PUMILIO, but not RBMX, binding is required for regulation of genomic stability by noncoding RNA NORAD
Figures
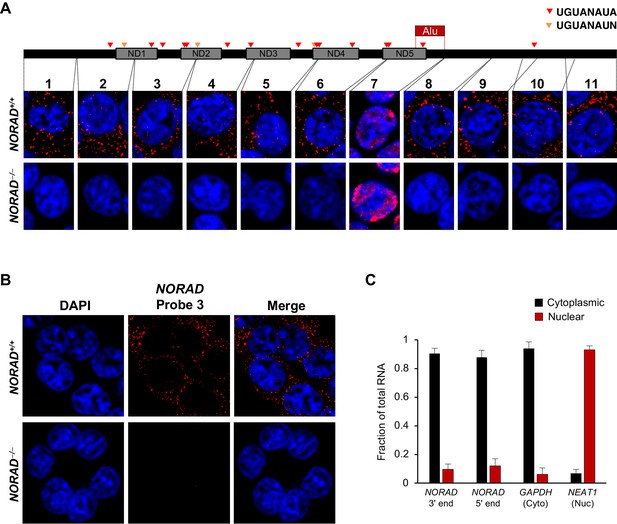
NORAD localizes predominantly to the cytoplasm.
(A) RNA FISH in HCT116 cells using a panel of 11 probes tiling the entire NORAD transcript reveals a predominantly cytoplasmic signal that is absent in NORAD–/– cells with all probes except probe 7, which produces a nonspecific signal likely due to the presence of an Alu repeat element. NORAD FISH signal in red, DAPI counterstain in blue. Locations of PREs indicated by arrowheads. ND1-ND5 represent repetitive NORAD domains, as previously described (Lee et al., 2016). (B) RNA FISH image using probe 3 showing a wider field of cells. (C) Subcellular fractionation followed by qRT-PCR in HCT116 cells using primers located at the 3′ or 5′ end of NORAD, in GAPDH (cytoplasmic control), or in NEAT1 (nuclear control). n = 3 biological replicates each with three technical replicates.
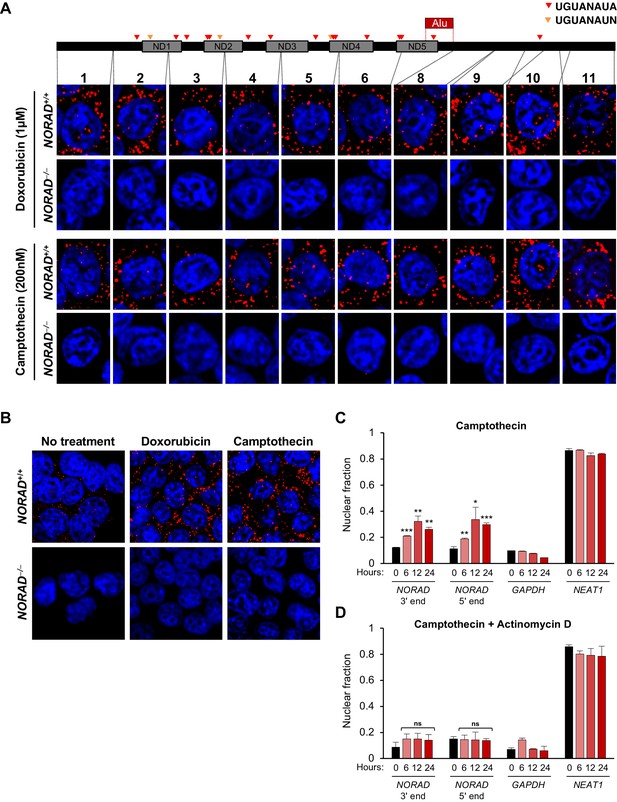
NORAD remains predominantly in the cytoplasm after treatment with DNA damaging agents.
(A) RNA FISH in HCT116 cells using the indicated NORAD probes following a 12 hr treatment with doxorubicin or camptothecin. (B) NORAD RNA FISH (probe 5) after the indicated drug treatments. Images captured with identical microscope settings. (C–D) Subcellular fractionation followed by qRT-PCR in HCT116 cells after treatment with camptothecin (C) or camptothecin plus actinomycin D (D) for the indicated number of hours. n = 3 biological replicates each with three technical replicates. ns, not significant; *p<0.05; **p<0.01; ***p<0.001; one-tailed t-test comparing each sample to the 0 hr time-point.
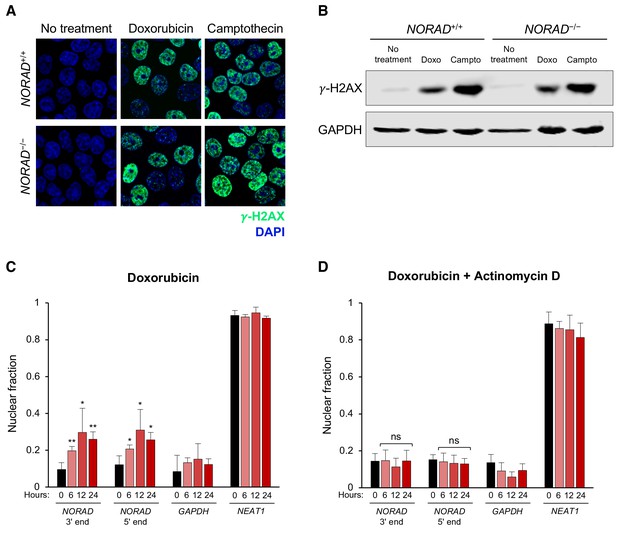
NORAD remains predominantly cytoplasmic following doxorubicin-induced DNA damage.
(A–B) Immunofluorescence (A) or western blot (B) analysis of the DNA damage marker γ-H2AX in HCT116 cells treated with or without doxorubicin (1 μM) or camptothecin (200 nM) for 12 hr. (C–D) Subcellular fractionation followed by qRT-PCR in HCT116 cells after treatment with doxorubicin (C) or doxorubicin plus actinomycin D (D) for the indicated number of hours. n = 3 biological replicates each with three technical replicates. ns, not significant; *p<0.05; **p<0.01; one-tailed t-test comparing each sample to the 0 hr time-point.
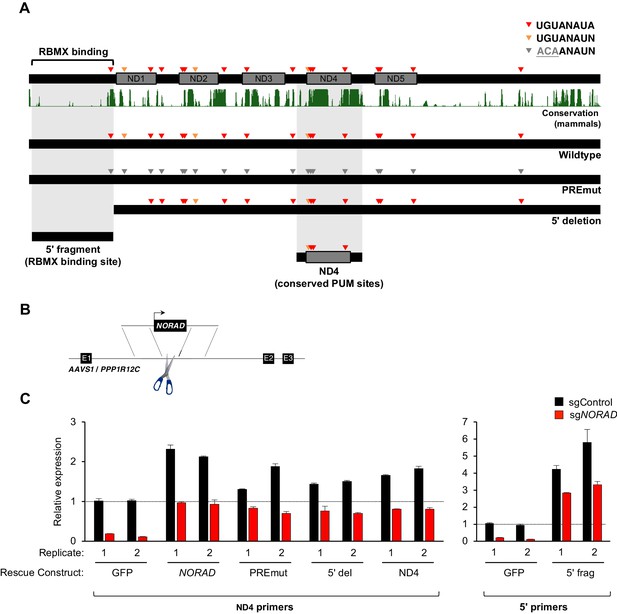
Generation and stable expression of NORAD constructs.
(A) Schematic depicting wild-type or mutant NORAD constructs. NORAD sequence conservation in mammals (UCSC Genome Browser Hg38 PhastCons track) highlights the strong conservation of the region of NORAD harboring PREs (arrowheads). PREmut contains 18 UGU to ACA mutations in PREs (gray arrowheads); 5′ deletion (5′ del) lacks the RBMX binding site (nt 1–898) (Munschauer et al., 2018); 5′ fragment (5′ frag) spans the RBMX binding site (nt 33–898); ND4 construct represents the most conserved segment of NORAD (nt 2494–3156). (B) Schematic depicting insertion of constructs into the AAVS1/PPP1R12C locus using TALENs. (C) qRT-PCR analysis of expression of each NORAD construct in HCT116 CRISPRi cells after infection with control or endogenous NORAD-targeting sgRNAs. Expression was normalized to endogenous NORAD level, represented by expression in AAVS1-GFP cells infected with sgControl (replicate 1 samples normalized to sgControl AAVS1-GFP replicate 1; replicate 2 samples normalized to sgControl AAVS1-GFP replicate 2). The data in the left graph were generated with a primer pair in ND4 that does not amplify the 5′ fragment, while the right graph used primers at the NORAD 5′ end. Replicates represent two independently-derived AAVS1 knock-in and sgRNA-infected cell lines. Values normalized to GAPDH expression. n = 3 technical replicates per sample.
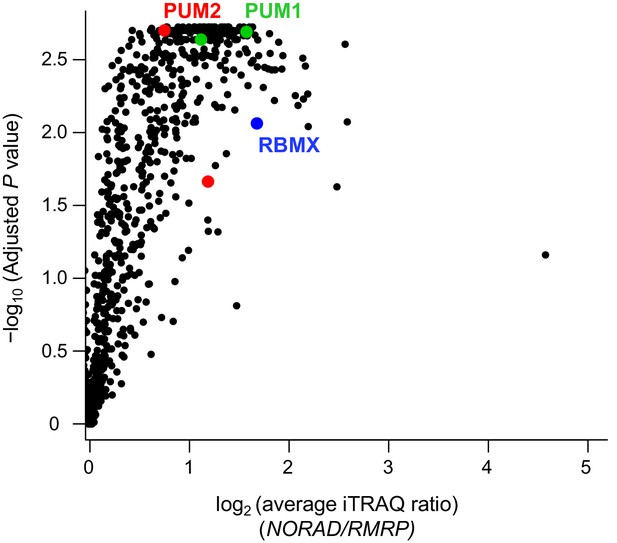
Reanalysis of NORAD RAP-MS data.
Analysis of previously published NORAD RAP-MS data (Munschauer et al., 2018) using a combined three search engine algorithm (MS Amanda, Sequest HT, Mascot) identifies isoforms of PUM1 (green), PUM2 (red), and RBMX (blue) as significantly enriched NORAD interactors. Volcano plot showing the average fold-change compared to control RMRP pull-down and significance from two biological replicates.
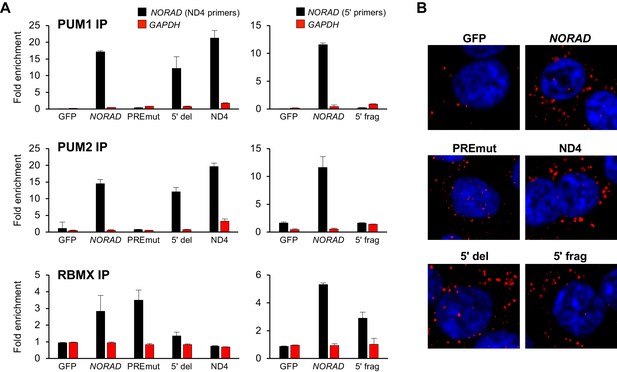
RNA immunoprecipitation and localization of NORAD constructs.
(A) UV crosslinking and RNA immunoprecipitation (RIP) was used to assess PUM1, PUM2, and RBMX interactions with GFP mRNA or the indicated NORAD constructs. After knock-in of the indicated constructs to the AAVS1 locus in HCT116 CRISPRi cells, endogenous NORAD was silenced with a lentivirally-expressed sgRNA. qRT-PCR was used to assess NORAD or GAPDH recovery in each RIP sample, expressed as fold-enrichment over pull-down with IgG. The data in the left graphs were generated with a primer pair in ND4 that does not amplify the 5′ fragment, while the right graphs used primers at the NORAD 5′ end. n = 2 biological replicates, each measured with three technical replicates. (B) Representative RNA FISH images of wild-type or mutant NORAD transcripts expressed from the AAVS1 locus in HCT116 CRISPRi cells after knockdown of endogenous NORAD. Probe 10 was used for full-length NORAD, PREmut, and 5′ del constructs; probe 1 was used for 5′ frag; and probe 6 was used for ND4.

Representative western blots of PUM1, PUM2, and RBMX in RIP experiments.
PUM1 migrates as a doublet in HCT116, possibly indicating post-translational modification or alternative splicing. We have confirmed that both bands represent PUM1 protein using siRNA knockdown experiments (data not shown).
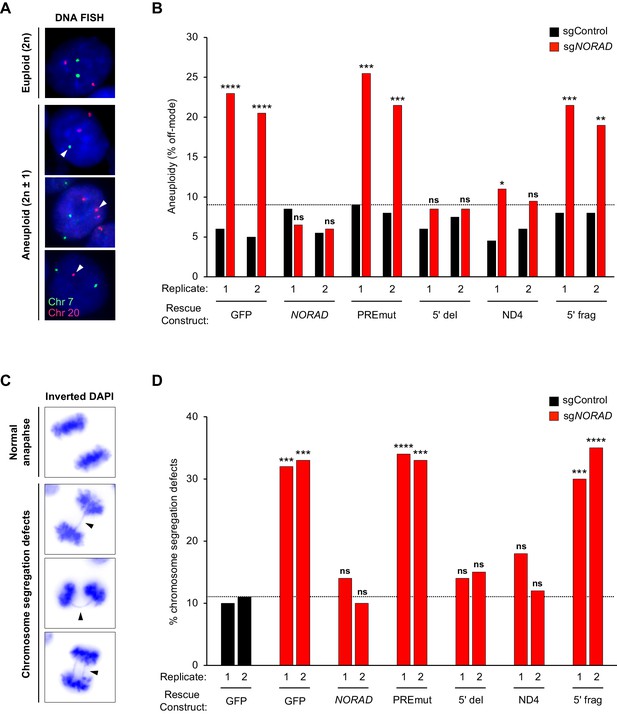
PUMILIO, but not RBMX, binding to NORAD is necessary for genome stability.
(A) Representative DNA FISH images for chromosome 7 (green) and 20 (red) showing examples of cells with modal (2 n) and non-modal (2n ± 1) chromosome numbers. Arrowheads indicate chromosome gain or loss. (B) HCT116 CRISPRi cells stably expressing the indicated AAVS1 knock-in construct were infected with lentivirus expressing control or endogenous NORAD-targeting sgRNA. Aneuploidy was assayed 18–21 days later using DNA FISH for chromosome 7 and 20, and the frequency of interphase cells exhibiting a non-modal (2 n) chromosome number was scored. Replicates represent two independently-derived AAVS1 knock-in and sgRNA-infected cell lines. 200 nuclei were scored per sample. The dotted line denotes the highest level of background aneuploidy observed in sgControl-infected cells. ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001, chi-square test comparing sgNORAD to sgControl for each replicate. (C) Representative images of anaphase cells with normal or abnormal (arrowheads) chromosome segregation in DAPI-stained HCT116 CRISPRi cells. (D) The frequency of mitotic cells exhibiting chromosome segregation defects was determined in both biological replicates of each cell population (100 anaphase cells assayed per sample). The dotted line denotes the highest percentage of chromosome segregation defects observed in sgControl-infected cells. ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001, chi-square test comparing all sgNORAD replicate 1 samples to sgControl GFP replicate 1 and all sgNORAD replicate 2 samples to sgControl GFP replicate 2.
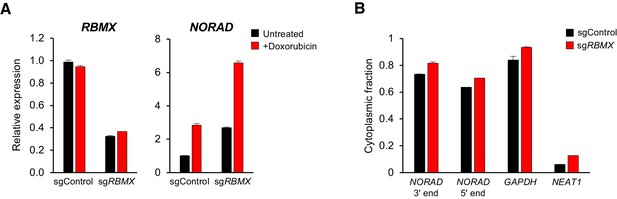
RBMX is not required for NORAD expression or localization.
(A) qRT-PCR analysis of RBMX and NORAD transcript levels in HCT116 CRISPRi cells after introduction of the indicated lentivirally-expressed sgRNA with or without doxorubicin treatment (1 μM for 24 hr). Quantification relative to GAPDH. n = 3 technical replicates. (B) Subcellular fractionation and qRT-PCR of NORAD, GAPDH (cytoplasmic control), or NEAT1 (nuclear control) following introduction of control or RBMX-targeting sgRNAs. n = 3 biological replicates each with three technical replicates.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | NORAD (LINC00657) | NA | Ensembl:ENSG00000260032 | |
| Cell line (Homo sapiens) | HCT116 | ATCC | CCL-247, RRID:CVCL_0291 | |
| Cell line (Homo sapiens) | NORAD-/- HCT116 | Lee et al., 2016 | ||
| Cell line (Homo sapiens) | HCT116 CRISPRi | this paper; see Materials and methods section Cell culture and generation of HCT116 CRISPRi cell line | ||
| Antibody | Anti-PUM1 (polyclonal rabbit) | Santa Cruz | sc-135049, RRID:AB_10610604 | RIP |
| Antibody | Anti-PUM2 (polyclonal goat) | Santa Cruz | sc-31535, RRID:AB_654939 | RIP |
| Antibody | Anti-RBMX (monoclonal rabbit) | Cell Signaling | #14794, RRID:AB_2798614 | RIP, WB (1:1000) |
| Antibody | Anti-PUM2 (monoclonal rabbit) | Abcam | ab92390, RRID:AB_10563318 | WB (1:1000) |
| Antibody | Anti-PUM1 (monoclonal rabbit) | Abcam | ab92545, RRID:AB_10563695 | WB (1:1000) |
| Antibody | Anti-GAPDH (monoclonal rabbit) | Cell Signaling | #2118, RRID:AB_561053 | WB (1:5000) |
| Antibody | Anti-phospho-histone H2A.X (Ser139) | Cell Signaling | #2577, RRID:AB_2118010 | IF (1:500), WB (1:1000) |
| Antibody | IRDye 800CW anti-rabbit (donkey) | Licor | 925–32213, RRID:AB_2715510 | WB (1:10000) |
| Antibody | Anti-Digoxigenin (monoclonal mouse) | Roche | 11333062910, RRID:AB_514495 | RNA FISH |
| Antibody | Anti-Mouse IgG, Cy3 (polyclonal goat) | EMD Millipore | AP124C | RNA FISH |
| Recombinant DNA reagent | AAVS1/PPP1R12C targeting vector | Addgene | #22072, RRID:Addgene_22072 | |
| Recombinant DNA reagent | hAAVS1 1L TALEN; hAAVS1 1R TALEN | Addgene | #35431, RRID:Addgene_35431; #35432, RRID:Addgene_35432 | |
| Recombinant DNA reagent | pU6-sgRNA EF1a-PuroR-T2A-BFP | Addgene | #60955, RRID:Addgene_60955 | CRISPRi-mediated knockdown |
| Sequence-based reagent | Chromosome enumeration Probe Chr. 7 (green) | Empire Genomics | CHR07-10-GR | DNA FISH |
| Sequence-based reagent | Chromosome enumeration Probe Chr. 20 (red) | Empire Genomics | CHR20-10-RE | DNA FISH |
| Software, algorithm | Prism 7 | GraphPad Software | ||
| Software, algorithm | Proteome Discoverer | Thermo Fisher | ||
| Software, algorithm | Limma package for R | Smyth, 2004 |
Additional files
-
Supplementary file 1
Sequences of oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.48625.011
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48625.012






