An evolutionary recent IFN/IL-6/CEBP axis is linked to monocyte expansion and tuberculosis severity in humans
Figures
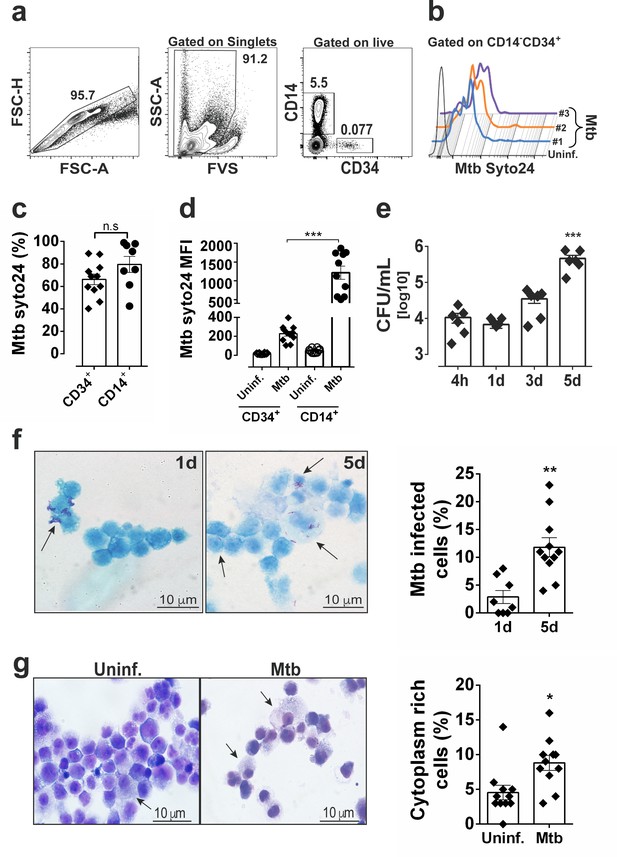
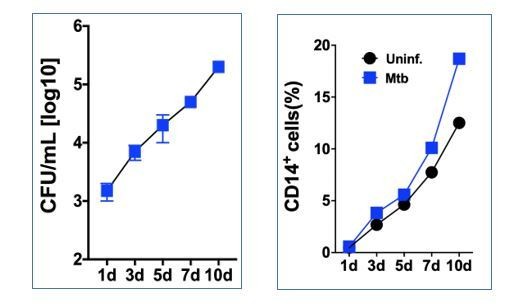
Mtb H37Rv infects human CD34+ cells and proliferates in cell cultures in vitro.
PBMC from healthy donors were exposed to syto24-labeled Mtb H37Rv (MOI3, Figure 1—figure supplement 1a) for 4 hr. (a) Representative flow cytometry contour plots of gating strategy to analyze Mtb syto24 association in FVS-negative (live) CD34+ events and CD14+ events. (b) Live CD34+Lin- events gated in a were analyzed for Mtb-Syto24 MFI. Black line: Uninfected control. Blue, orange and purple lines represent samples from three different donors. (c) Frequencies and (d) MFI of Mtb syto24+ events in CD34+ or CD14+ events gates from uninfected or Mtb syto24-exposed bulk PBMCs. Results are means ± SEM of data pooled from three independent experiments, n = 10 healthy donors. ***p≤0.001 between Mtb syto24 CD34+ vs CD14+ groups. (e,) Purified cord blood-derived CD34+ cells were exposed to Mtb H37Rv (MOI3) for different time points and CFUs from cell culture lysates were enumerated in 7H10 media. Results are means ± SEM of data pooled from five independent experiments, ***p≤0.001 between 5d vs 4 hr groups. (f) Kinyoun staining of CD34+ cells after 1d and 5d of infection and quantification, as described in the methodology section, shown in the right panel. Arrows indicate cells associated with bacilli. Experiments shown are representative of two performed. **p≤0.01 between 5d vs 1d groups. (g) Representative Giemsa staining of CD34+ cells of 5d-cultures and quantification, as described in the methodology section, shown in the right panel. Arrow indicates cytoplasm-rich cells in Mtb-infected cultures and uninfected cultures. Experiments shown are representative of two performed. *p≤0.05 between Mtb vs uninfected groups.
-
Figure 1—source data 1
Raw data from Figure 1.
- https://doi.org/10.7554/eLife.47013.005
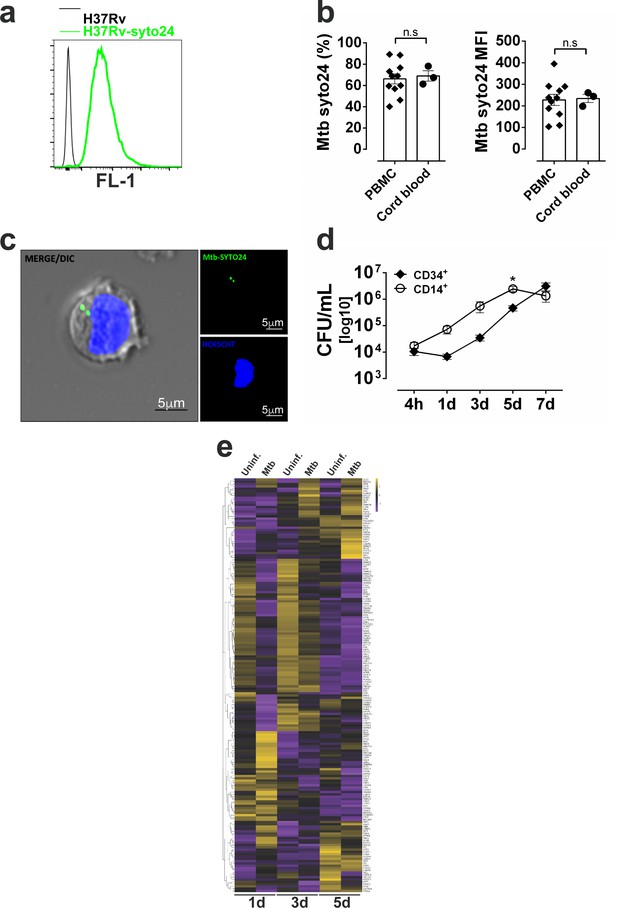
Mtb-CD34+ interactions and signaling pathways associated with HSPC differentiation.
(a) representative histogram of median fluorescence intensity (MFI) from H37Rv stained or not with syto24 (FL1 channel). PBMC or cord-blood derived purified CD34+ cells from healthy donors were exposed to syto24-labeled Mtb H37Rv (MOI3) for 4 hr. (b) Frequencies (left panel) and MFI (right panel) of Mtb syto24+ events in CD34+ cells from PBMC vs purified CD34+ cell cultures. Results are means ± SEM of data pooled from three independent experiments. (c) Confocal microscopy showing a CD34+ cell infected with Syto24-stained Mtb H37Rv. Nuclei = DAPI/blue. Mtb = Syto24/green. (d) Purified CD14+ or purified CD34+ cells were exposed to Mtb H37Rv (MOI3) for different time points and CFUs from cell culture lysates were enumerated in 7H10 media. Results are means ± SEM of data pooled from five independent experiments, *p≤0.01 between CD14+ vs. CD34+ groups 5d. (e) Heat map showing z-score values of 180 transcription factors (Novershtern et al., 2011) expressed by CD34+ cells exposed to Mtb (MOI3) at days 1,3 and 5 post-infection.
-
Figure 1—figure supplement 1—source data 1
Raw data from Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.47013.004
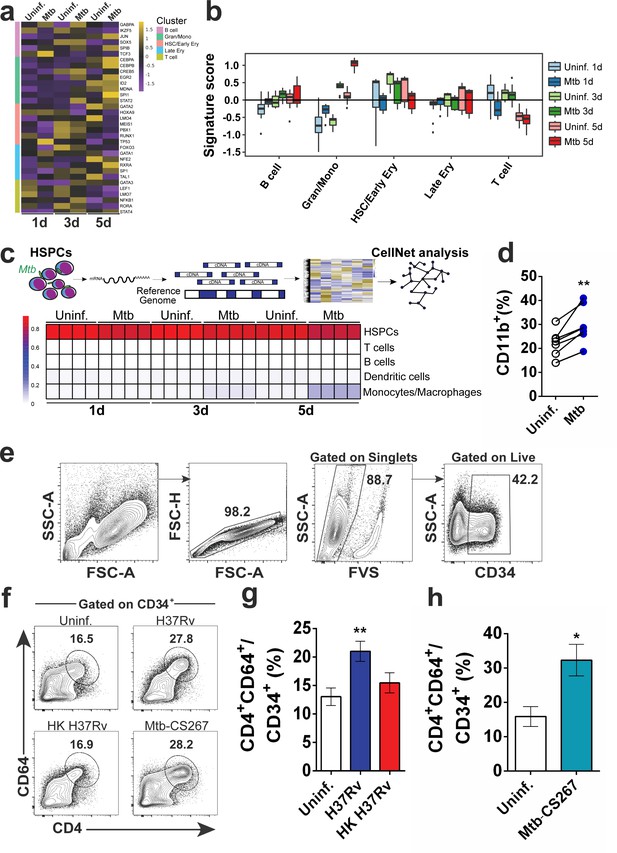
Live Mtb induces human CD34+ cells towards myeloid differentiation in vitro.
Purified CD34+ cells from healthy donors (n = 3) were exposed to Mtb H37Rv (MOI3) for different time points and mRNA-seq was performed as described in methodology section. (a) Heatmap of the mRNA expression (z-score) of transcription factors involved in cell lineage commitment (Novershtern et al., 2011). (b) Signature score of data from a) by employing CellRouter analysis. (c) Heatmap from mRNA data of uninfected vs Mtb infected cultures analyzed by CellNet. (d) Purified CD34+ cells were exposed to Mtb H37Rv (MOI3) for 5 days and flow cytometry was performed. Graph represents frequencies of CD11b+ events in uninfected (open circles) vs Mtb-infected cultures (blue circles) from four independent experiments. **p≤0.01 between Mtb and uninfected groups. (e) Purified CD34+ cells were exposed to Mtb H37Rv, Heat-killed (HK) Mtb H37Rv or Mtb clinical isolate 267 (Mtb-CS267) (MOI3) for 5 days and flow cytometry with the gating strategy was performed. (f) Representative contour plots show frequencies of CD4+CD64+ events in CD34+ events. CD34+CD4+CD64+ events of polled data from f) were plotted to generate bar graphs (g) and (h). Results are means ± SEM of data pooled from four independent experiments (g) and two independent experiments (h). (g) ** indicates p≤0.01 between H37Rv vs uninfected or HK H37RV groups. (h) * indicates p≤0.05 between Mtb-CS267 vs uninfected groups.
-
Figure 2—source data 1
Raw data from Figure 2.
- https://doi.org/10.7554/eLife.47013.009
-
Figure 2—source data 2
Counts matrix of RNAseq data of Mtb-exposed and control CD34+ cell transcriptomes.
- https://doi.org/10.7554/eLife.47013.010
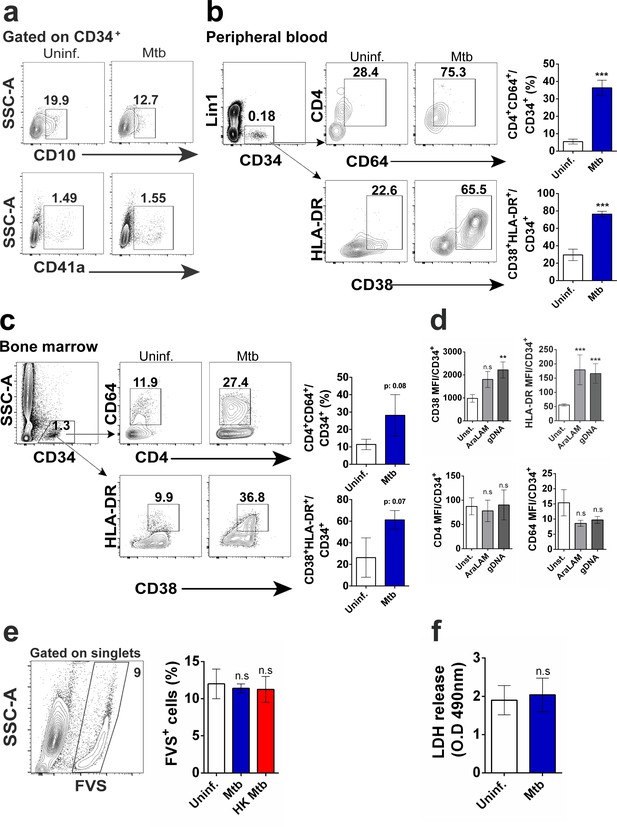
Myeloid differentiation by PBMC or bone marrow CD34+ cells exposed to Mtb, mycobacterial ligands and cell death analysis in vitro.
(a) Frequencies of CD10+CD34+ and CD41a+CD34+ cells in uninfected vs Mtb-infected cell cultures. Representative dot plots of (b) peripheral blood samples from four healthy individuals and (c) bone marrow obtained from two healthy subjects showing frequencies of CD4+CD64+CD34+ and CD38+HLADR+CD34+ cells in uninfected vs Mtb-infected cell cultures. ***p≤0.0001 between Mtb vs uninfected groups. (d) Purified CD34+ cells were exposed to AraLam (10 μg/ml) or gDNA (10 μg/ml) and 5 days later, CD38, HLADR, CD4 and CD64 MFI calculated within CD34+ events. Cell death analysis by means of (e) frequencies of cell permeability dye (FVS+) and (f) LDH quantities detected in the supernatants from uninfected or Mtb-exposed CD34+ cell cultures five dpi. **p≤0.001 and ***p≤0.0001 between mycobacterial ligands vs control groups.
-
Figure 2—figure supplement 1—source data 1
Raw data from Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.47013.008
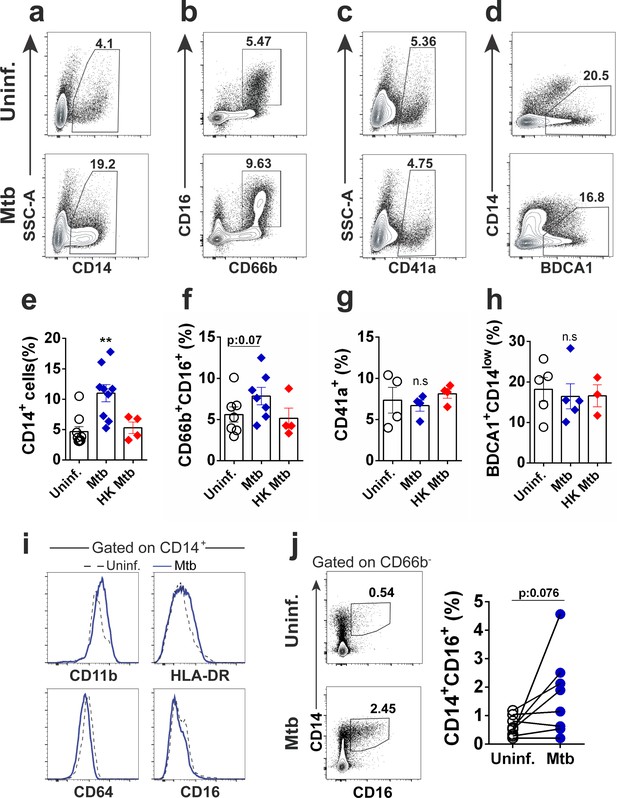
Mtb infection increases monocyte output from CD34+ cells in vitro.
Purified CD34+ cells were exposed to live Mtb H37Rv or HK Mtb H37Rv (MOI3) for 10 days and flow cytometry was employed to determine the mature cell frequencies in the cell cultures. Representative dot plots of (a) monocytes (CD14+), (b) neutrophils (CD16+CD66b+), (c) megakaryocytes/platelets (CD41a+) and (d) classical myeloid dendritic cells (BDCA1+CD14low) in uninfected and Mtb-infected CD34+ cell cultures. Graphs show frequencies of (e) CD14+, (f) CD16+CD66b+, (g) CD41a+ and (h) BDCA1+CD14low events in uninfected (open circles), live Mtb-infected (blue diamonds) or HK Mtb-exposed (red diamonds) cell cultures at day 10. Each symbol represents one individual experiment. Results are means ± SEM of data pooled from 3 to 9 independent experiments. **p≤0.01 between Mtb vs uninfected or HK Mtb groups. (i,) Histograms show the expression of CD11b, HLA-DR, CD64 and CD16 in CD14+ events from a). Black dashed lines: Uninfected control. Blue solid lines: Mtb-infected group. Data representative of 5 independent experiments. (j,) Frequency of CD14+CD16+ events in Mtb-exposed cell cultures after 10d. Contour dot plot of CD14+CD16+ frequencies from one representative donor. Open circles: Uninfected control. Blue circles: Mtb-infected group. Each symbol represents an individual experiment. Pooled data of eight independent experiments, n = 5 different donors. p=0.076 between Mtb vs uninfected groups.
-
Figure 3—source data 1
Raw data from Figure 3.
- https://doi.org/10.7554/eLife.47013.014
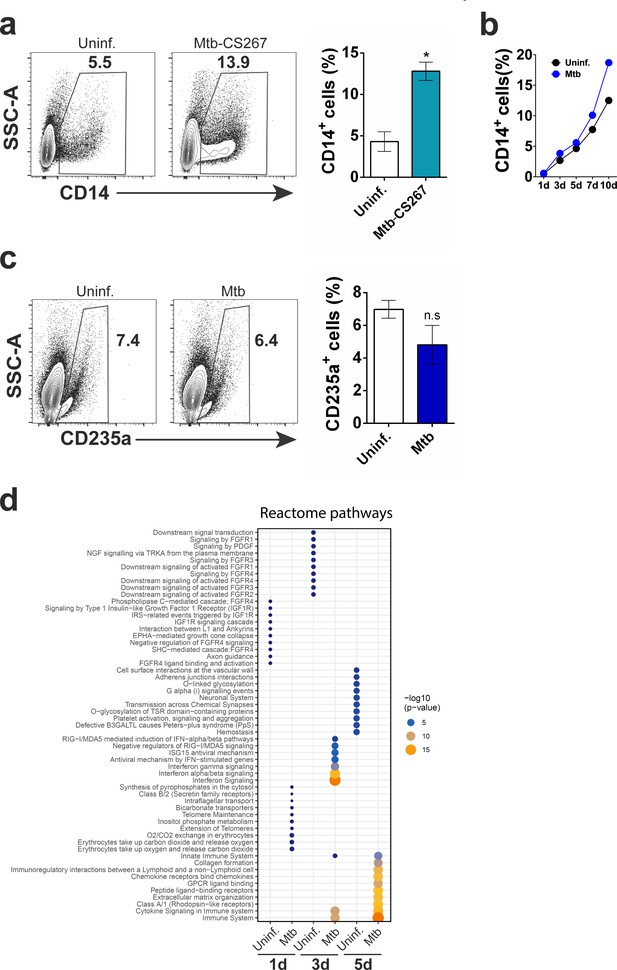
Monocyte differentiation and reactome pathways associated to Mtb-exposed CD34+ cells in vitro.
(a) Frequencies of CD14+ cells during 10 day exposure to a clinical isolate of Mtb. *p≤0.05 between Mtb-CS267 vs uninfected groups. (b) Purified CD34+ cells were exposed to Mtb H37Rv for 1, 3, 5, 7 and 10 days and flow cytometry was employed to determine the CD14+ monocyte frequency. (c) The erythroid cell marker CD235a was measured by flow cytometry in Mtb-exposed and uninfected purified CD34+ cells at 10 dpi. Results shown are representative from two experiments. (d) Enrichment of Reactome pathways based on gene signatures derived from each experimental condition. Gene signatures were composed by genes with log2 fold change >0.75 when comparing one experimental condition versus all others. The size and color of the circles are proportional to -log10 of the adjusted p-value.
-
Figure 3—figure supplement 1—source data 1
Raw data from Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.47013.013
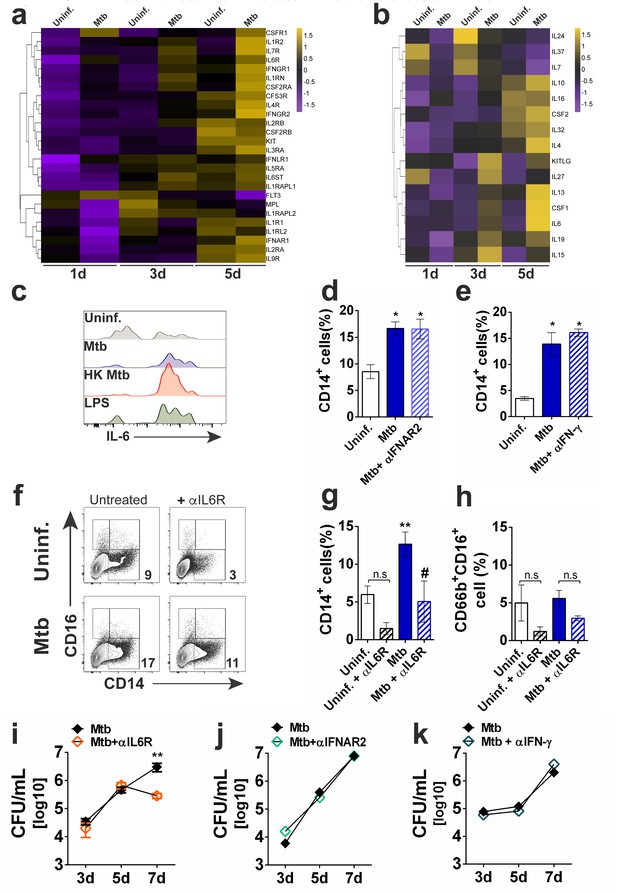
Mtb enhances IL-6R-mediated myeloid differentiation in vitro.
Purified CD34+ cells were exposed to Mtb H37Rv (MOI3) for different time points and mRNA-seq was performed as described in the methodology section. (a) Heatmap (z-score) of differentially expressed cytokine receptor genes. (b) Heatmap (z-score) of differentially expressed cytokine genes. Shown is the average mRNA expression of three different donors from two independent experiments. (c) PBMC from healthy donors were exposed to Mtb H37Rv, HK Mtb or LPS (100 ng/mL) for 24 hr and intracellular IL-6 was detected by flow cytometry. Live CD34+Lin- events gated as in Figure 1a were analyzed for IL-6 MFI. Representative histogram from two independent experiments. Purified CD34+ cells were treated with (d) α-IFNAR2 (1 µg/ml) or (e) α-IFN-γ (10 µg/ml) and then exposed to Mtb H37Rv (MOI3) during 10d for determination of CD14+ monocyte frequencies. Results are means ± SEM of data pooled from two independent experiments. *p≤0.05 between Mtb, α-IFNAR2 or α-IFN-γ vs uninfected groups. (f) Representative contour plots of CD14+ monocytes in CD34+ cell cultures exposed to Mtb, in the presence or absence of α-IL6R (Tocilizumab, 1 µg/ml) for 10d. (g) Results shown are means ± SEM of data pooled from three independent experiments from (f) **p≤0.01 between Mtb vs uninfected groups and #p≤0.05 between Mtb and Mtb+α-IL6R-treated groups. (h) Results shown are means ± SEM of data pooled from three independent experiments showing frequency of CD66+CD16+ neutrophils in Mtb-infected cell cultures in the presence or absence of α-IL6R. Purified CD34+ cell cultures were treated as in (d–f) with (i) α-IL6R, (j) α -IFNAR2 and (k) α-IFN-γ and then exposed to Mtb (MOI3) for different time points and CFU enumerated as described in the methodology section. Results are means ± SEM of data pooled from four independent experiments. **p≤0.01 between Mtb and Mtb+ α-IL6R at 7d.
-
Figure 4—source data 1
Raw data from Figure 4.
- https://doi.org/10.7554/eLife.47013.018
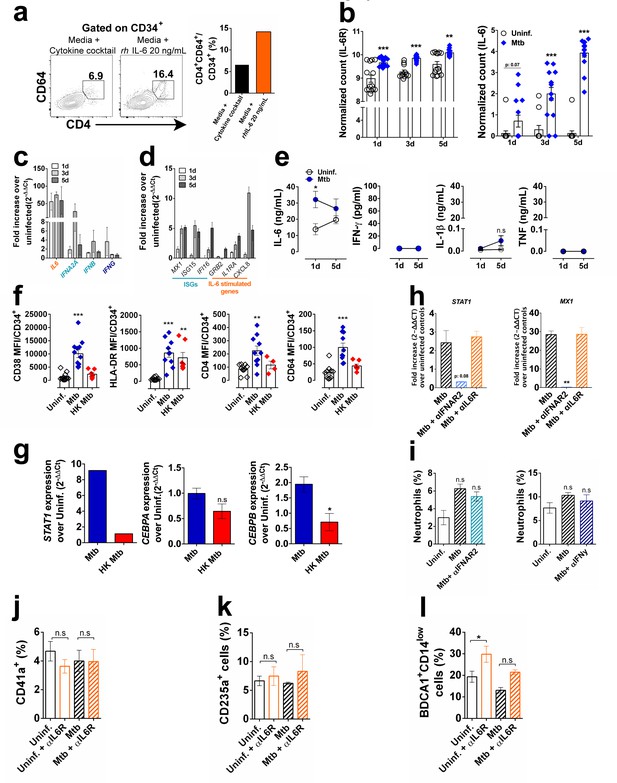
Gene expression and cytokine production during myeloid differentiation in vitro.
(a) CD34+ cells were stimulated with or without recombinant IL-6 (20 ng/mL) for 5 days and frequency of CD4+CD64+CD34+ cells were measured by flow cytometry. (b) normalized counts from RNA-seq data as determined by library size normalization. Results are means ± SEM of data pooled from two independent experiments (eight replicates). ***p≤0.001 between Mtb vs uninfected groups at different time points. CD34+ cells were exposed to Mtb (MOI3) and at days 1, 3 and 5 p.i. qPCR was performed for quantification of (c) IL6/IFN cytokines and (d) ISGs and IL-6-induced genes. (e) IL-6, IFN-γ, IL-1β and TNF measurements from unexposed or Mtb-exposed purified CD34+ cell culture supernatants at day 1 and 5 p.i. Open circle: uninfected control. Blue circle: Mtb-infected group. Results are means ± SEM of data pooled from 6 to 8 independent experiments. *p≤0.05 between 1d Mtb vs uninfected groups. (f) purified CD34+ cells were exposed to live or HK-Mtb (MOI3) and 5 days later, CD38, HLADR, CD4 and CD64 MFI calculated within CD34+ events. Results are means ± SEM of data pooled from five independent experiments. **p≤0.001; ***p≤0.0001 between Mtb or HK Mtb vs uninfected groups. (g) CD34+ cells were exposed to live or HK-Mtb (MOI3) and at days 5 p.i., qPCR was performed for quantification of STAT1, CEBPA and CEBPB. Results are means ± SEM of data pooled from three independent experiments. *p≤0.05 between Mtb vs HK Mtb groups. (h) CD34+ cells were exposed to live Mtb (MOI3) in the presence or absence of α-IFNAR2 or α-IL-6R and at day 5 p.i., qPCR was performed for quantification of STAT1 and MX1. **p≤0.001 between Mtb vs Mtb+αIFNAR2 groups. CD66b+CD16+ neutrophil frequencies in 10 day culture of Mtb-exposed CD34+ treated with (i) α-IFNAR2 (1 µg/ml) or α-IFN-γ (10 µg/ml) blocking antibodies. (j,) CD41+ megakariocytes; (k) CD235+ erythrocytes and (l) BDCA1+CD14low myeloid DC frequencies in 10 day culture of Mtb-exposed CD34+ treated with α-IL-6R blocking or control antibodies.
-
Figure 4—figure supplement 1—source data 1
Raw data from Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.47013.017
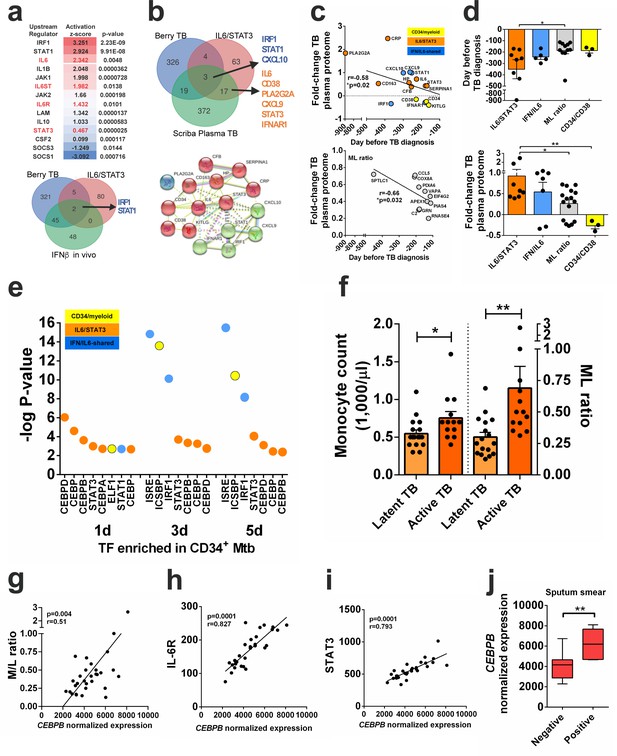
IL6/IL6R/CEBPB gene module is enriched in active TB transcriptome and proteome and correlates with monocyte expansion.
(a) Top panel: upstream regulators significantly enriched by causal Ingenuity Pathway Analysis (IPA) in monocyte transcriptomes from patients with active TB (GSE19443), ranked by activation z-score, p-values are corrected for genome-wide testing (FDR). Bottom panel: IRF1 and STAT1 are the top upstream regulators shared between the ‘Berry TB’ disease signature (Berry et al., 2010) (GSE19435, GSE19439, GSE19444), the ‘IL6/STAT3’ pathway (Hallmark GSEA) and the human ‘in vivo IFN-β” signature (GSEA HECKER_IFNB1_TARGETS). (b) Top panel: overlap between the ‘Berry TB’ disease signature, the ‘IL6/STAT3’ pathway and the ‘Scriba plasma TB’ proteomic signature (Scriba et al., 2017) identified ‘IFN/IL6-shared’ and ‘IL6/STAT3-specific’ signatures. Bottom panel: significant STRING protein-protein interaction network (p<10−16) for ‘IFN/IL6-shared’ genes (green marbles) and ‘IL6/STAT3’ genes (red marbles), clustering separately by k-means. (c) Top panel: significant linear increase over time before active TB diagnosis in plasma proteome (Scriba et al., 2017) for ‘CD34/myeloid’ (yellow), ‘IL6/STAT3’ (orange) and ‘IFN/IL6-shared’ (blue) clusters found in (b). Bottom panel: monocyte/lymphocyte (ML) ratio gene set members defined by Naranbhai et al. (2015) over time before active TB diagnosis in plasma proteome (Scriba et al., 2017).(d) Top panel: increased ‘IL6/STAT3’ cluster protein expression precedes monocyte expansion markers (ML ratio gene set) in the TB plasma proteome. Bottom panel: data as in d) shows significant higher fold-changes for ‘IL6/STAT3’ vs. ‘ML ratio’ or ‘CD34/myeloid’ cluster members. *p-value<0.05, ** p-value<0.01. (e) Transcription factor enrichment analysis (GSEA) of differentially expressed genes determined by RNA-seq in Mtb-exposed CD34+ cells in vitro (n = 3 donors). (f), monocyte count and ML ratio in samples from latent vs active TB patients from Berry et al. (2010) reanalysis. *p-value<0.05, ** p-value<0.01 between active TB vs latent TB groups. Transcriptional data of whole blood reanalysis from Berry et al. (2010) shows a significant correlation of CEBPB transcripts with g) M/L ratio; h) IL6R; i) STAT3 transcript levels, and j) mycobacterial positivity in sputum smears in patients with active TB. ** p-value<0.01 between positive vs negative groups.
-
Figure 5—source data 1
Raw data from Figure 5.
- https://doi.org/10.7554/eLife.47013.021
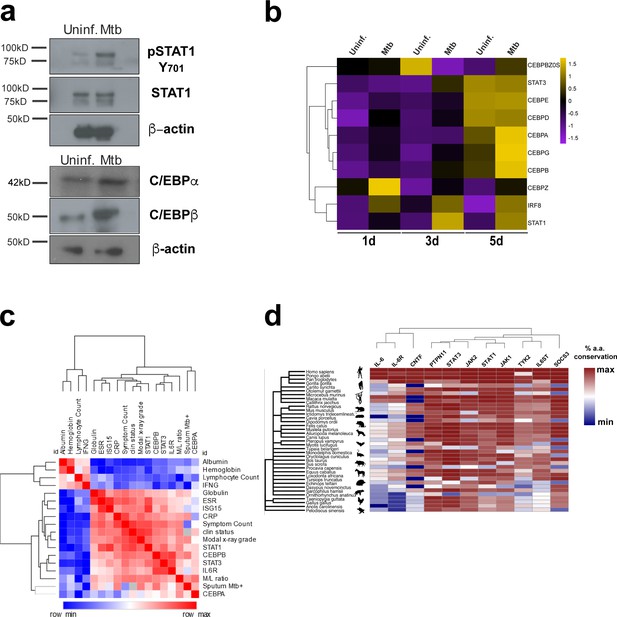
Gene expression and protein conservation of the IFN/IL6/CEBP gene module and correlation analysis to TB disease.
(a) Western blotting for pSTAT1 (Y701), total STAT1, C/EBPα, C/EBPβ and actin from uninfected or Mtb-infected CD34+ cells for 5 days as described in the materials and methods. (b) Heat maps showing z-score values of CEBP family members and e ISGs expressed by CD34+ cells exposed to Mtb (MOI3) at different time points. (c) Cluster dendrogram and heatmap of Spearman correlation coefficients between molecular and clinical data from a UK cohort of patients with latent and active TB (Berry et al., 2010) (n = 30, raw data were obtained from GXB, sputum smear only available for active TB patients). (d) Heat map showing IL6 network generated by STRING co-occurrence protein conservation scores across primates, mammals, birds and reptiles. Note highly conserved STAT1/STAT3 and other molecules found in the network (depicted in Figure 6a, STRING) throughout primate and mammalian evolution.
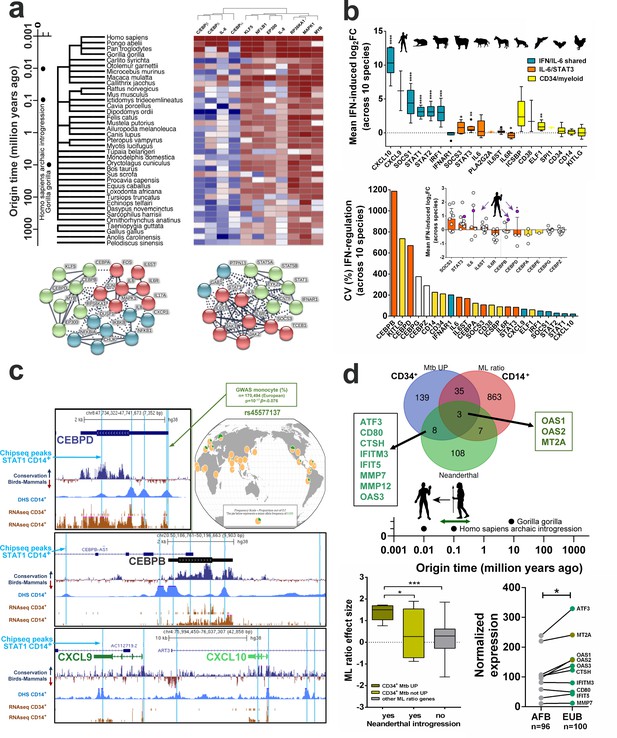
Evolutionary recent and human-specific genetic adaptation link IL6/IL6R/CEBP gene module with monocyte expansion and TB pathogenesis.
(a) Heat map showing CEBPB network generated by STRING co-occurrence protein conservation scores across primates, mammals, birds and reptiles. Note only CEBPB and CEBPA differ strongly among hominids, while CEBPA/CEBPB/CEBPD vary significantly throughout primate and mammalian evolution, as compared to highly conserved STAT1/STAT3 (Figure 5—figure supplement 1d). (b) Top panel: Highly conserved type I IFN upregulation of ‘IFN/IL6-shared’ genes from humans to birds (derived from http://isg.data.cvr.ac.uk/) (Shaw et al., 2017), as compared to ‘IL6/STAT3’ and ‘CD34/myeloid differentiation’ genes. Bottom panel: CEBPB and CEBPD displays highest variation, and CXCL10 the lowest variation in type I IFN transcriptional regulation across human-mammalian-bird evolution. Inset, CEBPB and CEBPD selectively acquired type I IFN upregulation in humans (filled circles); ** p-value<0.01 and * p-value<0.05 represent CEBPB and CEBPD values, respectively for humans versus the other species. (c) ChipSeq analysis of STAT1-binding peaks in CEBPD (top panel), CEBPB (middle panel), CXCL9 and CXCL10 (bottom panel) in IFN-stimulated human monocytes, corresponding to regions with active chromatin (DNase Hypersensitivity Sites, DHS) and correlating with increased downstream transcription in CD14+ monocytes, as compared to purified CD34+ cells. Conservation analysis among >40 vertebrates (phyloP [Pollard et al., 2010], from chicken to human, analogous to Figure 5b) indicates STAT1 peaks are mostly conserved in CXCL9/CXCL10 (6/7) but not in CEBPD/CEBPB (3/11). (d) Top panel: overlap between human genes with significant Neanderthal introgression (Enard and Petrov, 2018; Quach et al., 2016), genes differentially expressed in Mtb-exposed CD34+ cells (CD34+ Mtb UP) and the ‘ML ratio’ gene set. Bottom left panel: OAS1, OAS2 and MT2A transcripts presented significantly higher effect sizes upon ML ratios, corresponding to monocyte expansion, as compared to other introgressed genes (p<0.05) and to all other genes shown to regulate ML ratio in vivo (p<0.001). Bottom right panel: normalized expression of introgressed genes found in CD34+Mtb UP (Venn diagram) in TLR1/2 agonist-treated monocytes from a cohort of matched Belgian individuals of European (EUB) vs. African (AFB) descendance, with documented presence or absence of Neanderthal introgression (Quach et al., 2016), respectively. p-value<0.05, ** p-value<0.01, *** p-value<0.001, **** p-value<0.0001.
-
Figure 6—source data 1
Raw data from Figure 6.
- https://doi.org/10.7554/eLife.47013.025
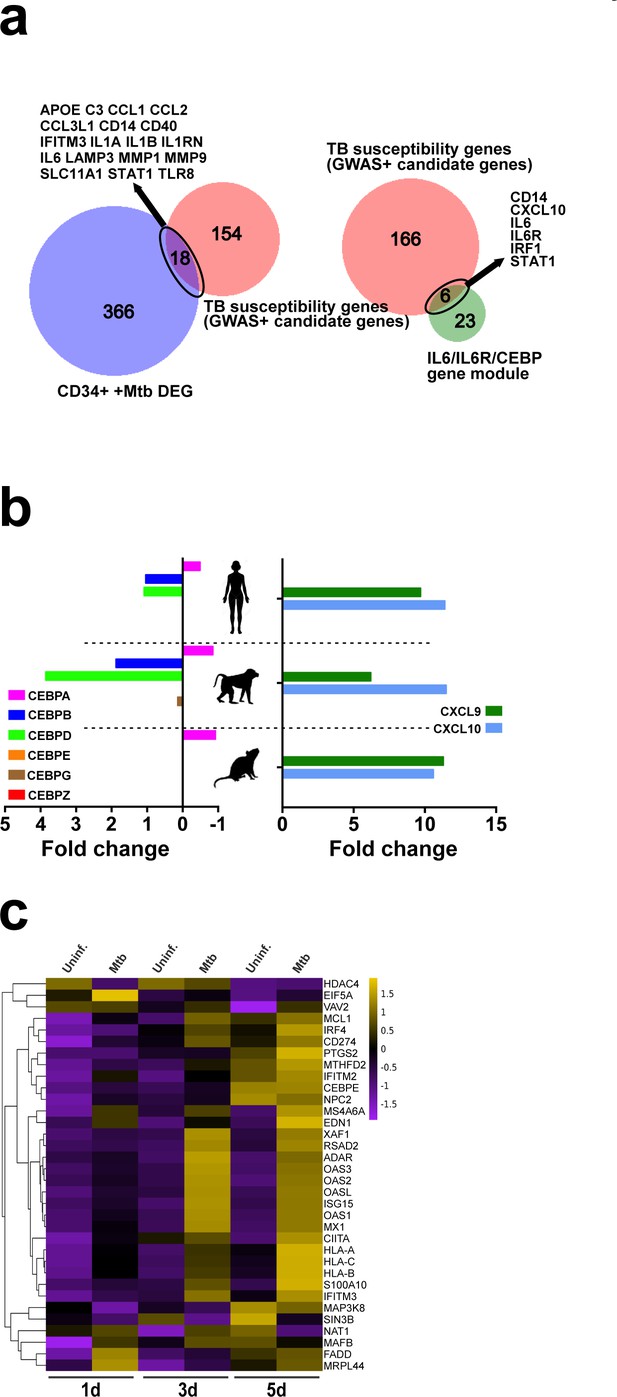
TB susceptibility genes of the IFN/IL6/CEBP gene module and ISG induction during myeloid differentiation in vitro.
(a,) Primary dermal fibroblasts from humans, macaques and mice were stimulated for 8 hr in vitro with dsRNA analog (polyI:C) and CEBP family members, CXCL9 and CXCL10 transcript levels were quantified by RNA-seq (expressed as fold-change over unstimulated cells). Raw data obtained from Hagai et al. (2018) (https://scb.sanger.ac.uk/#/base/main). (b) Significant overlap (hypergeometric test p<0.0001) between TB susceptibility genes (identified by GWAS or candidate gene studies) and differentially expressed genes (DEG) in CD34+ cells exposed to Mtb (left panel) as well as the IL6/IL6R/CEBP gene module (right panel). (c) Purified CD34+ cells were exposed to Mtb H37Rv (MOI3) for different time points. Heat map shows z-score values of ISGs expressed by CD34+ cells exposed to Mtb (MOI3) at different time points.
-
Figure 6—figure supplement 1—source data 1
Raw data from Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.47013.024
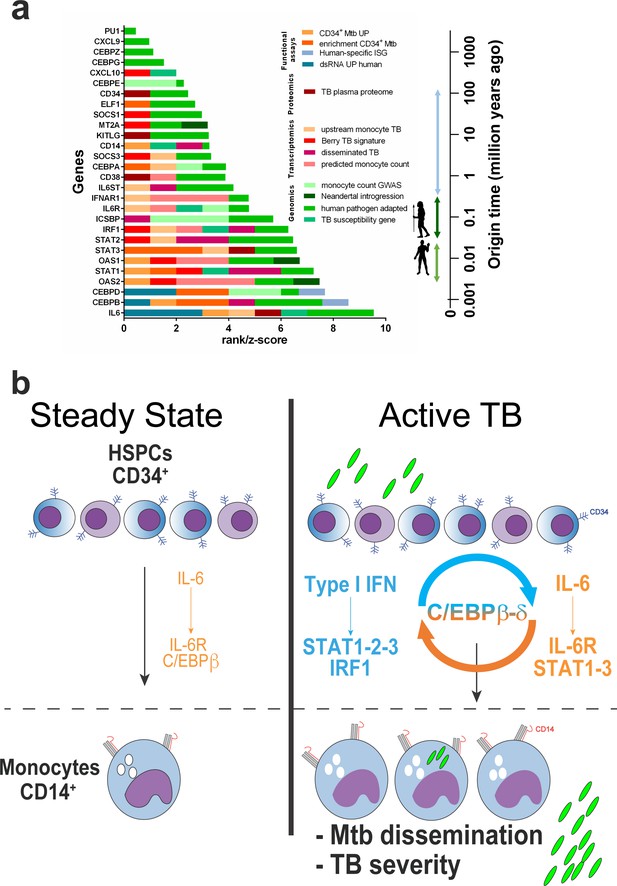
Compiled multi-level evidence for an IL6/IL6R/CEBP gene module linking CD34+ myeloid differentiation to TB pathogenesis and disease severity.
(a) Ranks and scores were determined as 0–1 (presence-absence in data set) or 0-1-2-3, according to enrichment analysis or differential gene expression (quartiles); z-scores were obtained from Daub et al.55 (b) Proposed model for C/EBPβ and C/EBPδ acting as a bridge in the type I IFN and IL-6 feed-forward loop exploited by Mtb to induce monocyte differentiation and TB disease severity (details in the text).
-
Figure 7—source data 1
Raw data from Figure 7.
- https://doi.org/10.7554/eLife.47013.027
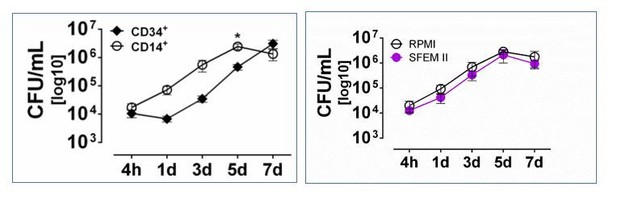
Comparative Mtb growth in purified CD14+ vs CD34+ cells.
Left panel, Mtb growth curve in sorted purified CD34+ or CD14+ cell cultures. Right panel, Mtb growth curve of sorted purified CD14+ cells cultivated in RPMI vs StemSpan SFEM II media.
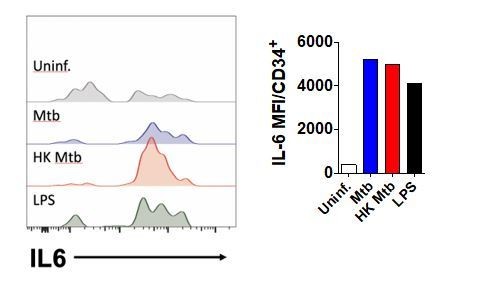
PBMC cultures were exposed to Mtb H37Rv (MOI=3), HK Mtb or LPS (100 ng/mL) for 24h and intracellular IL-6 was detected by flow cytometry.
Live CD34+Lin- events gated as in Figure 1A were analyzed for IL-6 MFI.
Tables
| Reagent type (species) or resources | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Mycobacterium tuberculosis) | H37Rv | ATCC | ||
| Strain (Mycobacterium tuberculosis) | Mtb-CS267 | Clinical Isolate | This study | |
| Cell Line (Homo sapiens) | Human Cord Blood (CB) purified CD34+ cells | STEMCELL Technologies | Catalog#70008.5 | Cell line maintained in StemSpan II Expansion Media - STEMCELL Technologies- Catalog#09605 |
| Cell (Homo sapiens) | Peripheral blood mononuclear cell (PBMC) | Cells maintened in RPMI 1640 complete, Sigma-Aldrich – Catalog#R8758 | ||
| Chemical Compound, drug | Middlebrook 7H10 agar | BD Biosciences | Catalog# 262710 | Supplemented with 10% Oleic Acid,Albumin, Dextrose, Catalase (Sigma-Aldrich – Catalog# M0678-1VL) |
| Chemical Compound, drug | L-Glutamine (200 mM) | Sigma-Aldrich | Catalog# 25030081 | |
| Chemical Compound, drug | Sodium Pyruvate (100 mM) | Life Technologies. | Catalog# 11360070 | |
| Biological Sample | Genomic DNA, Mycobacterium tuberculosis, Strain H37Rv | This study | Dosage: 10 μg/ml | |
| Chemical Compound, drug | Mycobacterium tuberculosis, Strain H37Rv, Purified Lipoarabinomannan (LAM) | BEI Resources | Catalog# NR-14848 | Dosage: 5 µg/mL |
| Peptide, Recombinant protein | Recombinant Human Interleukin 6 (rh IL-) | ImmunoTools | Catalog# 11340064 | |
| Antibody | FITC Anti-Lineage 1, human: CD3 clone SK7, CD16 clone 3G8, CD19 clone SJ25C1, CD20 clone L27, CD14 clone MφP9, CD56 clone NCAM. | BD Biosciences | Catalog# 340546 | (1:30) |
| Antibody | PE Anti-human CD34, clone 581. | BD Biosciences | Catalog# 555822 | (1:20) |
| Antibody | FITC Anti-human CD34, clone 8G12. | BD Biosciences | Catalog# 345801 | (1:50) |
| Antibody | PerCP Anti-human CD34, clone 581. | Biolegend | Catalog# 343519 | (1:20) |
| Antibody | PECy7 Anti-human HLA-DR, clone L243 | Biolegend | Catalog# 307615 | (1:200) |
| Antibody | Bv510 Mouse Anti-Human HLA-DR, clone G46-6 | BD Horizon | Catalog# 563083 | (1:100) |
| Antibody | APC Anti-human CD38, clone HIT2 | Biolegend | Catalog# 303510 | (1:100) |
| Antibody | Bv421 Anti-Human CD64, clone 10.1. | BD Biosciences | Catalog# 562872 | (1:100) |
| Antibody | FITC Anti-human CD10, clone HI10A | BD Biosciences | Catalog# 340925 | (1:50) |
| Antibody | Alexa Fluor 488 Anti-human CD14, clone M5E2. | Biolegend | Catalog# 301817 | (1:50) |
| Antibody | APCCy7 Anti-mouse/human CD11b, clone M1/70 | Biolegend | Catalog# 101226 | (1:100) |
| Antibody | V450 Anti-human CD14, clone MφP9 | BD Biosciences | Catalog# 560350 | (1:100) |
| Antibody | PE Anti-human CD66b, clone G10F5. | Biolegend | Catalog#305106 | (1:100) |
| Antibody | APCCy7 Anti-human BDCA1, clone L161. | Biolegend | Catalog# 331520 | (1:200) |
| Antibody | FITC Anti-human CD41a, clone 6C9. | ImmunoTools | Catalog# 21330413 | (1:50) |
| Antibody | APC Anti-human BDCA2, clone 201A | Biolegend | Catalog# 354205 | (1:50) |
| Antibody | Bv510 Anti-human BDCA3, clone 1A4. | BD Biosciences | Catalog# 563298 | (1:100) |
| Antibody | PE Anti-human CD123, clone 7G3. | BD Biosciences | Catalog# 554529 | (1:20) |
| Antibody | APC Anti-human CD16, clone 3G8. | BD BiosciencesCatalog# | 561248 | (1:50) |
| Antibody | V450 Anti-human CD64, clone 10.1. | BD Biosciences | Catalog# 561202 | (1:20) |
| Antibody | FITC Anti-human CD3, clone UCHT1. | Biolegend | Catalog# 300440 | (1:100) |
| Antibody | FITC Anti-human CD19, clone 4G7. | BD Biosciences | Catalog# 347543 | (1:50) |
| Antibody | Alexa Fluor 488 Anti-human CD14, clone M5E2. | BD Biosciences | Catalog# 561706 | (1:50) |
| Antibody | PerCP-Cy5.5 Anti-human CD34, clone 8G12. | BD Biosciences | Catalog# 347203 | (1:25) |
| Antibody | PE Anti-human IL-6, clone 8C9. | ImmunoTools | Catalog# 21670064 | (1:10) |
| Antibody | FITC Anti-human CD56, clone NCAM16.2. | BD Biosciences | Catalog# 345811 | (1:100) |
| Antibody | FITC Anti-human CD16, clone HI16a. | ImmunoTools | Catalog# 21810163 | (1:100) |
| Antibody | Monoclonal Anti-STAT1 (phospo Y701), clone M135. | Abcam | Catalog# ab29045 | (1:1000) |
| Antibody | Monoclonal Anti-STAT1, clone SM1. | Abcam | Catalog# ab3987 | (1:1000) |
| Antibody | Polyclonal Anti-C/EBPβ | Santa Cruz Biotechnology | Catalog# sc-150 | (1:250) |
| Antibody | Neutralizing Anti-human IFNAR2, clone MMHAR-2 | PBL Assay Science | Catalog# 21370–1 | Dosage: 1 µg/mL |
| Antibody | Monoclonal, Anti-IFN-γ, clone B27. | ImmunoTools | Catalog# 21853531 | Dosage: 10 µg/mL |
| Antibody | Anti-IL6, Tocilizumab. | Roche | Dosage: 1 µg/mL | |
| Antibody | Monoclonal Anti-beta actin | Abcam | Catalog# mAbcam 8226 | (1:5000) |
| Chemical Compound, drug | Flexible Viability Stain 450 | BD Horizon | Catalog#562247 | (1:1000) |
| Chemical Compound, drug | Carbol Fuchsin | Sigma-Aldrich | Catalog# C4165 | |
| Chemical Compound, drug | Methylene Blue | Sigma-Aldrich | Catalog# 03978 | |
| Chemical Compound, drug | Hoechst 33342 | Immunochemistry technologies | Catalog# 639 | |
| Commercial assay, kit | M-PER Mammalian Protein Extraction Reagent | Thermo Fisher Scientific | Catalog# 78501 | |
| Commercial assay, kit | cOmplete ULTRA Tablets, Mini, EASYpack Protease Inhibitor Cocktail | Roche | Catalog# 05 892970001 | |
| Commercial assay, kit | High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Catalog# 4368814 | |
| Chemical Compound, drug | TRIzol LS Reagent | Invitrogen | Catalog# 10296010 | |
| Commercial assay, kit | NuGEN - Trio low input RNA-seq | NuGEN | Catalog#0507–08 | |
| Software, algorithm | FlowJo software v. 10.1 | TreeStar | FlowJo, RRID:SCR_008520_ | https://www.flowjo.com/ |
| Software, algorithm | GraphPad Prism 6 Software | GraphPad | GraphPad Prism, RRID:SCR_002798 | https://www.graphpad.com/ |
Additional files
-
Supplementary file 1
Reactome Pathways analysis of Mtb-exposed and control CD34+ cell transcriptomes.
- https://doi.org/10.7554/eLife.47013.028
-
Supplementary file 2
Systems analysis (Ingenuity Pathway Analysis and Gene Set Enrichment Analysis) of cohorts of healthy controls, patients with latent TB, active TB, disseminated TB, overlap with IL6/STAT3 signaling and myeloid development.
- https://doi.org/10.7554/eLife.47013.029
-
Supplementary file 3
Human adaptation z-scores for IL6/IL6R/CEBP CD34 myeloid gene module and Gene set enrichment of Top500 human adaptation genes.
- https://doi.org/10.7554/eLife.47013.030
-
Supplementary file 4
List of statistical methods used in the manuscript.
- https://doi.org/10.7554/eLife.47013.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47013.032