Metabolic and non-metabolic liver zonation is established non-synchronously and requires sinusoidal Wnts
Figures
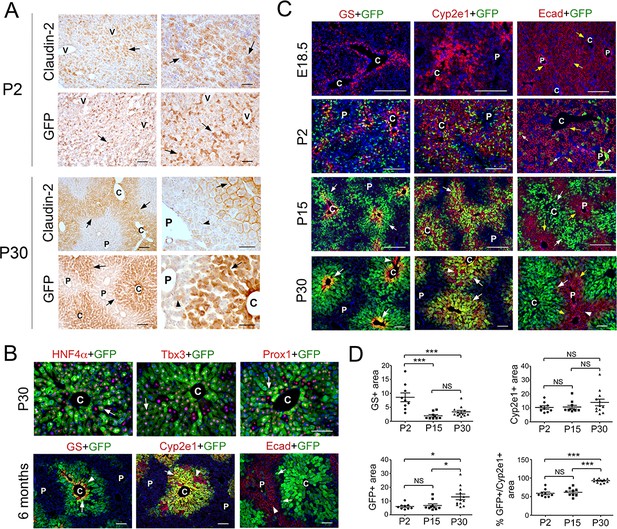
Claudin-2 expression becomes zonated during liver maturation.
(A) Immunohistochemistry analysis of claudin-2 and claudin-2/GFP expression showing identical distribution of these proteins in hepatocytes (arrows) at P2 and P30. Few hepatocytes near the portal veins (arrowheads) also express low claudin-2 or claudin-2/GFP at P30. Wildtype specimens were used for claudin-2 analysis and Cldn2-EGFP specimens for claudin-2/GFP analysis. Scale bars: 50 μm (right); and 100 μm (left). (B) Double-immunofluorescence analysis showing co-expression of claudin-2/GFP (arrows) with the hepatocyte transcription factors HNF4α, Tbx3 and Prox1 around the central veins in P30 Cldn2-EGFP livers. Scale bars: 100 μm. (C) Double-immunofluorescence results show lack of claudin-2/GFP hepatic expression late in gestation (E18.5), numerous claudin-2/GFP+ hepatocytes (arrows) distributed throughout the liver after birth (P2), and high claudin-2/GFP expression (arrows) restricted to Zone 3 (GS+) and Zone 2/3 (Cyp2e1+) hepatocytes in juvenile (P15) and adult (P30) livers (a few pericentral/perivenous hepatocytes [arrowheads] do not express GFP at P30). E-cadherin expression (yellow arrows) is detected in all hepatocytes at E18.5 and P2, and is restricted to periportal hepatocytes that lack high claudin-2/GFP expression at P15-P30. Scale bars: 200 μm (E18.5, P2, P15) and 100 μm (P30). (D) Quantification of GS+, Cyp2e1+ and claudin-2/GFP+ areas and the relative abundance of claudin-2/GFP+ hepatocytes in Zone 2 in postnatal (P2), juvenile (P15) and adult (P30) livers. 3–4 representative fields from three individual livers of each genotype were used for quantification. Statistical difference was determined by one-way ANOVA with Bonferroni’s multiple comparisons test. NS, not significant (p>0.05), *p<0.05, ***p<0.001. (A–C) Each image represents 3–4 individual livers. (C), central vein. (P), portal vein. (V), vein. Related data can be found in Figure 1—figure supplements 1–3.
-
Figure 1—source data 1
Quantification of GS+, Cyp2e1+ and claudin-2/GFP+ areas and the relative abundance of claudin-2/GFP+ hepatocytes in Zone two in P2, P15 and P30 livers.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig1-data1-v2.xlsx
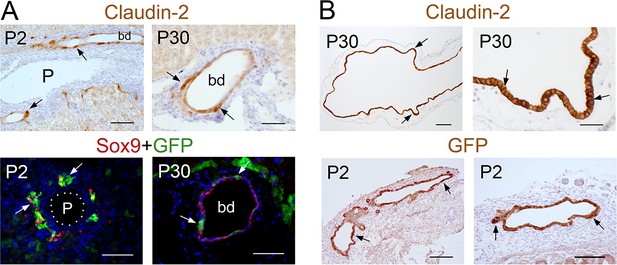
Claudin-2 and GFP proteins have identical expression in intrahepatic and extrahepatic biliary tissues of wildtype and Cldn2-EGFP mice.
(A) Immunostaining results showing claudin-2 expression (arrows) in cells lining the intrahepatic bile ducts of wildtype newborn (P2) and adult (P30) mice (top panels), and GFP (arrows) co-expression with the cholangiocyte marker Sox9 in intrahepatic bile ducts of P2 and P30 Cldn2-EGFP mice (bottom panels). Scale bars: 100 µm (left, top), 50 μm (right, top and bottom panels). (B) Claudin-2 is highly expressed (arrows) in the gall bladder epithelium (top panels) of an adult wildtype mouse. GFP (arrows) is similarly expressed in the gall bladder (bottom, left) and the common bile duct (bottom, right) of a Cldn2-EGFP P2 mouse. Scale bars: 200 µm (left, top), 25 μm (right, top), 100 µm (bottom panels).
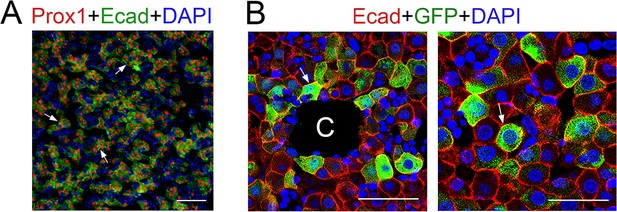
E-cadherin and claudin-2/GFP are expressed throughout the newborn liver.
(A) All hepatocytes express Prox1 (red) and E-cadherin (green, arrows) in P2 wildtype livers. (B) All Claudin-2/GFP+ hepatocytes (arrows) located in perivenous (left) and parenchymal (right) areas express E-cadherin in P2 Cldn2-EGFP livers. C = central vein. Scale bars: 50 μm (A), 25 µm (B).
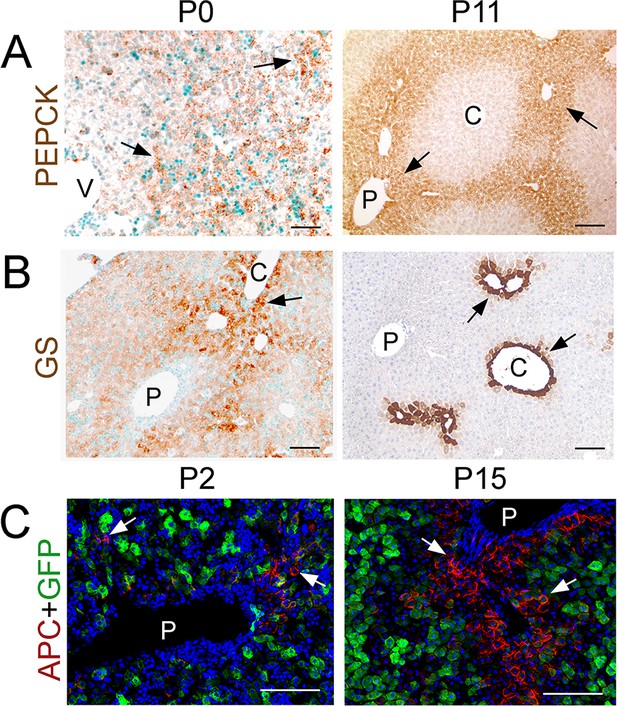
PEPCK expression is not restricted to periportal hepatocytes at birth.
(A) Immunohistochemistry analysis of PEPCK shows broad expression of this protein at birth (P0, arrows) and restricted periportal expression at P11 (arrows). (B) In contrast, GS shows restricted expression in pericentral hepatocytes at birth (P0, arrows) and P11 (arrows). (C) Immunofluorescence analysis of APC (red) shows expression of this protein in very few periportal hepatocytes at P2 (arrows) and in numerous periportal hepatocytes at P15 (arrows). Claudin-2/GFP hepatocytes express very low or no APC proteins at both P2 and P15. ‘C’ is central vein, ‘P’ is portal vein. Scale bars: 100 μm.
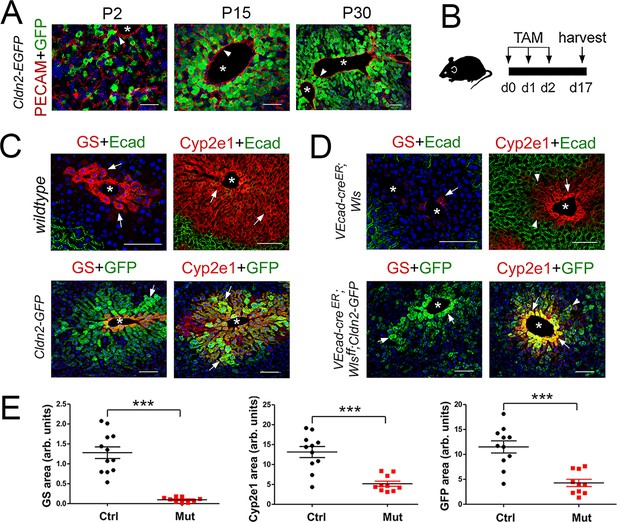
Wls deletion in HECs severely reduces the expression of claudin-2 and Cyp2e1 in perivenous hepatocytes in adult livers.
(A) Immunofluorescence results showing that pericentral claudin-2/GFP+ hepatocytes physically contact the central vein endothelium (PECAM+, arrowheads) in newborn (P2) juvenile (P15) and adult (P30) Cldn2-EGFP livers. (B) Schematic of tamoxifen administration and tissue harvesting. (C) Tamoxifen injection does not affect the zonated distribution of GS, Cyp2e1 and E-cadherin in wildtype livers (top, arrows), or claudin-2/GFP, GS and Cyp2e1 (bottom, arrows) in Cldn2-GFP livers. (D) Wls deletion in endothelial cells causes near depletion of GS+ hepatocytes (top left, arrows), decreases both Cyp2e1 (right panels, arrows) and claudin-2/GFP perivenous expression (bottom panels, arrows; arrowheads show distal perivenous hepatocytes expressing low GFP), and expands E-cadherin expression into the margins of Zone 2 (top right, arrowheads). (E) Quantification of Zone 3 (GS+) and Zone 2 (Cyp2e1+/GFP+) areas in adult livers with or without endothelial Wls. Statistical difference was determined by two-tailed unpaired Student’s t-test (***p<0.001, 3–4 representative fields from three individual livers of each genotype were used for quantification). Each image represents three individual livers. Asterisks indicate central vein lumens. Scale bars: 50 μm (A), 100 μm (C,D). Related data can be found in Figure 2—figure supplement 1.
-
Figure 2—source data 1
Quantification of Zone three and Zone two areas in adult livers with or without endothelial Wls.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig2-data1-v2.xlsx
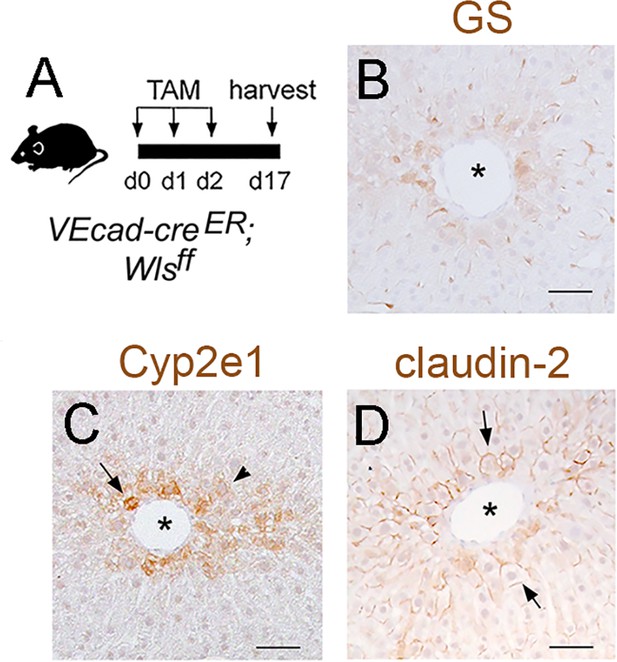
Wls deletion in adult HECs disrupts zonated protein expression in hepatocytes.
(A) Schematic of tamoxifen administration and tissue harvesting. (B–D) Immunohistochemistry results showing that Wls endothelial deletion reduces the expression of GS, Cyp2e1 and Claudin-2 proteins in perivenous (asterisks) hepatic areas. Each image represents 2–3 individual livers. Scale bars: 50 µm.
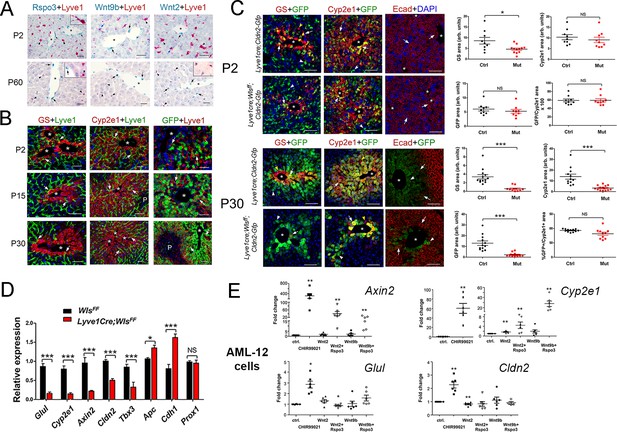
Lack of Wnt ligand secretion from LSECs impairs adult zonation maintenance.
(A) Double in situ hybridization for Rspo3 (green), Wnt9b (green) and Wnt2 (green) showing a few LSCEs (Lyve1+, red) expressing those transcripts (blue arrows and inset) in P2 livers. Only Wnt2 transcripts are detected in some LSECs (inset) in P60 livers. Arrows indicate central vein endothelial cells and arrowheads indicate LSECs. Scale bars: 25 μm. Each image is representative of 3 individual mice (n = 3). (B) Double-immunofluorescence results show that hepatic Zones 3 (GS+) and 2/3 (Cyp2e1+) are densely irrigated by the hepatic sinusoids (Lyve1+, arrows) in P2, P15 and P30 wildtype livers. The sinusoidal vasculature (arrows) is also in direct contact with claudin-2/GFP+ hepatocytes in P2, P15 and P30 Cldn2-EGFP livers. Scale bars: 50 μm. Each image is representative of 2–4 individual mice (n = 2–4). (C) P2, Quantitative double immunofluorescence results show that Zone 2 (Cyp2e1+, claudin-2/GFP+) is relatively unchanged and Zone 3 (GS+) is significantly reduced in Lyve1-Cre;Wlsf/f;Cldn2-GFP livers at P2. E-cadherin expression (arrows) is indistinguishable in P2 livers with or without endothelial Wls-deletion. P30, Similar quantitative results demonstrate that GS+ hepatocytes are nearly absent, Zone 2 (Cyp2e1+/GFP+) is significantly reduced and restricted to pericentral areas, and E-cadherin expression (arrows, arrowheads are GFP+ hepatocytes) is expanded towards the central veins in P30 Lyve1-Cre;Wlsf/f;Cldn2-GFP livers. 3–4 representative fields from three individual livers of each genotype were used for quantification. p values were determined by two-tailed unpaired Student’s t-test, NS, not significant (p>0.05), *p<0.05, ***p<0.001. Arrows indicate GFP-double positive hepatocytes, white arrowheads are GFP+ hepatocytes and yellow arrowheads are GFP– hepatocytes. Scale bars: 100 µm (D) Q-PCR results demonstrate reduced expression of Zone 2/3 transcripts, increased expression of Zone 1 transcripts, and normal levels of the hepatocyte transcript Prox1, in adult Lyve1-Cre;Wlsf/f livers (n = 3). p values were determined by two-way ANOVA, NS, not significant (p>0.05), *p<0.05, ***p<0.001. (E) Q-PCR results showing the effects of culturing AML-12 mouse hepatic cells with CHIR99021, Wnt2, Wntb9, or Wnt2/Wnt9b plus Rspo3 on Axin2, Cyp2e1, Glul and Cldn2 expression. p values from two-tailed unpaired Student’s t-test, *p<0.05, ***p<0.01; n = 6. (A–C) Asterisks indicate central vein lumens. Related data can be found in Figure 3—figure supplements 1–3.
-
Figure 3—source data 1
Quantification of GS+, Cyp2e1+ and claudin-2/GFP+ areas and the relative abundance of claudin-2/GFP+ hepatocytes in P2 and P30 Lyve1-Cre;Wlsf/f;Cldn2-GFP livers, and Quantification of Wnt/β-catenin target genes expression of P30 Lyve1-Cre;Wlsf/f;Cldn2-GFP livers.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig3-data1-v2.xlsx
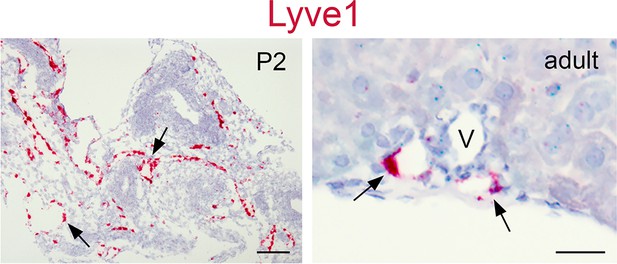
The Lyve-1 probe used for in situ hybridization in LSECs stains lymphatic endothelial cells.
RNA in situ hybridization showing Lyve-1 transcript expression in peritoneal lymphatic vessels (arrows, left) from a newborn mouse and intrahepatic periportal lymphatic vessels (arrows, right) from an adult mouse. V, blood vessel. Scale bars: 100 µm (left), 25 µm (right).
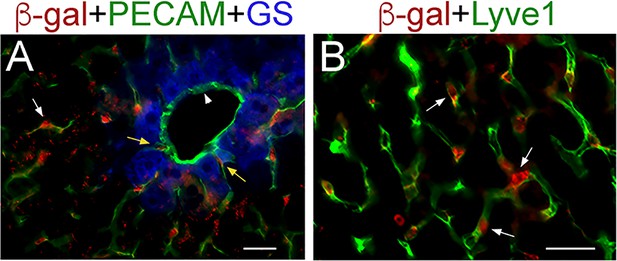
Lineage tracing results demonstrate selective β-gal expression in LSECs in Lyve1-Cre;ROSA-LacZ livers.
(A, B) Immunofluorescence results showing expression of the lineage tracer β-gal in PECAM+ endothelial cells connecting to the central vein (A, yellow arrows) and in parenchymal PECAM+/Lyve1+ LSECs (white arrows in (A and B). β-gal is not expressed in PECAM+ central vein endothelial cells (A, arrowhead; the central vein is surrounded by GS+ hepatocytes). Images are representative of 2 individual livers. Scale bars: 25 μm.
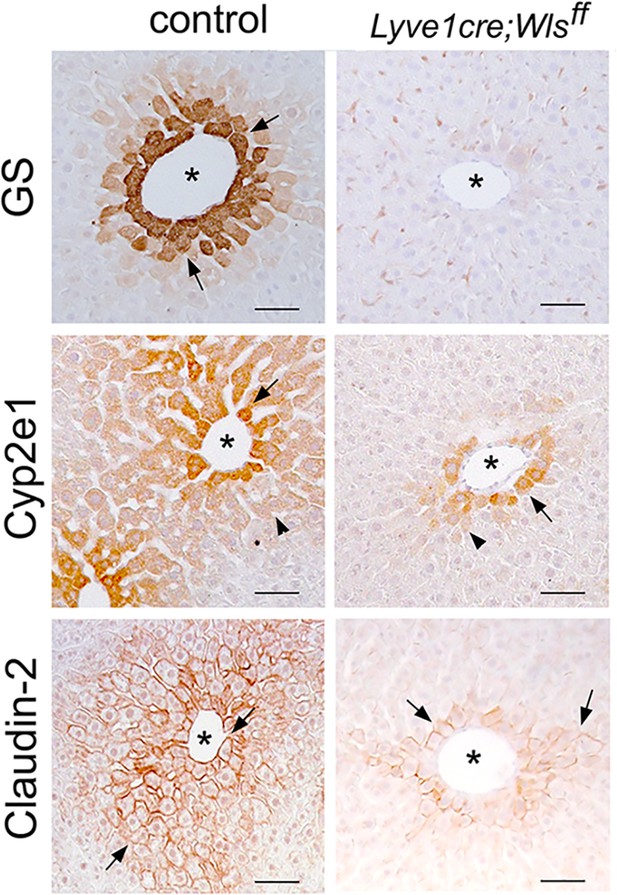
Wls deletion in the adult endothelium disrupts liver zonation.
Immunohistochemistry results showing reduced expression of GS, Cyp2e1 and Claudin-2 proteins in perivenous (asterisks) hepatic areas following sinusoidal (Lyve1-Cre) Wls deletion. Each image represents 2–3 individual livers. Scale bars: 50 µm.
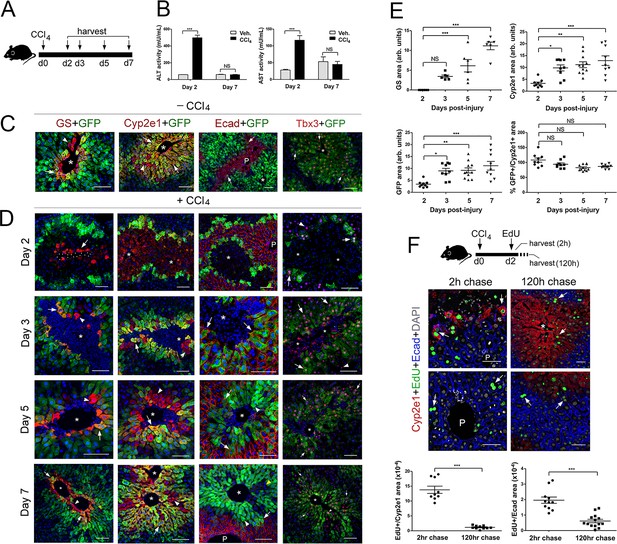
Proliferation and not de novo expression restore Claudin-2+ and Cyp2e1+ hepatocytes in the CCl4-injured liver.
(A) Experimental strategy for Cldn2-EGFP mice injected with CCl4. (B) ALT/AST serum levels demonstrate liver damage 2 days post-CCl4 (n = 3). p values were determined by two-way ANOVA, NS, not significant (p>0.05), ***p<0.001. (C) Expression of Zone 3 (GS+), Zone 2/3 (Cyp2e1+, claudin-2/GFP+) and Zone 1 (Ecad+) markers and Tbx3 in the corn oil-injected Cldn2-EGFP adult liver. (D) Spatial distribution of the markers indicated above during the recovery phase that follows a CCl4 bolus (see text for details). (Day 2, arrow, enucleated GS+ cell. Days 3–7, arrows, GFP double-positive hepatocytes; arrowheads, GFP– hepatocytes. Asterisks indicate central vein lumens.) (E) Quantification of GS+, Cyp2e1+ and claudin-2/GFP+ areas, and the GFP+/Cyp2e1+ ratio, 2–7 days post-CCl4. 3–4 representative fields from three individual livers dissected at the indicated time points were used for quantification. p values were determined by one-way ANOVA with Bonferroni’s multiple comparisons test, NS, not significant (p>0.05), *p<0.05, **p<0.01, ***p<0.001. (F) (Top) Schematic of EdU administration and tissue harvesting post-CCl4. (Middle) Immunofluorescence results show EdU incorporation (arrows) in Cyp2e1+ hepatocytes located in the Zone 2 remnant and the undamaged Zone 1 (Ecad+) 2 days post-CCl4. Both Cyp2e1+ hepatocytes (arrows, top) and Ecad+ hepatocytes located at the margins of restored Zone 2 (arrows, bottom) retain the EdU label after a 5 day chase. (Bottom) Quantification of EdU+ hepatocytes in Zone 2/3 and Zone 1. 3–4 representative fields from three individual livers dissected at the indicated time points were used for quantification. p values were determined by two-tailed unpaired Student’s t-test, ***p<0.001. (C,D,F) asterisks and dots indicate central vein lumens and images are from 3 to 4 individual livers. (B, F) n = 3. NS, not significant (p>0.05), *p<0.05, **p<0.01, ***p<0.001. P, portal veins. Scale bars: 100 μm (C, D) and 50 μm (F). Related data can be found in Figure 4—figure supplements 1 and 2.
-
Figure 4—source data 1
Quantitation of ALT, AST, zonal markers and EdU+ hepatocytes in CCl4-injured liver.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig4-data1-v2.xlsx
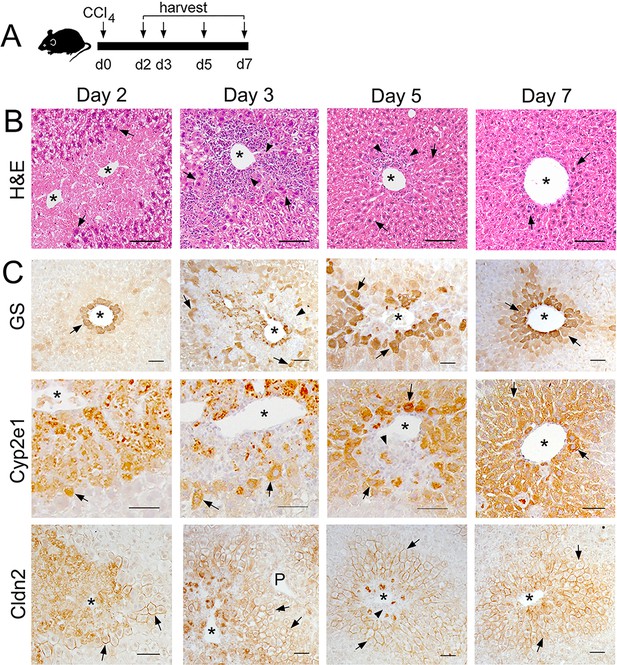
Changes in liver histology and Zone 2/3 protein expression after a CCl4 bolus.
(A) Schematic of CCl4 administration and tissue harvesting. (B) H and E staining reveals broad areas of necrosis separating the central vein (asterisks) from the hepatocyte area at day 2, abundant perivenous immune infiltrates at day 3, reduced immune infiltrates and partially restored parenchyma at day 5, and normal liver architecture at day 7, following acute CCl4 administration. (Arrow show hepatocytes, arrowheads indicate immune infiltrates). (C) Immunohistochemistry analysis reveals very low GS expression (arrows) remaining around the central veins at day 2, a few GS+ nucleated cells separated from the central vein by a cellular infiltrate (arrowheads) at day 3, increased GS+ cells in perivenous areas at day 5, and normal GS expression in Zone 3 at day seven post-CCl4. Expression of Cyp2e1 and claudin-2 proteins is limited to few hepatocytes located at the margins of the perivenous immune infiltrate at days 2–3. The Cyp2e1+ and claudin-2+ cell population continues to expand pericentrally (day 5) and looks restored 7 days post- CCl4 administration. Each image represents 2–3 individual livers. Scale bars: 50 μm (B, C), 100 μm (A).
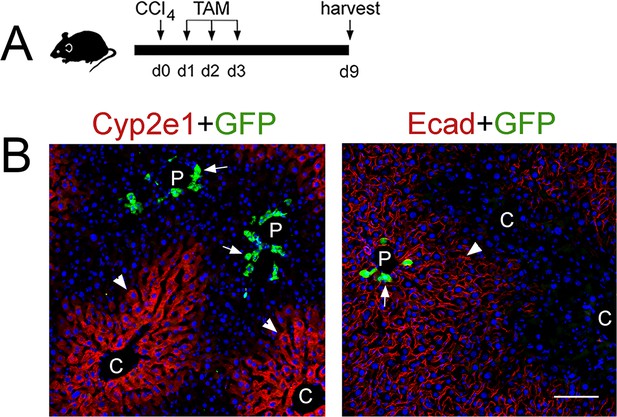
Sox9+ hepatocytes do not contribute to Zone 2/3 restoration after CCl4 induced acute liver injury.
(A) Schematic of CCl4 administration and tissue harvesting. (B) Double-immunofluorescence showing expression of the lineage tracer GFP (arrows) in hepatocytes surrounding the portal veins (P) and located in Zone 1 (E-cadherin+, arrowhead, right). GFP is not expressed in the restored Zone 2/3 (Cyp2e1+, arrowheads, left) up to 9 days after CCl4-induced injury. Scale bar: 100 μm.
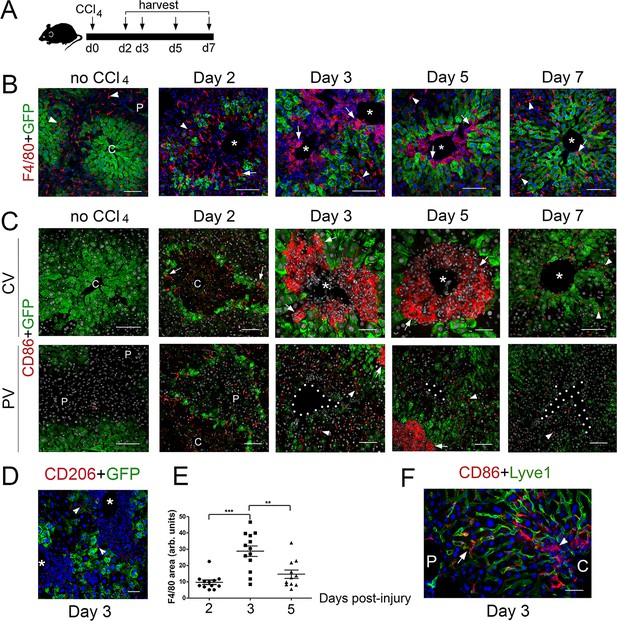
M1 macrophages infiltrate the perivenous space in CCl4-acutely injured livers.
(A) Experimental strategy for Cldn2-EGFP mice injected with CCl4. (B) (No CCl4) Results of double-immunofluorescence analysis show expression of the pan-macrophage marker F4/80+ in Kupffer cells (arrowheads) in a control adult liver (no CCl4 injection). (Day 2–5) In the CCl4-injected Cldn2-EGFP liver, macrophages (arrows) start to infiltrate the necrotic perivenous area at day two and form a physical barrier between the central vein endothelium and the expanding GFP+ Zone 2 at days 3 and 5. Resolution of macrophage perivenous infiltrates and restoration of claudin-2/GFP expression occur 7 days post-CCl4 injection (arrows are Kupffer cells). (C) (No CCl4) M1 macrophages (CD86+) are not detected in perivenous or periportal regions in Cldn2-EGFP control mice (C is central vein, P is portal vein). (Day 2–5, top) Immunofluorescence images of CCl4-injected Cldn2-EGFP livers showing increasingly abundant infiltrates of CD86+ macrophages (arrows) 2 days post-CCl4 and persistence of these cells around the central veins at days 3 and 5. CD86+ macrophages are no longer present in perivenous areas at day 7 (CV, central vein region). (Day 2–5, bottom) CD86+ macrophages are very scarce in periportal areas in CCl4-injected livers (PV, portal vein region). (The exposure in the green channel was decreased for better visualization of the red [CD86] signal. (D) Macrophages infiltrating the perivenous region do not express the M2 macrophage marker CD206 3 days post-CCl4 injection (arrowheads indicate low expression of this marker in LSECs). (E) Quantification of F4/80+ immunofluorescence distribution 2-, 3- and 5 days post-CCl4. p values were determined by one-way ANOVA with Bonferroni’s multiple comparisons test. **p<0.01, ***p<0.001. 3–4 representative fields from three individual livers dissected at the indicated time points were used for quantification. Asterisks and C, central veins. P, portal veins. CV, central vein area. PV, portal vein area. (F) A few CD86+ macrophages are seen within the Lyve1+ hepatic sinusoids in periportal areas of the CCl4 injected liver. CD86+ macrophages are overabundant in perivenous areas (right, ‘C’, arrowhead) in comparison to periportal areas (‘P’). Each image represents 3–4 individual livers. Scale bars: 100 μm (B), (C) [no CCl4, Days 2,7]) Scale bars: 50 μm (C) [Days 3,5], (D,F).
-
Figure 5—source data 1
Quantification of F4/80+ immunofluorescence distribution post-CCl4 injection.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig5-data1-v2.xlsx
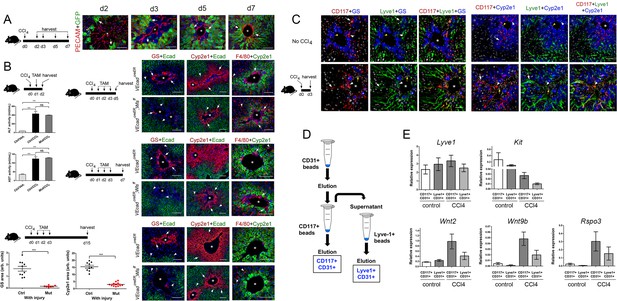
Endothelial Wnt ligand secretion reestablishes metabolic zonation in the CCl4-acutely injured liver.
(A) (Left) Schematic of CCl4 administration and tissue harvesting using Cldn2-EGFP mice. (Right) Double-immunofluorescence results show physical association of claudin-2/GFP+ hepatocytes (arrows) with hepatic endothelial cells (PECAM+) throughout the recovery period that follows CCl4-acute injury. (B) (Top, left) Schematic of the experimental strategy and tissue harvesting. ALT/AST serum levels demonstrate that CCl4 promotes liver damage in Cdh5-CreERT2 and Cdh5-CreERT2;Wlsf/f mice injected with tamoxifen. (n = 3). p values were determined by one-way ANOVA with Bonferroni’s multiple comparisons test, NS, not significant (p>0.05), ***p<0.001. (Right and bottom) Diagrams indicate the experimental strategy and tissue harvesting. Double-immunofluorescence and quantitative results show progressive expansion and full restoration of Zone 3 (GS+, arrows) and Zone 2 (Cyp2e1+, arrows) and transient macrophage infiltrates (F4/80+, arrows; arrowheads are Kupffer cells) in the liver of Cdh5-CreERT2 mice after acute CCl4 administration. In contrast, Zone 3 is nearly absent, Zone 2 is significantly smaller, and Zone 1 is expanded, in Cdh5-CreERT2;Wlsf/f livers 5-, 7- and 15 days post-CCl4. Perivenous macrophage infiltrates are observed in both Cdh5-CreERT2 and Cdh5-CreERT2;Wlsf/f livers 5–7 days post-CCl4 but not 15 days after CCl4 administration. p values were determined by two-tailed unpaired Student’s t-test, ***p<0.001, (n = 3). (C) (Left) Schematic of CCl4 administration and tissue harvesting. (Right) Triple-immunofluorescence results show that CD117 proteins are restricted to the Lyve-1+/Lyve-1LOW sinusoidal endothelium traversing Zones 2/3 in the normal and CCl4-injected (day 3) adult liver. (D) Schematic of CD117+ and Lyve1+ HECs isolation. Nonparenchymal liver cells (NPCs) were isolated using a two-step collagenase perfusion method and incubated with CD31-coated Dynabeads. The eluted CD31+ (HEC) fraction was incubated with CD117-coated Dynabeads to isolate CD117+CD31+ (‘pericentral/perivenous’) HECs. The unbound fraction from this step was incubated with Lyve-1-coated Dynabeads to separate Lyve+CD31+ hepatic sinusoidal cells from other HECs. (E) QRT-PCR results show comparable Lyve1 transcript expression in CD117+/CD31+ and Lyve1+/CD31+ isolates from saline-injected (‘control’) or CCl4-injected (day 3) livers, lower Kit expression in LSECs from injured livers compared to control livers, and higher Wnt2, Wnt9b and Rspo3 expression in LSECs from injured livers compared to control livers. (Three individual livers per condition were used to isolate LSECs.) (A–C) Each image represents 2–4 individual livers. Asterisks: central veins. (B) NS, not significant (p>0.05), ***p<0.001, (n = 3). Scale bars: 50 μm (C), 100 μm (A, B).
-
Figure 6—source data 1
Quantification of ALT/AST serum levels, GS, Cyp2e1 immunofluorescence in Cdh5-CreERT2;Wlsf/f livers post-CCl4 injection.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig6-data1-v2.xlsx
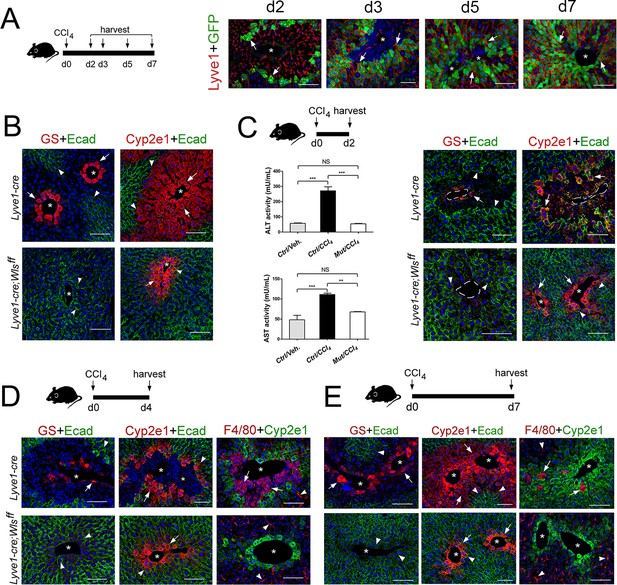
Lyve1-Cre;Wlsf/f mice are refractory to CCl4-induced hepatotoxicity.
(A) (Left) Schematic of CCl4 administration and tissue harvesting using Cldn2-EGFP mice. (Right) Double-immunofluorescence results show that claudin-2/GFP+ hepatocytes (arrows) are in close proximity to the sinusoidal endothelium (Lyve1+, arrows) throughout the recovery period that follows CCl4 acute injury. (B) GS (Zone 3), Cyp2e1 (Zone 2/3) and E-cadherin (Zone 1) expression in control and Lyve1-Cre;Wlsf/f livers without CCl4 treatment. (C–E) Top: Schematics of tissue harvesting post-CCl4 administration. (C) (Left) ALT/AST serum levels indicate liver damage in Lyve1-Cre (control) mice and no liver damage in Lyve1-Cre;Wlsf/f mice, 2 days post-CCl4. p values were determined by one-way ANOVA with Bonferroni’s multiple comparisons test. NS, not significant (p>0.05), **p<0.01, ***p<0.001 (n = 3). (C–E) (top panels): Immunostaining results show that in Lyve1-Cre livers injected with CCl4, Zone 3 is nearly undetected (GS, arrows) and Zone 2 (Cyp2e1, arrows) is severely destroyed at day 2 (C); a few enucleated cells express GS (arrow) around the central vein, Zone 1 (‘E-cad’, arrowheads) is expanded, and Zone 2 (‘Cyp2e1’, arrow) is separated from the central vein by macrophage infiltrates (‘F4/80’, arrows; arrowheads are Kupffer cells) at day 4; and Zones 1–3 (‘E-cad’/‘Cyp2e1’/‘GS‘, arrows) look nearly restored and macrophage infiltrates are scarce (arrows) at day 7. (C–E) (bottom panels): GS+ cells and macrophage infiltrates are undetected and both, Cyp2e1 and E-cadherin expression are unchanged, in Lyve1-Cre;Wlsf/f livers 2–7 days post-CCl4. Each image represents 3–4 individual livers. Asterisks: central veins. Scale bars: 100 μm. Related data can be found in Figure 7—figure supplements 1 and 2.
-
Figure 7—source data 1
Quantification of ALT/AST serum levels in in Lyve1-Cre;Wlsf/f mice post-CCl4 injection.
- https://cdn.elifesciences.org/articles/46206/elife-46206-fig7-data1-v2.xlsx
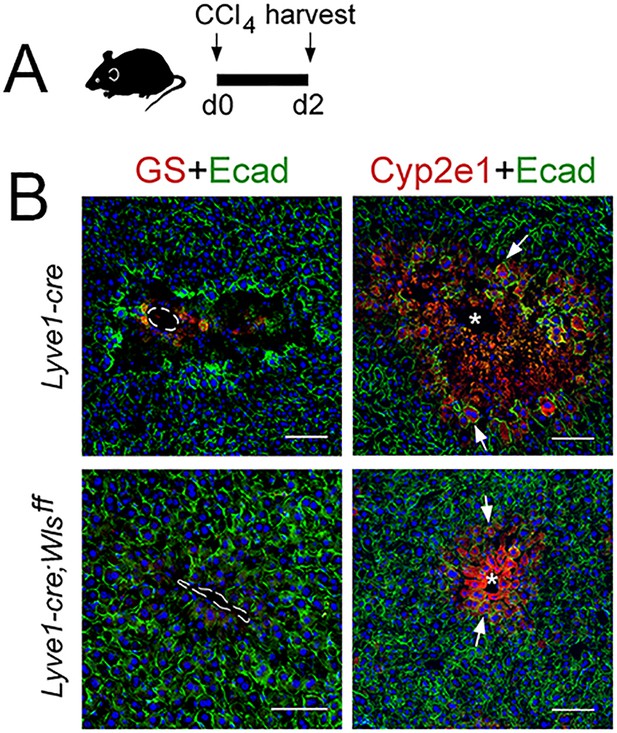
A CCl4-bolus does not induce hepatotoxicity in Lyve1-Cre;Wlsf/f mice.
(A) Diagram of tissue harvesting post-CCl4 administration. (B) Zone 3 (GS+) is nearly absent and Zone 2 (Cyp2e1+, arrows) looks damaged and reduced in Lyve1-Cre livers 2 days post-CCl4 administration. In contrast, GS+ cells are completely absent, Zone 2 (Cyp2e1+, arrows) is very reduced and looks unperturbed, and Zone 1 (E-cadherin+, arrows) is expanded pericentrally in Lyve1-Cre;Wlsf/f livers 2 days post-CCl4. Images are representative of 2 individual livers. Scale bars: 100 μm.
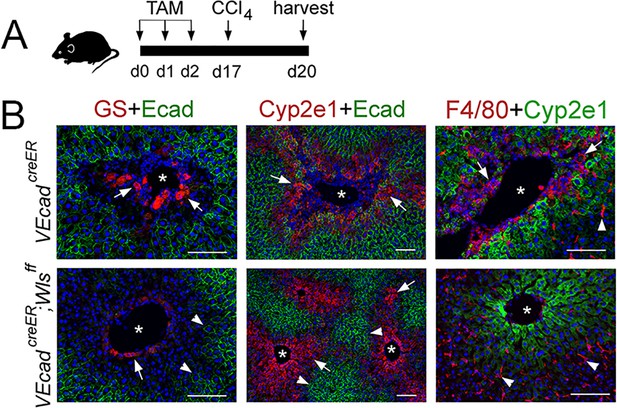
Wls ablation using VE-cadherincreER disrupts zonation and prevents CCl4-hepatotoxicity.
(A) Schematic of the experimental strategy and tissue harvesting. (B) Immunostaining results show destruction of Zones 3 (GS, arrows) and 2 (Cyp2e1, arrows) and macrophage infiltration (F4/80+, arrows; arrowhead is a Kupffer cell), in the liver of VE-cadherincreER mice injected with tamoxifen and then CCl4. This analysis shows almost no Zone 3 hepatocytes (GS, arrow), a small but relatively intact Zone 2 (Cyp2e1, arrows), an intact Zone 1 (E-cadherin, arrowheads), and no macrophage infiltrates (F4/80+, arrowheads indicate Kupffer cells) in the liver of VE-cadherincreER;Wlsf/f mice following similar tamoxifen-CCl4 treatment. Images are representative of 2–3 individual livers. Asterisks: central vein lumen. Scale bars: 100 μm.
Tables
| Reagent type (species) or resource | Designation | Source/reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Commercial assay or kit | Click-iT EdU Imaging Kit | Invitrogen | Invitrogen: C10338 | |
| Commercial assay or kit | PureLink RNA Mini Kit | Invitrogen | Invitrogen: 12183018A | |
| Commercial assay or kit | iScript cDNA Synthesis Kit | Biorad | Biorad: 170–8891 | |
| Commercial assay or kit | ALT Activity Assay | Sigma | Sigma: MAK052 | |
| Commercial assay or kit | AST Activity Assay | Sigma | Sigma: MAK055 | |
| Commercial assay or kit | ABC reagent | Vector Laboratories | Vector: PK-6100 | |
| Commercial assay or kit | DAB solution | Vector Laboratories | Vector: SK-4105 | |
| Commercial assay or kit | Pierce ECL Plus Western blotting Substrate | Pierce | Pierce: 32132 | |
| Commercial assay or kit | RNAscope 2.5 HD Duplex manual assay | Advanced cell diagnostics | Cat. #: 322436 | |
| Commercial assay or kit | DynabeadsTM FlowCompTM MouseCD4 kit | Invitrogen | Cat. #: 11461D | |
| Probe | RNAscope Probe Mm-Wnt2 | Advanced cell diagnostics | Cat #: 313601 | NM_023653.5, region 857–2086 |
| Probe | RNAscope Probe Mm-Rspo3-O2 | Advanced cell diagnostics | Cat. #: 483781 | NM_028351.3, region 717–2099 |
| Probe | RNAscope Probe Mm-Wnt9b | Advanced cell diagnostics | Cat.#: 405091 | NM_011719.4, region 706–1637 |
| Probe | RNAscope Probe Mm-Lyve1-C2 | Advanced cell diagnostics | Cat. #: 42451-C2 | NM_053247.4, region 2–952 |
| Probe | RNAscope Probe Positive Control Probe | Advanced cell diagnostics | Cat. #: 320761 | Mm-Polr2a, NM_001291068.1, region 3212–4088 |
| Probe | RNAscope 2-Plex Negative Control | Advanced cell diagnostics | Cat. #: 320751 | DapB, CP015375.1, region 2252107–2252555 |
| Antibody | anti-mouse GFP (chicken polyclonal) | Abcam | Cat. #: ab13970; RRID:AB_300798 | IF, IHC (1:1000) |
| Antibody | anti-mouse GS (rabbit polyclonal) | Abcam | Cat. #: ab49873; RRID:AB_880241 | IF, IHC (1:5000) |
| Antibody | anti mouse Cyp2e1 (rabbot polyclonal) | Abcam | Cat. #: ab28146; RRID:2089985 | IF, IHC (1:500) |
| Antibody | anti-mouse E-cadherin (rat monoclonal) | Novex | Cat. #: 13–1900; RRID:AB_2533005 | IF (1:5000) |
| Antibody | anti-mouse HNF-4α (goat polyclonal | Santa Cruz Biotechnology | Cat. #: sc-6556; RRID:AB_2117025 | IF (1:50) |
| Antibody | anti-mouse Prox1 (rabbit polyclonal) | Proteintech | Cat. #: 11067–2-AP; RRID:AB_2268804 | IF (1:1000) |
| Antibody | anti-mouse Tbx3 (rabbit polyclonal) | Abcam | Cat. #: ab99302; RRID:AB_10861059 | IF (1:100) |
| Antibody | anti-mouse APC (rabbit polyclonal) | Abcam | Cat. #: ab52223; RRID:AB_867687 | IF (1:50) |
| Antibody | anti-mouse F4/80 (rat monoclonal) | Abcam | Cat. #: ab6640; RRID:AB_1140040 | IF (1:1000) |
| Antibody | anti-mouse PECAM-1 (rat monoclonal) | BD Pharmingen | Cat. #: 550274; RRID:AB_393571 | IF (1:100) MI (2 µg/25 µL) |
| Antibody | anti-mouse Lyve1 (goat polyclonal) | R and D Systems | Cat. #: BAF2125; RRID:AB_2138529 | IF (1:250) |
| Antibody | anti-mouse Claudin-2 (rabbit polyclonal) | Invitrogen | Cat. #: 51–6100; RRID:AB_2533911 | IHC (1:250) |
| Antibody | anti-mouse CD86 (rat monoclonal) | SouthernBiotech | Cat. #: 1735–01; RRID:AB_2795211 | IF (1:100) |
| Antibody | anti-mouse PCK1 (rabbit polyclonal) | Abcam | Cat. #: ab28455; RRID:AB_777191 | IHC (1:100) |
| Antibody | anti-mouse Beta-gal (chicken polyclonal) | Abcam | Cat. #: ab9361; RRID:AB_307210 | IF (1:2000) |
| Antibody | anti-mouse Lyve1 (rat monoclonal) | R and D systems | Cat. #: MAB215, RRID:AB_2138528 | MI (2 µg/25 µL) |
| Antibody | anti-mouse CD117 (rat monoclonal) | R and D Systems | Cat. #: MAB1356; RRID:AB_2131131 | IF (1:50) MI (2 µg/25 µL) |
| Antibody | Cy3 Anti-Rabbit IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 705-165-152; RRID:AB_2307443 | IF (1:250) |
| Antibody | Cy3 Anti-Goat IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 705-165-147; RRID:AB_2307351 | IF (1:250) |
| Antibody | Cy3 Anti-Rat IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 712-165-153; RRID:AB_2340667 | IF (1:250) |
| Antibody | Alexa Fluor 488 Anti-Chicken IgY (IgG) (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 703-545-155; RRID:AB_2340375 | IF (1:250) |
| Antibody | Alexa Fluor 488 Anti-Rat IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 712-545-153; RRID:AB_2340684 | IF (1:250) |
| Antibody | Alexa Fluor 488 Anti-Goat IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 705-545-147; RRID:AB_2336933 | IF (1:250) |
| Antibody | Biotin-SP (long spacer) Anti-Rabbit IgG (H+L) (donkey polyclonal) | Jackson ImmunoResearch | Cat. #: 711-065-152; RRID:AB_2340593 | IHC (1:250) |
| Sequencebased reagent | Glul_F | This paper | PCR primers | TGAACAAAGGCATCAAGCAAATG |
| Sequence-based reagent | Glul_R | This paper | PCR primers | CAGTCCAGGGTACGGGTCTT |
| Sequence- based reagent | Cyp2e1_F | This paper | PCR primers | CGTTGCCTTGCTTGTCTGGA |
| Sequence-based reagent | Cyp2e1_R | This paper | PCR primers | AAGAAAGGAATTGGGAAAGGTCC |
| Sequence- based reagent | Axin2_F | This paper | PCR primers | TGACTCTCCTTCCAGATCCCA |
| Sequence- based reagent | Axin2_R | This paper | PCR primers | TGCCCACACTAGGCTGACA |
| Sequence-based reagent | Cldn2_F | This paper | PCR primers | CAACTGGTGGGCTACATCCTA |
| Sequence- based reagent | Cldn2_R | This paper | PCR primers | CCCTTGGAAAAGCCAACCG |
| Sequence-based reagent | Tbx3_F | This paper | PCR primers | AGATCCGGTTATCCCTGGGAC |
| Sequence based reagent | Tbx3_R | This paper | PCR primers | CAGCAGCCCCCACTAACTG |
| Sequence-based reagent | Cdh1_F | This paper | PCR primers | CCAAGCACGTATCAGGGTCA |
| Sequence- based reagent | Cdh1_R | This paper | PCR primers | ACTGCTGGTCAGGATCGTTG |
| Sequence-based reagent | Prox1_F | This paper | PCR primers | AAGCGCAATGCAGGAAGGGCT |
| Sequence- based reagent | Prox1_R | This paper | PCR primers | ACCACTTGATGAGCTGCGAGG |
| Sequence- based reagent | Actb_F | This paper | PCR primers | AGATCAAGATCATTGCTCCTCCT |
| Sequence-based reagent | Actb_R | This paper | PCR primers | ACGCAGCTCAGTAACAGTCC |
| Sequence- based reagent | Apc_F | This paper | PCR primers | CTTGTGGCCCAGTTAAAATCTGA |
| Sequence- based reagent | Apc_R | This paper | PCR primers | CGCTTTTGAGGGTTGATTCCT |
| Sequence- based reagent | Wnt2_F | This paper | PCR primers | TCCGAAGTAGTCGGGAATCG |
| Sequence- based reagent | Wnt2_R | This paper | PCR primers | GCCCTGGTGATGGCAAATAC |
| Sequence- based reagent | Wnt9b_F | This paper | PCR primers | GGGCATCAAGGCTGTGAAGA |
| Sequence- based reagent | Wnt9b_R | This paper | PCR primers | AACAGCACAGGAGCCTGACA |
| Sequence- based reagent | Rspo3_F | This paper | PCR primers | ACACCTTGGAAAGTGCCTTGA |
| Sequence- based reagent | Rspo3_R | This paper | PCR primers | GCCTCACAGTGTACAATACTGACACA |
| Sequence- based reagent | Pecam1_F | This paper | PCR primers | AGCCTAGTGTGGAAGCCAAC |
| Sequence- based reagent | Pecam1_R | This paper | PCR primers | GGAGCCTTCCGTTCTTAGGG |
| Sequence- based reagent | Kit_F | This paper | PCR primers | CAGGAGCAGAGCAAAGGTGT |
| Sequence-based reagent | Kit_R | This paper | PCR primers | GGGCCTGGATTTGCTCTTTG |
| Sequence-based reagent | Lyve1_F | This paper | PCR primers | GCCAACGCGGCCTGTAAGAT |
| Sequence- based reagent | Lyve1_R | This paper | PCR primers | CCCAGGTGTCGGATGAGTTG |
| Sequence- based reagent | Dll4_F | This paper | PCR primers | TGTGATTGCCACAGAGGTATAAGG |
| Sequence- based reagent | Dll4_R | This paper | PCR primers | GCAATGTAAACAATGCAGAAGGAA |
| Cell line (Mus musculus) | Mus musculus AML12 Cell line | ATCC | Cat. #: CRL-2254; RRID:CVCL_0140 | |
| Cell culture media | DMEM F12 | Gibco | Cat. #: 11320–033 | |
| Chemical compound, drug | Dexamethasone | Sigma-Aldrich | Cat. #: D4902 | (40 ng/ml) |
| Chemical compound, drug | 10 mg/ml insulin,5.5 mg/ml transferrin,5 ng/ml selenium | Gibco | Cat. #: 41400045 | (1X) |
| Chemical compound, drug | CHIR99021 | Sigma Aldrich | Cat. #: SML1046 | (3 µM) |
| Chemical compound, drug | CCl4 | Sigma Aldrich | Cat. #: 319961 | |
| Chemical compound, drug | Tamoxifen | Sigma Aldrich | Cat. #: T5648 | |
| Chemical compound, drug | Corn oil | Sigma Aldrich | Cat. #: C8267 | |
| Chemical compound, drug | Mayer's Hematoxylin | ScyTek Laboratories | Cat. #: HMM500 | |
| Chemical compound, drug | DAPI | Life Technologies | Cat. #: D1306 | (1 µg/mL) |
| Chemical compound, drug | TRIzol Reagent | Thermo Fisher | Cat. #: 15596026 | |
| Chemical compound, drug | cOmpleteTM, Mini EDTA-free protease inhibitor | Roche | Cat. #: 11836170001 | |
| Chemical compound, drug | DynabeadsTM Sheep Anti-Rat IgG | Invitrogen | Cat. #: 11035 | 25 µl/sample |
| Chemical compound, drug | Colagenase H from clostridium histolyticum | Millipore Sigma | Cat. #: 11074059001 | 0.5 mg/mL |
| Recombinant protein | Rspo3 | R and D Systems | Cat. #: 4120-RS-025 | (500 ng/ml) |
| Recombinant protein | Wnt2 | Abnova | Cat. #: H00007472-P01 | (500 ng/ml) |
| Recombinant protein | Wnt9b | R and D Systems | Cat. #: 3669-WN-025 | (500 ng/ml) |
| Strain Mus musculus | B6.129P2-Lyve1tm1.1(EGFP/cre)Cys/J | The Jackson Laboratory | Cat. #: JAX:012601; RRID:IMSR_JAX:012601 | |
| Strain Mus musculus | Cdh5CreERT2 | Ralf H. Adams, Max Planck Institute for Molecular Biomedicine, Münster, Germany Wang et al., 2010 | ||
| Strain Mus musculus | Tg[Cldn2-EGFP]OU78Gsat/Mmucd | Mutant Mouse Regional Resource Center [MMRRC], University of California, Davis Gong et al., 2003 | ||
| Strain Mus musculus | 129S-Wlstm1.1Lan/J | The Jackson Laboratory | Cat. # JAX:012888; RRID:IMSR_JAX:012888 | |
| Strain Mus musculus | RoB6.129S4-Gt(ROSA)26Sortm1Sor/Jsa | The Jackson Laboratory | Cat. # JAX:004077; RRID:IMSR_JAX:004077 | |
| Strain Mus musculus | Sox9CreERT2 (Tg(Sox9-cre/ERT2)1Msan/ | The Jackson Laboratory | Cat. # JAX:018829; RRID:IMSR_JAX:018829 | |
| Software | Adobe Photoshop CC | Adobe Systems | RRID:SCR_014199 | |
| Software | ImageJ | NIH https://imagej.net/ | RRID:SCR_003070 | |
| Software | GraphPad Prism 5.0 software | GraphPad Software;http://www.graphpad.com | RRID:SCR_002798 |





