), nephrocytes become swollen and contain enlarged Rab7 endosomes. Strikingly, no Vps41-9xHA is detected on these vesicles. Moreover, no Vps41-9xHA can be detected on Rab7+ vesicles in vps18 class C RNAi cells (D), and these late endosomes are fragmented due to the simultaneous loss of miniCORVET and HOPS. (E) Late endosomes are also fragmented and colocalize with Vps41-9xHA in vps8 miniCORVET RNAi cells. (A’– E’) Averaged scatter plots (generated from 18 control and 16 Vps8↑, 16 vps39 RNAi, 15 vps18 RNAi and 14 vps8 RNAi cells) show the intensity correlation profile of Vps41-9xHA (labeled with anti-HA) with endogenous Rab7. Pearson correlation coefficients shown at the top of panels A’ and E’ indicate substantial colocalization, which is lost in Vps8 overexpressing, vps39 and vps18 RNAi cells (B’–D’). Quantification of colocalization data, including data from Figure 6—figure supplement 1. The median and the average of Pearson correlation coefficients are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, and significant differences are indicated in panels. ns: not significant difference, ***: p<0.001.
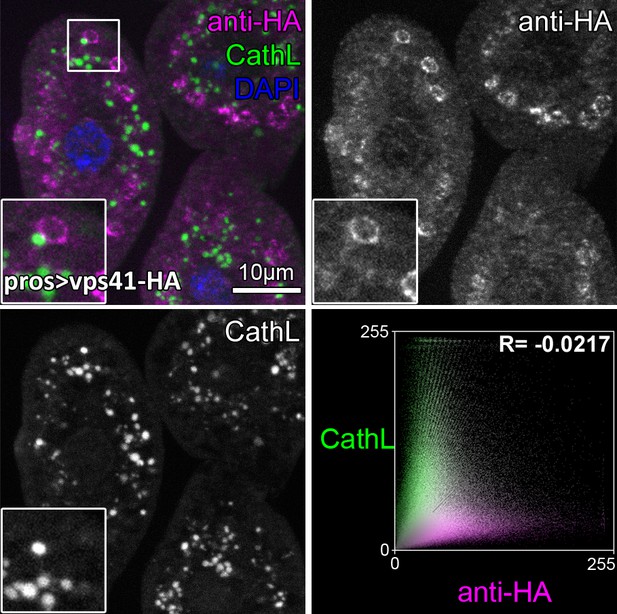
Vps41-9xHA is absent from Cathepsin L-containing lysosomes.
No overlap is seen between Vps41-9xHA endosomes and CathL-containing lysosomes in garland nephrocytes. Note that adjacent but separate endosomes and lysosomes are often seen (inset). Averaged scatter plot (generated from 18 cells) shows the intensity correlation profile of Vps41-9xHA (labeled with anti-HA) with CathL. The negative Pearson correlation coefficient shown at the top of the plot indicates the lack of colocalization.
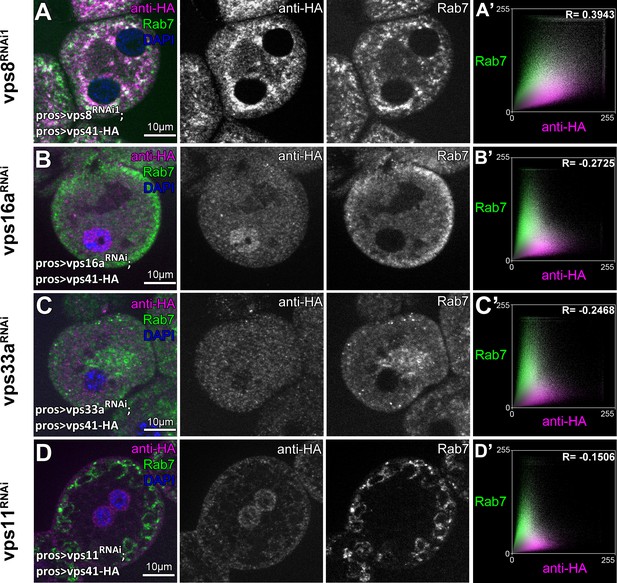
Additional Vps41-9xHA data.
All images show nephrocytes expressing Vps41-9xHA on different genetic backgrounds, stained with anti-HA (magenta) and anti-Rab7 (green). (A) Vps41-9xHA sustains its late endosomal localization in garland nephrocytes undergoing vps8 RNAi. (B–D) Vps41-9xHA is dispersed throughout the cytoplasm in cells undergoing vps16a (B), vps33a (C) or vps11 (D) RNAi. Note that late endosomes are small and fragmented in miniCORVET (A) or class C (B,C) RNAi cells, while they are enlarged in HOPS (D) depleted cells, as expected. Averaged scatter plots (generated from 17 vps8 RNAi1, 16 vps16a RNAi, 16 vps33a RNAi and 17 vps11 RNAi cells) show the intensity correlation profile of Vps41-9xHA (labeled with anti-HA) with endogenous Rab7. Pearson correlation coefficient shown at the top of panel A’ indicate substantial colocalization, which is lost in vps16a, vps33a and vps11 RNAi cells.
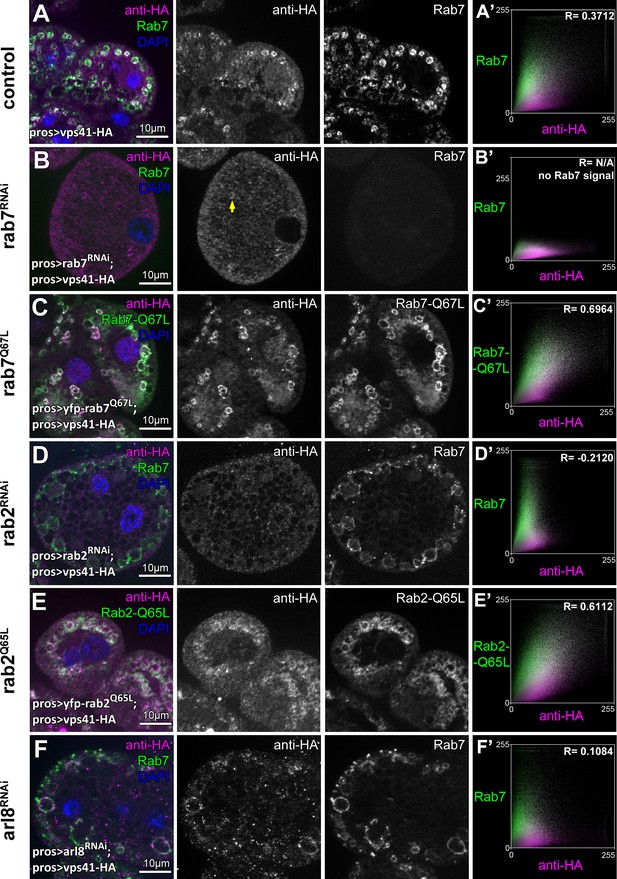
The late endosomal recruitment of Vps41 requires Rab2 and Rab7 but not Arl8.
All images show nephrocytes expressing Vps41-9xHA on different genetic backgrounds, stained with anti-HA (magenta) and anti-Rab7 or YFP (green). (A) A subset of Rab7 endosomes are positive for Vps41-9xHA. (B) Vps41-9xHA is mostly dispersed throughout the cytoplasm in cells undergoing rab7 RNAi. Interestingly, occasional Vps41-9xHA rings are still observed (arrow). (C) The colocalization of Vps41-9xHA with Rab7 increases on enlarged endosomes in cells expressing the constitutively active mutant form of Rab7. (D) Knockdown of rab2 results in the swelling of nephrocytes that contain enlarged Rab7 endosomes with no Vps41-9xHA signal. (E) Vps41-9xHA is recruited to vesicles positive for GTP-locked Rab2. (F) The overlap of Vps41-9xHA with enlarged Rab7 vesicles decreases, but it is still obvious in cells undergoing arl8 RNAi. Averaged scatter plots (generated from 13 rab7 RNAi, 11 Rab-Q67L, 15 rab2 RNAi, 11 Rab2-Q65L and 12 arl8 RNAi cells) show the intensity correlation profiles of Vps41-9xHA (labeled with anti-HA) with Rab7 (B’, D’. (F’) or Rab7-Q67L (C’) or Rab2-Q65L (E’). Pearson correlation coefficient was not determined in B’ as no punctate Rab7 signal was detected. Pearson correlation coefficients shown at the top of panels A’, B’ and D’ indicate substantial colocalization, no overlap in panel C’, and still detectable colocalization in panel E’. Please note that since the experiments shown here and in Figure 6 were carried out in parallel, the same averaged plot and Pearson correlation coefficient value is shown for controls (A’) in both Figures.
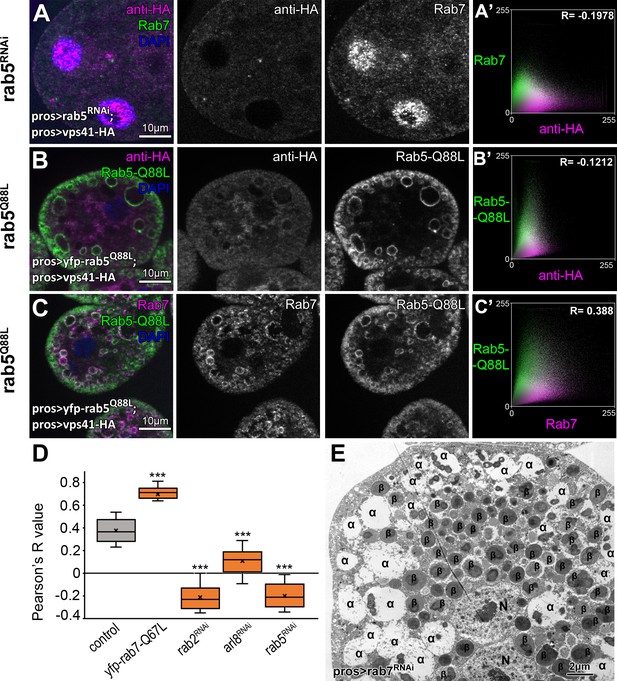
Additional Vps41-9xHA data.
(A) Vps41-9xHA looses its endosomal localization in garland nephrocytes undergoing rab5 RNAi as these cells lack Rab7+ endosomes. Vps41-9xHA is dispersed throughout the cytoplasm in cells expressing GTP-locked Rab5-Q88L (B), even though some of these Rab5-Q88L positive rings are Rab7 positive (C), indicating the formation of hybrid early and late endosomes. (A’,B’) Averaged scatter plots (generated from 13 rab5 RNAi, and 17 Rab5-Q88L cells from) show the intensity correlation profile of Vps41-9xHA (labeled with anti-HA) with endogenous Rab7 (A’) or Rab5-Q88L (B). Pearson correlation coefficient shown at the top of panels A’ and B’ indicate no colocalization. C’ Averaged scatter plot (generated from 10 Rab5-Q88L cells from) shows the intensity correlation profile of Rab7 with Rab5-Q88L. Pearson correlation coefficient shown at the top of panel C’ indicate substantial colocalization. (D) Quantification of Vps41-9xHA and Rab7 colocalization data from panel A and from Figure 7A,C,D,F. The median and the average of the measured Pearson correlation coefficients are indicated as a horizontal black line and x within the boxes, respectively. Bars show the upper and lower quartiles, and significant differences are indicated in panels. ***: p<0.001 (E) Ultrastructural analysis of nephrocytes undergoing rab7 RNAi revealed the presence of enlarged late endosomes (α-vacuoles – indicated by α in the panels) and aberrant lysosomes (β: β-vacuoles), please compare to data in Figure 1 panels F-I. N: nucleus.
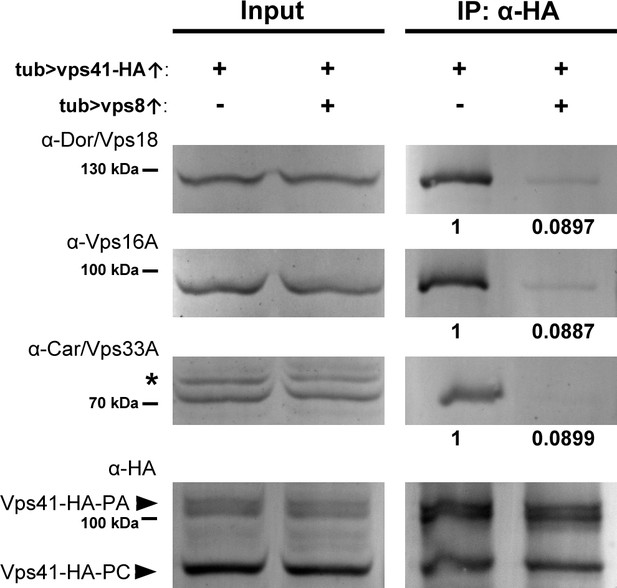
Vps8 overexpression strongly decreases Vps41 binding to the class C core.
Endogenous Dor/Vps18, Vps16A and Car/Vps33A class C proteins coprecipitate with Vps41-9xHA isoforms (marked by PA and PC, respectively) from larval lysates. The amount of coprecipitated class C proteins decreases to less than 9% in lysates of larvae overexpressing Vps8. Asterisk marks a nonspecific band. Numbers under the bands in IP lanes refer to class C protein levels normalized to Vps41-9xHA PA+PC based on densitometry.
Tables
| Reagent type (species) or resources | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | UAS-Vps8 | This study. | ||
| Genetic reagent (D. melanogaster) | UAS-Vps41-9xHA | This study. | ||
| Genetic reagent (D. melanogaster) | vps81: vps8[1] | (Lőrincz et al., 2016a) | FBal0320420 | |
| Genetic reagent (D. melanogaster) | ltll: lt[LL07138] | (Lőrincz et al., 2016a) | FBal0320422 | obtained from: DGGR |
| Genetic reagent (D. melanogaster) | vps11LL: vps11[LL06553] | (Takáts et al., 2014) | FBal0296360 | obtained from: DGGR |
| Genetic reagent (D. melanogaster) | vps16ad32 | (Takáts et al., 2014) | FBal0296357 | |
| Genetic reagent (D. melanogaster) | pros-Gal4 | FBtp0129317 | obtained from: Bruce Edgar | |
| Genetic reagent (D. melanogaster) | tub-Gal4 | FBtp0020111 | obtained from: BDSC | |
| Genetic reagent (D. melanogaster) | gen-vps8-HA | (Lőrincz et al., 2016a) | FBal0320419 | |
| Genetic reagent (D. melanogaster) | vps39RNAi: vps39 [GD12152] | (Lőrincz et al., 2016a) | FBal0205346 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps11RNAi: vps11 [KK102566] | (Lőrincz et al., 2016a) | FBal0231866 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps16aRNAi: vps16 [GD13782] | (Lőrincz et al., 2016a) | FBal0208987 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps33aRNAi: car [GD1397] | (Lőrincz et al., 2016a) | FBal0209225 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps18RNAi: dor [KK102176] | (Lőrincz et al., 2016a) | FBal0231650 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | rab7RNAi: rab7 [GD40337] | (Lőrincz et al., 2016a) | FBal0208211 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps8RNAi1: vps8 [KK100319] | This study. | FBal0230675 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | vps8RNAi2: vps8 [10144 R-1] | This study. | FBal0270342 | obtained from: NIG-Fly |
| Genetic reagent (D. melanogaster) | rab2RNAi: rab2 [GD34767] | (Lőrincz et al., 2017b) | FBal0208203 | obtained from: VDRC |
| Genetic reagent (D. melanogaster) | arl8RNAi: arl8 [7891 R-2] | (Boda et al., 2019) | FBal0275763 | obtained from: NIG-Fly |
| Genetic reagent (D. melanogaster) | rab5RNAi: rab5 [JF03335] | (Lőrincz et al., 2016a) | FBal0241752 | obtained from: BDSC |
| Genetic reagent (D. melanogaster) | UAS-YFP-Rab5-Q88L | FBal0215394 | obtained from: BDSC | |
| Genetic reagent (D. melanogaster) | UAS-YFP-Rab7-Q67L | FBal0215400 | obtained from: BDSC | |
| Genetic reagent (D. melanogaster) | UAS-Rab2-Q65L | FBal0215385 | obtained from: BDSC | |
| Genetic reagent (D. melanogaster) | dLamp-3xmCherry | (Hegedűs et al., 2016) | FBal0325101 | |
| Genetic reagent (D. melanogaster) | 3xmCherry-Atg8a | (Hegedűs et al., 2016) | FBal0325100 | |
| Genetic reagent (D. melanogaster) | UAS-Lamp1-GFP | (Pulipparacharuvil et al., 2005) | FBal0221465 | obtained from: Helmut Krämer |
| Genetic reagent (D. melanogaster) | fkh-Gal4 | (Csizmadia et al., 2018) | FBtp0013253 | |
| Genetic reagent (D. melanogaster) | Sgs3-DsRed | (Csizmadia et al., 2018) | FBal0268258 | |
| Genetic reagent (D. melanogaster) | Sgs3-GFP | (Csizmadia et al., 2018) | FBal0119388 | |
| recombinant DNA reagent | EST AT14809 | FBcl0024753 | obtained from: DGRC | |
| recombinant DNA reagent | EST LD33620 | FBcl0304050 | obtained from: DGRC | |
| Antibody | mouse monoclonal anti-Rab7 | DSHB: Rab7 (Riedel et al., 2016) | RRID:AB_2722471 | IHC: 1:10 |
| Antibody | rabbit polyclonal anti-CathL | Abcam: ab58991 | RRID:AB_940826 | IHC: 1:100 |
| Antibody | rat polyclonal anti-Atg8a | (Takáts et al., 2013) | IHC: 1:300 | |
| Antibody | rabbit polyclonal anti-Atg8a | (Takáts et al., 2013) | WB: 1:5000 | |
| Antibody | rabbit polyclonal anti-p62/Ref(2)p | (Pircs et al., 2012) | RRID:AB_2569199 | IHC: 1:1000 WB: 1:4000 |
| Antibody | mouse monoclonalanti-tubulin | DSHB: AA4.3 | RRID:AB_579793 | WB: 1:2000 |
| Antibody | rabbit polyclonal anti-Car/Vps33A | (Sevrioukov et al., 1999) | RRID:AB_2569524 | WB: 1:1000 |
| Antibody | rabbit polyclonal anti-Vps16A | (Pulipparacharuvil et al., 2005) | RRID:AB_2569229 | WB: 1:2000 |
| Antibody | rabbit polyclonal anti-Dor/Vps18 | (Pulipparacharuvil et al., 2005) | RRID:AB_2569230 | WB: 1:1000 |
| Antibody | rat monoclonal anti-HA | Roche: 3F10 | RRID:AB_2314622 | IHC: 1:80 WB: 1:2000 |
| Antibody | rabbit polyclonal anti-Rab5 | Abcam: ab31261 | RRID:AB_882240 | IHC: 1:100 |
| Antibody | chicken polyclonal anti-GFP | Invitrogen: A10262 | RRID:AB_2534023 | IHC: 1:1500 |
| Antibody | rabbit polyclonal anti-HA | Sigma-Aldrich: H6908 | RRID:AB_260070 | IHC: 1:100 |
| Antibody | rat polyclonal anti-Rbsn-5 | (Tanaka and Nakamura, 2008) | RRID:AB_2569807 | IHC: 1:1000 |
| Antibody | mouse monoclonal anti-HA-Agarose | Sigma-Aldrich: A2095 | RRID:AB_257974 | |
| Chemical compound, drug | LysoTracker Red | ThermoFisher Scientific: L7528 | 1:1000 | |
| Software, algorithm | SPSS 17 | IBM | RRID:SCR_002865 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 |
Additional files
-
Supplementary file 1
Genotype of animals used in this study.
- https://doi.org/10.7554/eLife.45631.022
-
Supplementary file 2
Additional table showing statistical tests, N and p-values.
- https://doi.org/10.7554/eLife.45631.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45631.024





