Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection
Figures
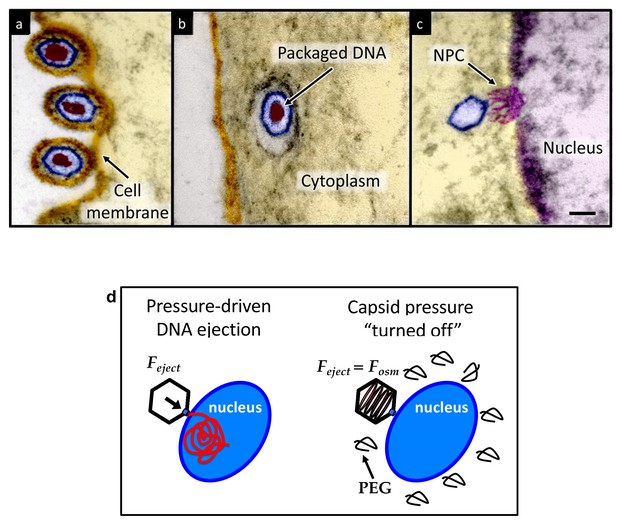
Ultrathin-sectioning EM visualization of the HSV-1 infection process showing viral DNA ejection from HSV-1 capsid into a host nucleus.
Ultrathin Epon sections of Vero cells infected with HSV-1 at an MOI of 300 PFU/cell. Artificially colored electron micrographs of HSV-1 at the cell membrane (A), in transport to the nucleus (B), and bound at a nuclear pore complex (NPC) embedded within the nuclear envelope (C). The dsDNA genome appears as an electron-dense region within the capsid, which is visible in (A) and (B), but absent in (C) due to DNA ejection upon NPC binding. Scale bar, 50 nm. Adapted for clarity from our earlier publication (Bauer et al., 2013). (D) Illustration of the osmotic suppression experiment. DNA ejection from a virus capsid into a reconstituted host nucleus is completed successfully without osmolyte addition. However, viral DNA ejection is fully suppressed, when the capsid pressure is ‘turned off’ with an external osmotic pressure, created by PEG, that matches the pressure of the packaged DNA in the capsid.
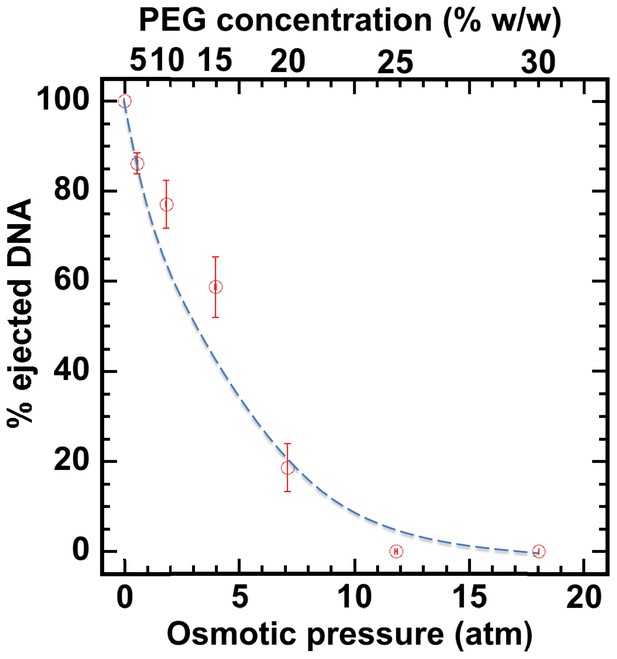
Percentage of viral genome ejected from HSV-1 capsids as a function of the external osmotic pressure.
DNA ejection from the capsids in vitro is triggered by mild trypsin treatment, which cleaves the portal protein (UL6) without degrading the major capsid protein (VP5) or causing morphological damage to capsids (Bauer et al., 2013). Figure shows that DNA ejection is progressively suppressed with increasing PEG concentration at 37°C with PEG 8 kDa. DNA ejection is completely suppressed at 18 atm external osmotic pressure, which matches and therefore ‘turns off’ the DNA pressure in the capsid. PEG is added to CBB buffer (capsid binding buffer: 20 mM HEPES-KOH with pH of 7.3, 80 mM K-acetate, 2 mM DTT, 1 mM EGTA, 2 mM Mg-acetate, 1 mM PMSF, and 1X CLAP cocktail), required for capsid binding to nuclei. (PEG concentration was converted to osmotic pressure using the relation in ref Evilevitch et al., 2003). Vertical error bars represent the standard error of the gel band intensity profile (see Materials and methods Section). Horizonal error bars representing standard error in weighted PEG concentration are negligibly small. Dashed line is drawn to guide the eye.
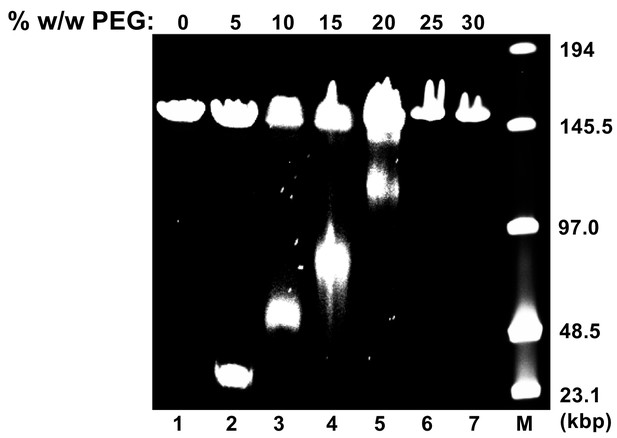
Osmotic suppression of HSV-1 genome ejection.
DNA ejection from HSV-1 capsids is initiated by trypsin digestion in the presence (or absence) of PEG 8 kDa and DNase I at 37°C in CBB buffer. Nonejected DNA was extracted from capsids by sodium dodecyl sulfate (SDS) and protease K treatment and analyzed by pulse field gel electrophoresis (PFGE). PFGE of osmotically suppressed DNA remaining inside viral capsids in the presence of varying concentrations of PEG shown in lanes 2–7. Lane M shows DNA ladder. For all experiments, DNA within unopened capsids resulted in an additional band of approximately 151 kbp corresponding to full length HSV-1 genome. Similar results were obtained in three independent experiments and a representative experiment is shown.
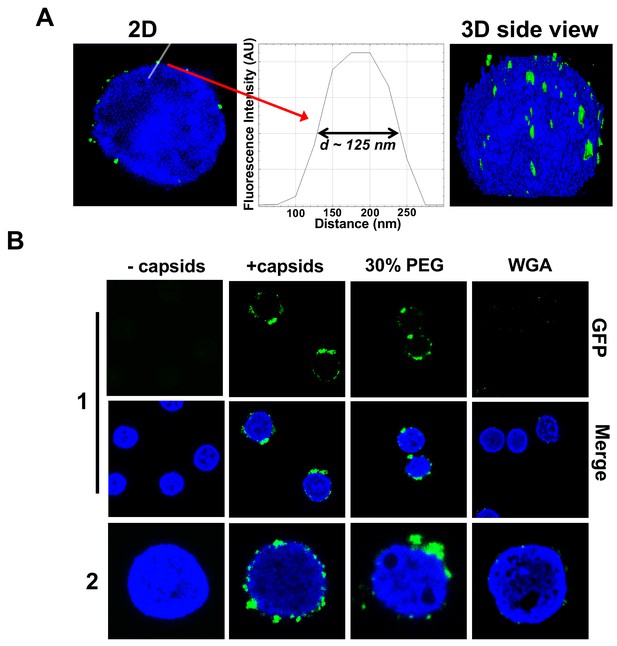
Imaging of reconstituted capsid-nuclei system confirms specific capsid binding to the NPCs at the nuclear membrane with and without PEG 8 kDa present.
(A) Representative super-resolution SIM image showing GFP-HSV-1 C-capsids (green) bound to isolated reconstituted rat liver nuclei (blue DAPI stain). A histogram of a capsid cross-section profile for a capsid GFP signal along the white line shows that individual C-capsids are resolved (HSV-1 C-capsid diameter ≈ 125 nm). (B) Confocal fluorescence microscopy images show that binding of GFP-HSV-1 C-capsids (green) to DAPI-stained isolated nuclei (blue), in the presence of cytosol (no ATP-regeneration system was added since it is not required for capsid binding [Ojala et al., 2000]), is not inhibited by the addition of 30% w/w PEG 8 kDa. The addition of wheat germ agglutinin (WGA) prevents most of the capsid binding to nuclei, which demonstrates that capsids bind specifically to NPCs as opposed to binding anywhere on the nuclear membrane [WGA associates with the specific glycoproteins within the NPC and competes with capsid binding (Ojala et al., 2000; Finlay et al., 1987)]. The images at the bottom of row two are a zoom-in of the individual nuclei.
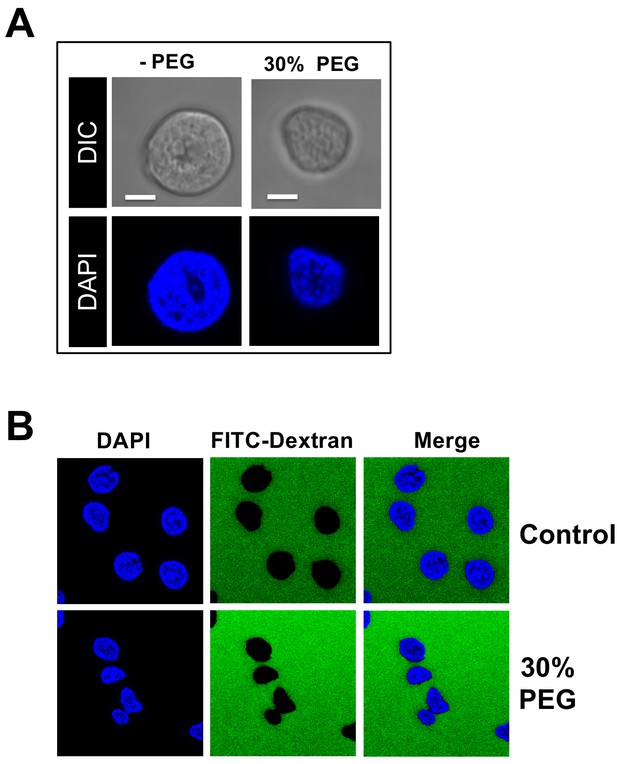
Effect of osmotic pressure on morphology and permeability of nuclei.
(A) Purified rat liver nuclei incubated in CBB buffer containing 30% w/w PEG 8 kDa. Nuclei were stained with DAPI (blue) and visualized by confocal fluorescence microscopy (second row). A DIC image of each field was also obtained (first row). Scale bar in all images: 5 μm. Under hyperosmotic conditions (~18 atm), the nuclei slightly shrunk but the sub-nuclear structure of heterochromatin DNA was essentially unchanged. (B) Isolated rat liver nuclei were incubated at 37°C for 40 min in CBB buffer containing a fluorescently tagged 70 kDa dextran (FITC-Dextran, green) and 30% w/w PEG 8 kDa. The nuclei were stained with DAPI (blue). The integrity of the nuclei was not affected by the addition of PEG 8 kDa since fluorescently labeled 70 kDa dextran was excluded from the nuclei interior with nuclei remaining intact and structured. Representative images are shown.
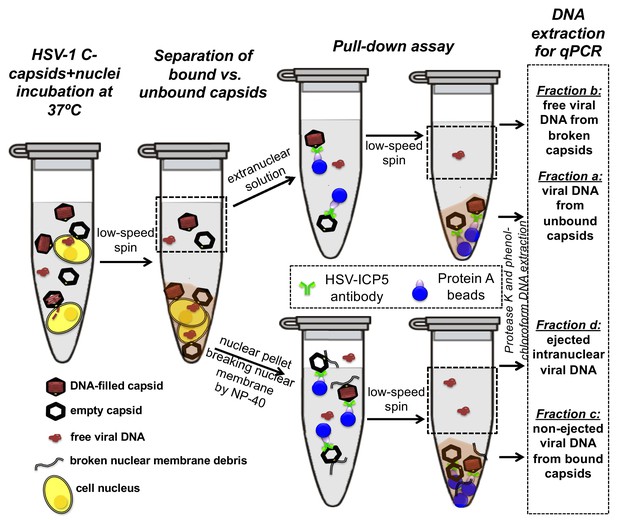
Schematic of the pull-down assay for quantification of the amount of DNA injected from HSV-1 capsids into cell nuclei when the capsid pressure is 'on' or 'off', modulated by PEG addition.
HSV-1 C-capsids were incubated with reconstituted rat liver cell nuclei in CBB buffer, with and without 30% w/w PEG 8 kDa (osmolyte is not shown in the sketch). This pull-down assay successfully separates four fractions of HSV-1 DNA originating from: (a) DNA extracted from capsids that failed to bind to NPCs, (b) free DNA in the extranuclear solution from broken capsids, (c) DNA retained inside the capsids that were bound to nuclei but did not eject DNA, and (d) DNA ejected from capsids into the nucleoplasm. Viral DNA in each fraction was further purified using phenol-chloroform extraction prior to qPCR quantification. Note that the anti-HSV-1/2 ICP5 antibody attached to Protein A beads for the immunoprecipitation step has multiple binding sites on the capsid but only one antibody bound to a capsid is shown for clarity of presentation. Nuclear chromosomal DNA present in fraction c is not shown.
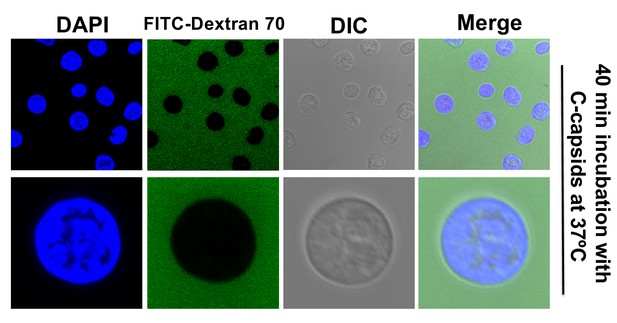
The integrity of isolated nuclei is not disrupted by DNA ejection from bound HSV-1 capsids.
Reconstituted rat liver nuclei were incubated with HSV-1 C-capsids at 37°C for 40 min in CBB buffer, allowing DNA ejection from capsids into nuclei, in the presence of fluorescently labeled 70 kDa dextran (green). The nuclei were stained with DAPI (blue). DIC images of each field are also shown. Complete dextran exclusion demonstrates that viral DNA ejection into nuclei does not affect the integrity of the nuclei.
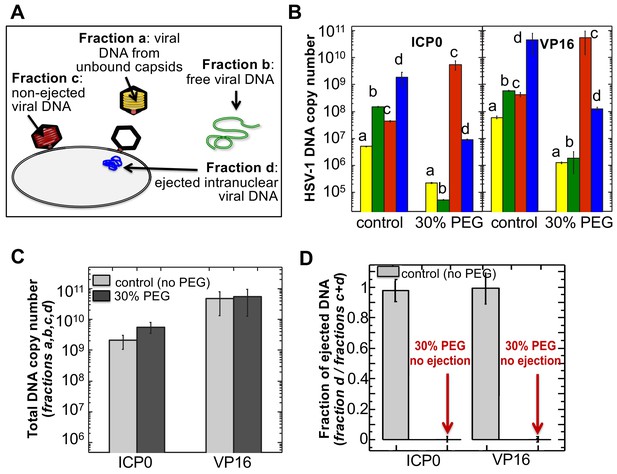
The amounts of HSV-1 DNA released into the nucleoplasm from capsids bound to NPCs quantified by qPCR after extraction from each fraction (a,b,c, and d).
Specific HSV-1 primers for VP16/UL48 and ICP0 genes were used as amplicons for the qPCR assays of viral DNA copy numbers.(A). Schematic of viral DNA fractions separated from the reconstituted capsid-nuclei system with pull-down assay. (B) Histograms show the DNA copy number for each of the four viral DNA-containing fractions. (C) Histograms show the total DNA copy number from fractions a, b, c, and d in the reconstituted capsid-nuclei system. (D) Fraction of DNA ejected from nuclei-bound capsids (fraction d/fractions c+d). After nuclei incubation with HSV-1 C-capsids at 37°C for 40 min, without osmolyte addition,~98% of all nuclei bound capsids ejected their DNA into nuclei. When the capsid pressure is turned off at 18 atm of external osmotic pressure (generated by addition of 30% w/w PEG 8 kDa), the ejection of DNA from capsids bound to nuclei is completely suppressed (fraction d/fractions c+d ~ 0.2%). qPCR DNA copy number quantification is based on a standard curve generated by serial dilution of a wild-type HSV-1 DNA with known DNA copy number. ICP0 and VP16 HSV-1 genes were quantified using specific primers. Error bars in B are standard deviations in DNA copy numbers from three independent qPCR reactions repeated at the same conditions. Error bars in C and D are progressed standard deviations.
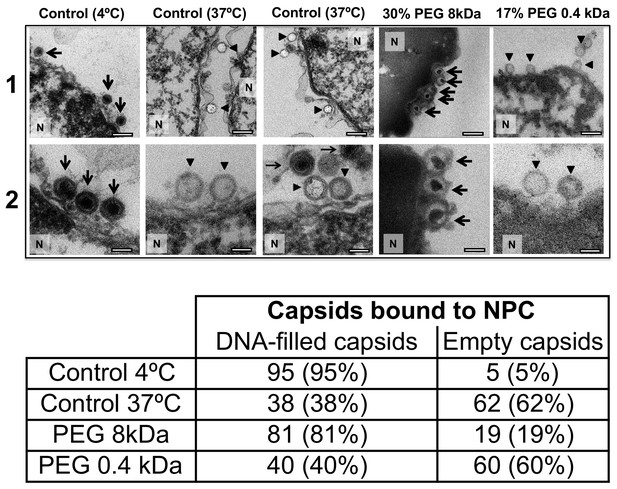
Ultrathin-sectioning EM visualization of complete osmotic suppression of DNA ejection from HSV-1 capsids into reconstituted cell nuclei when capsid pressure is 'turned off' by 18 atm osmotic pressure generated by PEG 8 kDa.
Negative control at 4°C without added PEG and without ATP-regenerating system, shows that no ejection from nuclei bound C-capsids occurs. Positive control at 37°C shows complete DNA ejection from C-capsids bound to isolated cell nuclei supplemented with cytosol and ATP-regenerating system. EM images show that capsids can bind to the nuclear membrane as individual capsids or in multilayer clusters. Consequentially, only capsids in the first layer that are bound to the NPCs are able to eject their DNA. EM shows that the addition of 30% PEG 8 kDa to reconstituted capsid-nuclei system inhibits DNA ejection from HSV-1 C-capsids into host nuclei through the NPC. In all samples, capsids and nuclei were incubated for 40 min. Thin arrows show DNA-filled capsids, and bold arrows show empty capsids that ejected DNA. 1. Bar 500 nm. 2. Bar 90 nm. Representative EM images are shown. At least 100 capsids bound to NPCs were counted for each sample’s statistical analysis, shown in the table below.
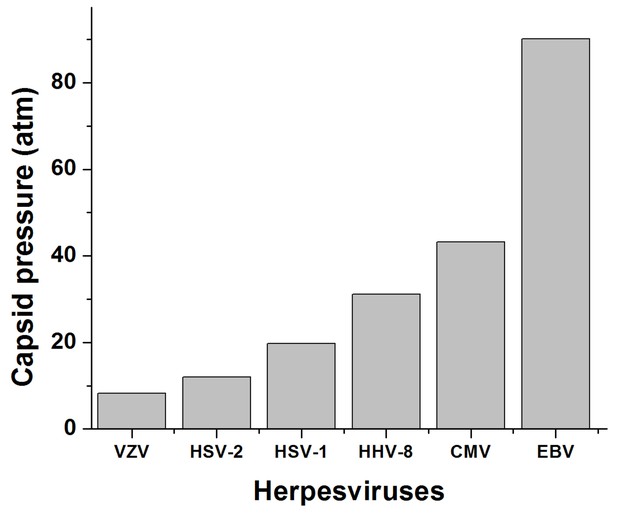
Calculated DNA pressures in herpesvirus capsids.
The analytical expression used to calculate pressures inside virus capsids is based on a theoretical model from refs. (Grayson et al., 2006; Purohit et al., 2005; Purohit et al., 2003; Tzlil et al., 2003) that involves forces generated by DNA-DNA electrostatic repulsive force and DNA bending stress, which depend on the DNA packing density. DNA packing density in the capsid was estimated with values for packaged DNA length and inner capsid radius derived from previous Cryo-EM data (Booy et al., 1991; Germi et al., 2012).
Additional files
-
Supplementary file 1
qPCR data for DNA copy numbers in each fraction obtained with the pull-down assay.
*The efficiency of pull-down assay was separately assessed by DNase protection method. This was shown by adding DNase I to Fraction b after immunoprecipitation step. DNase I digests all free DNA from broken capsids, but DNA inside the capsids, which were not immunoprecipitated, remains intact (DNase does not permeate the capsid shell). Protease K and phenol-chloroform extraction release the encapsidated DNA, which is then quantified with qPCR and reflects the number of capsids that were not pulled-down. This fraction constitutes only 1–6% of the total HSV-1 DNA amount in Fraction b prior to immunoprecipitation. Standard deviations in DNA copy numbers were obtained from three independent qPCR reactions repeated at the same conditions for each sample.
- https://doi.org/10.7554/eLife.47212.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47212.013





