Synaptic mitochondria regulate hair-cell synapse size and function
Figures
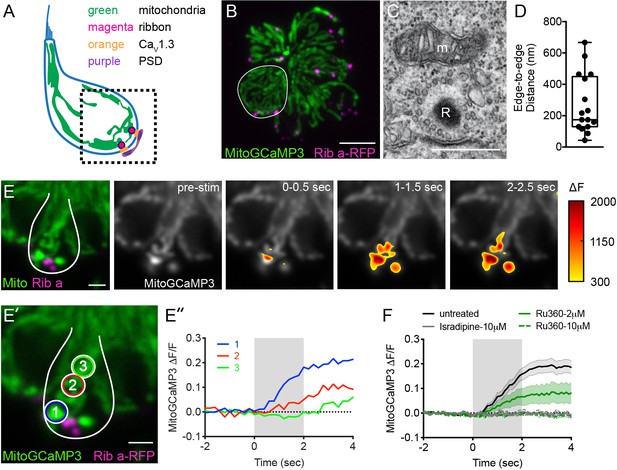
Mito-Ca2+ uptake initiates adjacent to ribbons.
(A) Cartoon illustration of a lateral-line hair cell containing: an apical mechanosensory bundle (blue), mitochondria (green), presynaptic ribbons (magenta), CaV1.3 channels (orange) and postsynaptic densities (purple). (B) Airyscan confocal image of 6 live hair cells (1 cell outlined in white) expressing MitoGCaMP3 (mitochondria) and Ribeye a-tagRFP (ribbons) in a developing neuromast at 2 dpf. Also see Figure 1—figure supplement 1. (C) A representative TEM showing a mitochondrion (m) in close proximity to a ribbon (R) at 4 dpf. (D) Quantification of mitochondrion to ribbon distance in TEM sections (n = 17 ribbons). (E) Side-view of a hair cell (outlined in white) shows the spatio-temporal dynamics of evoked mito-Ca2+ signals during a 2 s stimulation at 6 dpf. The change in MitoGCaMP3 signal (∆F) from baseline is indicated by the heatmap and are overlaid onto the pre-stimulus grayscale image. (E’-E’’) Circles 1–3 (1.3 μm diameter) denote regions used to generate the normalized (∆F/F0) temporal traces of mito-Ca2+ signals in E’’: adjacent to the presynapse (‘1’), and midbody (‘2’ and ‘3’) in the same cell as E. (F) Average evoked mito-Ca2+ response before (solid black) and after 30 min treatment with 10 μM Ru360 (dashed green), 2 μM Ru360 (solid green), or 10 μM isradipine (gray) (3–5 dpf, n ≥ 9 cells per treatment). Error bars in D are min and max; in F the shaded area denotes SEM. Scale bar = 500 nm in C, 5 µm in B and 2 µm in E and E’.
-
Figure 1—source data 1
Summary of quantified TEM data and mito-Ca2+ trace data.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig1-data1-v2.xlsx
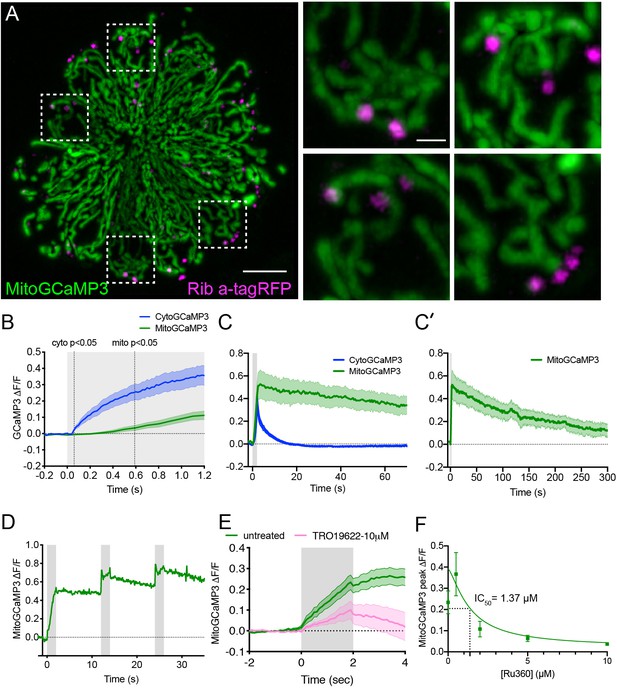
The time course of mechanically-evoked mito-Ca2+ signals are longer-lasting than cyto-Ca2+ signals.
(A) Airyscan confocal image of a live neuromast expressing MitoGCaMP3 (mitochondria) and Ribeye a-tagRFP (ribbons) at 6 dpf. Insets show the base of 4 individual hair cells from the neuromast in A (dashed white boxes). (B) Average cyto-Ca2+ (blue) and mito-Ca2+ (green) ∆F/F GCaMP3 signals during the onset of a 2 s stimulus. Mito-Ca2+ signals rise with a delay compared to cyto-Ca2+ signals, 3–6 dpf, n ≥ 18 cells. (C-C’) Average cyto-Ca2+ and mito-Ca2+ ∆F/F GCaMP3 signals during and after a 2 s stimulation shows that cyto-Ca2+ signals return to baseline shortly after stimulation (C) while mito-Ca2+ remains elevated up to 5 min after stimulation (C–C’), 3–6 dpf, n ≥ 7 cells. (D) A series of 3, evoked 2 s stimuli initiated at: t = 0–2, 12–14 and 24–26 s. A rise in MitoGCaMP3 can be detected during each stimulus, prior to MitoGCaMP3 signals returning to baseline. (E) 10 µM of the VDAC inhibitor TRO 19622 partially blocks evoked MitoGCaMP3 signals, 5 dpf, n = 15 cells. (F) A dose response curve indicates that Ru360 blocks evoked MitoGCaMP3 signals with an IC50 value of 1.37 µM at 5 dpf, n ≥ 9 cells per dose. Error in panel (B-C’) E and F represent SEM. Scale bar = 5 µm in A and 2 µm in inset.
-
Figure 1—figure supplement 1—source data 1
Summary CytoGCaMP3 and MitoGCaMP3 traces and MitoGCaMP3 data used to generate Ru360 dose response curve.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig1-figsupp1-data1-v2.xlsx
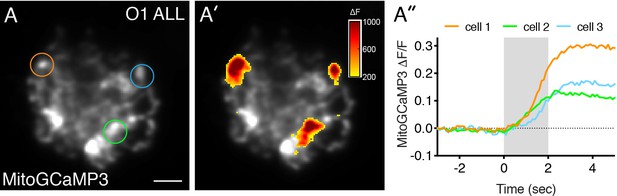
Mito-Ca2+ uptake occurs in anterior lateral-line hair cells.
(A) A live Image of an anterior-lateral line (ALL) neuromast viewed top-down, expressing the mito-Ca2+ sensor MitoGCaMP3 at 5 dpf. (A’) shows the spatio-temporal dynamics of evoked mito-Ca2+ signals during a 2 s stimulation. The MitoGCaMP3 signals during the stimulation (∆F) are indicated by the heatmap overlaid onto the baseline grayscale image. (A’’) Temporal traces of evoked mito-Ca2+ signals were generated from three regions denoted by three circles in A. Scale bar = 5 µm in A.
-
Figure 1—figure supplement 2—source data 1
MitoGCaMP3 traces in anterior lateral line hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig1-figsupp2-data1-v2.xlsx
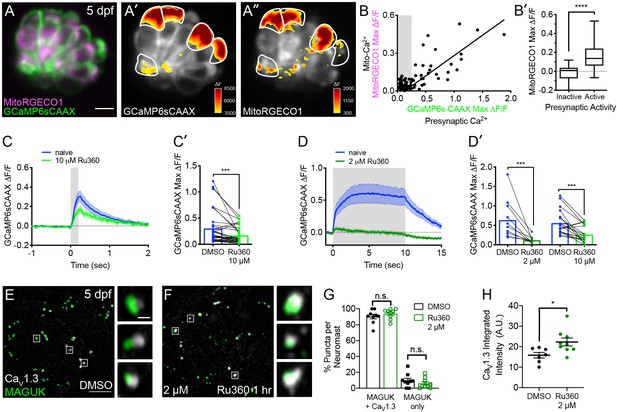
Mito-Ca2+ uptake can impact presynaptic-Ca2+ signals.
(A) A live Image of a neuromast viewed top-down, expressing the presynaptic-Ca2+ sensor GCaMP6sCAAX (green) and mito-Ca2+ sensor MitoRGECO1 (magenta) at 5 dpf. A’-A’’, GCaMP6sCAAX (A’) and MitoRGECO1 (A’’) signals (∆F) from baseline during a 2 s stimulation are indicated by the heatmaps and occur in the same cells (white outline). (B) Scatter plot with linear regression of peak presynaptic- and mito-Ca2+ response for individual cells at 4–5 dpf, n = 136 cells. Gray background in graph denotes presynaptic-Ca2+ signals below 0.25, a threshold used as a cutoff for presynaptic activity (below inactive, above active). (B’) Plot of mito-Ca2+responses segregated based on the activity threshold in B. (C-D’) Presynaptic-Ca2+ response (example in Figure 2—figure supplement 1C–C’) averaged per cell before (blue) and after a 30 min treatment with 10 μM Ru360 (light green) or 2 μM Ru360 (dark green), n ≥ 10 cells per treatment. C and D show averaged traces while C’ and D’ show before-and-after dot plots of the peak response per cell. (E-F) Representative images of mature neuromasts (5 dpf) immunostained with CaV1.3 (white, calcium channels) and MAGUK (green, postsynapses) after a 1 hr incubation in 0.1% DMSO (E) or 2 μM Ru360 (F). G-H, Scatter plots show percentage of postsynapses that pair with CaV1.3-channel clusters (CaV1.3 + MAGUK) and orphan postsynapses (MAGUK only) (G). The integrated intensity of CaV1.3-channel immunolabel at presynapses is lower in control compared to treatment group (H), n ≥ 7 neuromasts per treatment. Whiskers on plots in B’ represent min and max; the shaded area in plots C and D and the error bars in C’, D’ and G-H denotes SEM. Mann-Whitney U test was used in B’; Wilcoxon matched-pairs signed-rank test was used in C’ and D’. Welch’s unequal variance t-test was used in G-H. *p<0.05, ***p<0.001, ****p<0.0001. Scale bar = 5 µm in A and E.
-
Figure 2—source data 1
Summary of MitoRGECO1 and GCaMP6sCAAX and synapse quantification data.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig2-data1-v2.xlsx
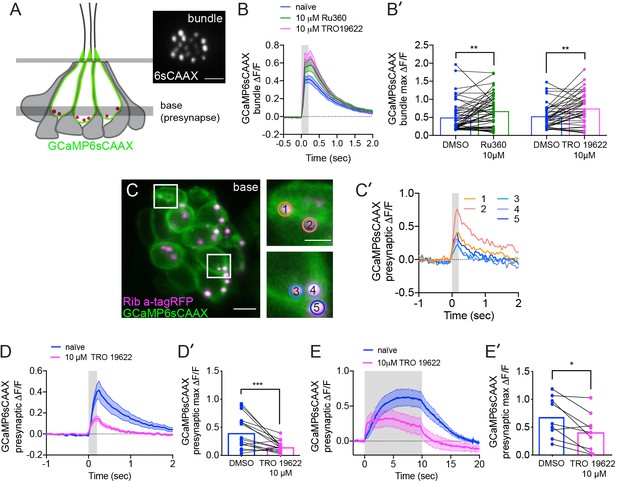
The effects of MCU and VDAC block on mechanotransduction and the effect of VDAC block on presynaptic-Ca2+ signals.
(A) Illustration of a neuromast and the imaging planes used to study the mechanotransduction in hair-bundles and the presynaptic-Ca2+ influx at ribbons. Localization of the membrane-localized Ca2+ sensor GCaMP6sCAAX shown in green. Inset in A shows an example top-down view of GCaMP6sCAAX bundle plane (6sCAAX) at 5 dpf. (B-B’) Bundle-Ca2+ signals before (blue) and after a 30 min treatment with 10 μM Ru360 (green) or 10 μM TRO 19622 (magenta), n ≥ 39 bundles per treatment. Average traces are shown in B while dot plots of the peak response per bundle are shown in B’. (C) Double-transgenic hair cells expressing GCaMP6sCAAX (At presynaptic membranes) and Ribeye a-tagRFP (Labels ribbons) at 5 dpf. Example cells in presynaptic imaging plane are boxed in white and duplicated in right insets. (C’) Example cells show evoked presynaptic-Ca2+ signals at ribbons during a 0.2 s stimulation. Circles 1–5 (1.3 μm diameter) in insets in C denote regions at ribbons used to generate the temporal traces of presynaptic-Ca2+ signals at each ribbon in C’. Similarly-colored traces of presynaptic-Ca2+ signals originate from different presynapses of the same cell. (D-E’) Presynaptic-Ca2+ signals averaged per cell before (blue) and after a 30 min 10 μM TRO 19622 (magenta), n ≥ 9 cells per treatment. D and E show averaged traces while D’ and E’ show before-and-after treatment dot plots of the peak response per cell. Error in panel B-B’, D-E’ represent SEM. A Wilcoxon matched-pairs signed-rank test was used in B’,D’ and E’. *p<0.05, **p<0.01, ***p<0.001. Scale bar = 5 µm in A and C and 2 µm in C inset.
-
Figure 2—figure supplement 1—source data 1
Mechanosensitive and presynaptic GCaMP6sCAAX traces and quantification after Ru360 and TRO 19622 application.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig2-figsupp1-data1-v2.xlsx
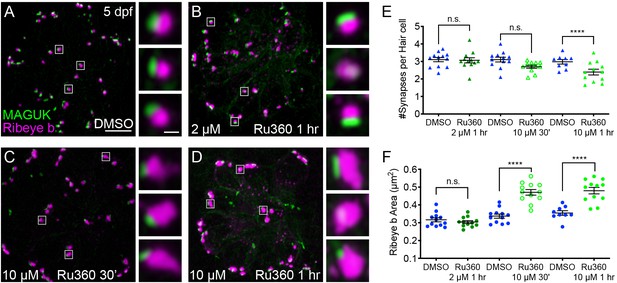
Mito-Ca2+ is important for ribbon size and synapse integrity in mature hair cells.
(A-D) Representative images of mature neuromasts (5 dpf) immunostained with Ribeye b (magenta, ribbons) and MAGUK (green, postsynapses) after a 1 hr 0.1% DMSO (A), a 1 hr 2 μM Ru360 (B), a 30 min 10 μM Ru360 (C), or a 1 hr 10 μM Ru360 (D) treatment. Insets show three example synapses (white squares). E-F, Scatter plots show synapse counts (E), and ribbon area (F) in controls and in treatment groups. Ribbon areas, synapse numbers, and hair-cell counts are unaffected after a 1 hr 2 µM Ru360 treatment. Ribbon areas are larger and there are fewer synapses without significant loss of hair cells after a 30 min treatment with 10 µM Ru360 (F). After a 1 hr 10 µM Ru360 treatment there is an increase in ribbon area and a decrease in synapse (E) and hair-cell counts. N ≥ 9 neuromasts per treatment. Error bars in E-F represent SEM. An unpaired t-test was used in E and a Welch’s unequal variance t-test was used in F. ****p<0.0001. Scale bar = 5 µm in A, and 2 µm in inset.
-
Figure 3—source data 1
Summary of synapse number and ribbon area after Ru360.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig3-data1-v2.xlsx
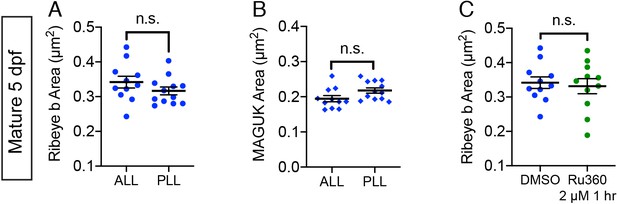
Ribbon and postsynapse size in mature ALL neuromasts.
(A-B) Scatter plots show that ribbon areas (A) and postsynaptic density areas (B) within the same fish are similar between mature anterior lateral-line (ALL) and posterior lateral-line (PLL) neuromasts. (C) Scatter plots show ribbon areas in controls and after a 1 hr treatment with 2 µM Ru360 are similar in mature hair cells within the ALL. Ribbon sizes of untreated anterior lateral-line hair cells are from the same data as A and (C) n ≥ 10 neuromasts per treatment; error bars represent SEM; and a Welch’s unequal variance t-test was used for comparisons.
-
Figure 3—figure supplement 1—source data 1
Summary of data comparing anterior and posterior lateral-line synapses in mature hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig3-figsupp1-data1-v2.xlsx
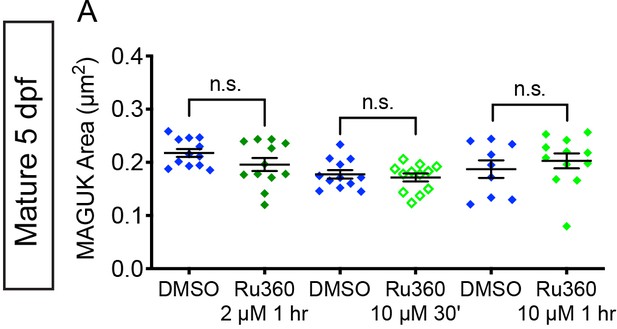
MCU block does not impact postsynapse size in mature hair cells.
(A) Quantification of postsynapse size assayed by MAGUK immunolabel in mature neuromasts indicate the treatments with 2 µM Ru360 and 10 µM Ru360 do not significantly alter postsynapse size compared to controls, n ≥ 9 neuromasts per treatment. Error bars represent SEM. A Welch’s unequal variance t-test was used for comparisons.
-
Figure 3—figure supplement 2—source data 1
Summary of MAGUK area measurements after Ru360 treatments in mature hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig3-figsupp2-data1-v2.xlsx
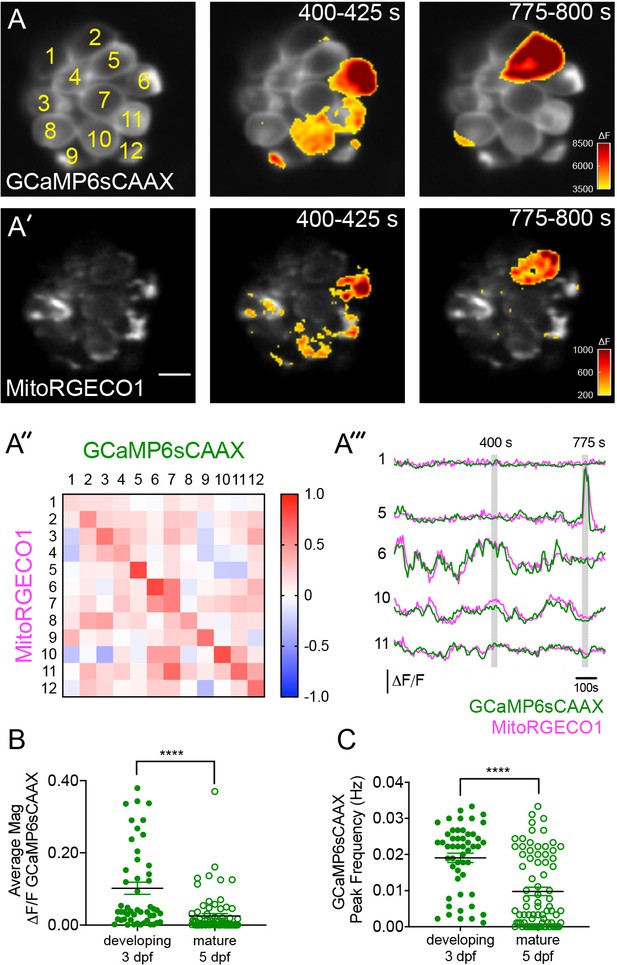
Spontaneous presynaptic- Ca2+ influx and mito-Ca2+ uptake are linked.
(A-A’) A live Image of an immature neuromast viewed top-down, expressing the presynaptic-Ca2+ sensor GCaMP6sCAAX (A) and mito-Ca2+ sensor MitoRGECO1 (A’) at 3 dpf. Example GCaMP6sCAAX (A’) and MitoRGECO1 (A’) signals during two 25 s windows within a 900 s acquisition are indicated by the ∆F heatmaps and occur in the same cells. (A’’) A heatmap of Pearson correlation coefficients comparing GCaMP6sCAAX and MitoRGECO1 signals from the cells in A-A’. (A’’’) Example GCaMP6sCAAX (green) MitoRGECO1 (magenta) traces during the 900 s acquisition from the 5 cells numbered in A, also see Video 2. (B) Scatter plot showing the average magnitude of GCaMP6sCAAX signals in developing and mature hair cells, n = 6 neuromasts per age. (C) Scatter plot showing frequency of GCaMP6sCAAX events in developing and mature hair cells, n = 6 neuromasts. Error bars in B-C represent SEM. A Mann-Whitney U test was used in B and C. ****p<0.0001. Scale bar = 5 µm in A’.
-
Figure 4—source data 1
Summary of the magnitude and frequency of spontaneous GCaMP6s-CAAX signals.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig4-data1-v2.xlsx
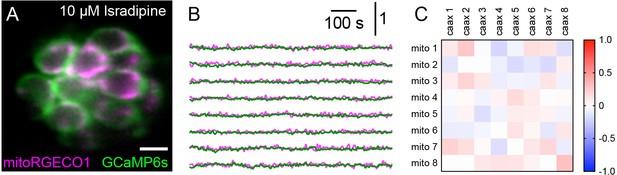
Spontaneous presynaptic and mito-Ca2+ signals are abolished by CaV1.3 channel antagonist isradipine.
(A) A live Image of a neuromast viewed top-down, expressing the presynaptic-Ca2+ sensor GCaMP6sCAAX (green) and mito-Ca2+ sensor MitoRGECO1 (magenta) at 6 dpf. (B) Representative GCaMP6sCAAX (green) and MitoRGECO1 (magenta) traces from the cells in A during a 900 s continuous image acquisition in the absence of stimuli and 10 µM isradipine. (C) There is no correlation between GCaMP6sCAAX and MitoRGECO1 signals within each cell from B in the presence of isradipine. Scale bar = 5 µm in A.
-
Figure 4—figure supplement 1—source data 1
Summary of MitoRGECO and GCaMP6s traces used to generate correlation plot.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig4-figsupp1-data1-v2.xlsx
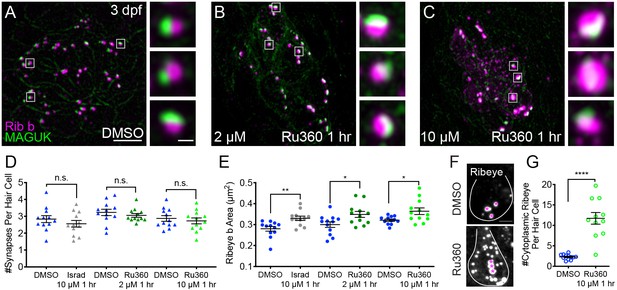
Mito-Ca2+ regulates ribbon formation.
(A-C) Representative images of immature neuromasts (3 dpf) immunostained with Ribeye b (magenta, ribbons) and MAGUK (green, postsynapses) after a 1 hr 0.1% DMSO (A), 2 μM Ru360 (B) or 10 µM Ru360 (C) treatment. Insets show three representative synapses (white squares) for each treatment. D-E, Scatter plot show quantification of synapse number (D), and ribbon area (E) in controls and in treatment groups. (F) Side-view of 2 hair cells (white outline) shows synaptic ribbon (three magenta asterisks in each cell) and extrasynaptic Ribeye b aggregates after a 1 hr 0.1% DMSO or 10 μM Ru360 treatment. (G) Quantification of extrasynaptic Ribeye puncta. N ≥ 12 neuromasts per treatment. Error bars in D-E and G represent SEM. An unpaired t-test was used in D and a Welch’s unequal variance t-test was used in E and G, *p<0.05, **p<0.01, ****p<0.0001. Scale bar = 5 µm in A, 2 µm in insets and F.
-
Figure 5—source data 1
Summary of synapse number and ribbon area after Ru360 application in developing hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig5-data1-v2.xlsx
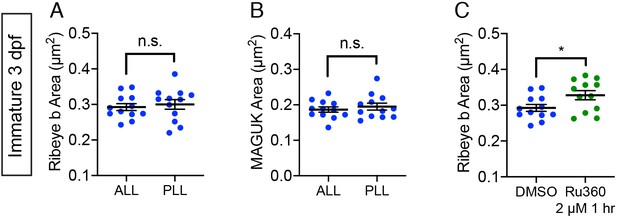
Ribbon and postsynapse size in immature ALL neuromasts.
(A-B) Scatter plots show that ribbon areas (A) and postsynaptic density areas (B) within the same fish are similar between immature anterior lateral-line (ALL) and posterior lateral-line (PLL) neuromasts. (C) Scatter plots show ribbon areas in controls and after a 1 hr treatment with 2 µM Ru360 are larger in immature hair cells within the ALL. Ribbon sizes of untreated anterior lateral-line hair cells are from the same data as A and C, n ≥ 10 neuromasts per treatment; error bars represent SEM; and a Welch’s unequal variance t-test was used for comparisons. *p<0.05.
-
Figure 5—figure supplement 1—source data 1
Summary of data comparing anterior and posterior lateral-line synapses in developing hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig5-figsupp1-data1-v2.xlsx
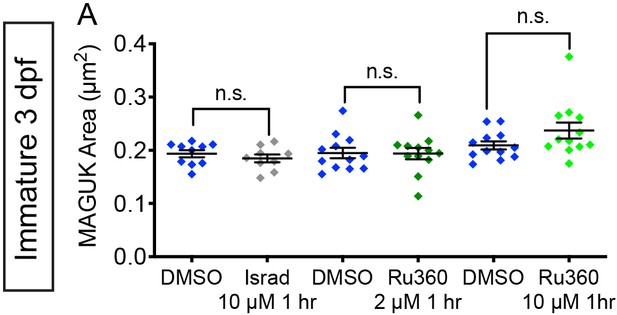
MCU and CaV1.3 block do not impact postsynapse size.
(A) Quantification of postsynapse size assayed by MAGUK immunolabel in mature neuromasts indicate the treatments with 10 µM isradipine, 2 µM Ru360 and 10 µM Ru360 do not significantly alter postsynapse size compared to controls, n ≥ 9 neuromasts per treatment. Error bars represent SEM. A Welch’s unequal variance t-test was used for comparisons.
-
Figure 5—figure supplement 2—source data 1
Summary of MAGUK area measurements after Ru360 treatment in developing hair cells.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig5-figsupp2-data1-v2.xlsx
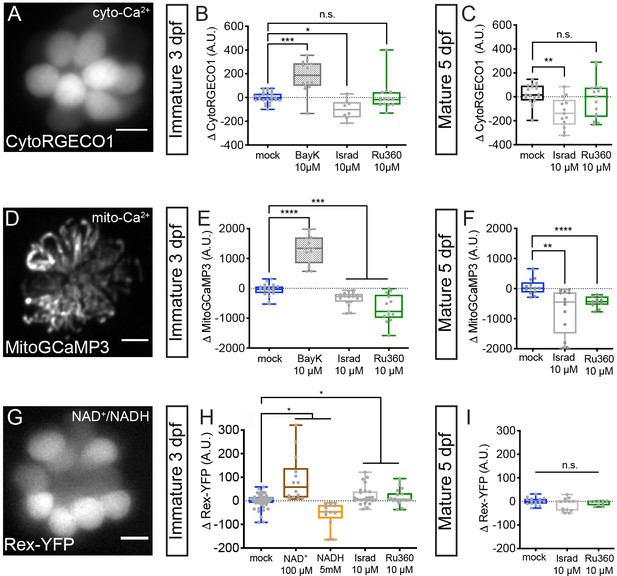
Cyto-Ca2+, mito-Ca2+ and NAD+/NADH redox baseline measurements.
Live hair cells expressing RGECO1 (A), MitoGCaMP3 (D), or Rex-YFP (G) show resting cyto-Ca2+, mito-Ca2+ or NAD+/NADH levels respectively. (B-C) RGECO1 baseline measurements before and after a 30 min mock treatment (0.1% DMSO) or after a 30 min 10 μM Bay K8644 (BayK), 10 μM isradipine, or 10 μM Ru360 treatment. (E-F) MitoGCaMP3 baseline measurements before and after a 30 min mock treatment (0.1% DMSO) or after a 10 μM BayK, 10 μM isradipine, or 10 μM Ru360 treatment. (H-I) Rex-YFP baseline measurements before and after 30 min mock treatment (0.1% DMSO) or after a 30 min 100 μM NAD+, 5 mM NADH, 10 μM isradipine, or 10 μM Ru360 treatment. All plots are box-and-whiskers plot that show median, min and max. N ≥ 9 neuromasts per treatment. A one-way Brown-Forsythe ANOVA with Dunnett’s T3 post hoc was used to calculate the difference in (B-C), (E-F), and a one-way Brown-Forsythe and Welch ANOVA with Holm-Sidak’s post hoc was used in H-I, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Horizontal lines in E, H, and I indicate that both conditions had similar p values compared to mock treatment. Scale bar = 5 μm in A, D and G.
-
Figure 6—source data 1
Summary of baseline CytoRGECO1, MitoGCaMP3 and Rex-YFP measurements.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig6-data1-v2.xlsx
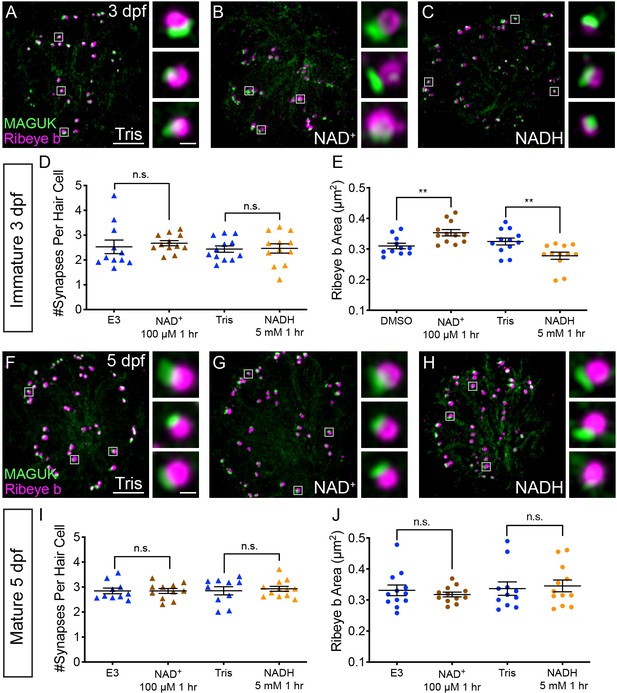
NAD+ and NADH directly influence ribbon formation.
Representative images of immature (A-C, 3 dpf) and mature (G-H, 5 dpf) neuromasts immunostained with Ribeye b (magenta, ribbons) and MAGUK (green, postsynapses) after a 0.1% Tris-HCl (A, F), 100 μM NAD+ (B, G) or 5 mM NADH treatment (C, H). Insets show three example synapses (white squares). D-E and I-J, Scatter plots show synapse count (D, I) and ribbon area (E, J) in controls and treatments groups. N ≥ 10 neuromasts per treatment. Error bars in B-C represent SEM. An unpaired t-test was used for comparisons in D and I and a Welch’s unequal variance t-test was used for comparisons in E and J, **p<0.01. Scale bar = 5 µm in A and F, 2 µm in insets.
-
Figure 7—source data 1
Summary of synapse number and ribbon area measurements after NAD+ and NADH application.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig7-data1-v2.xlsx
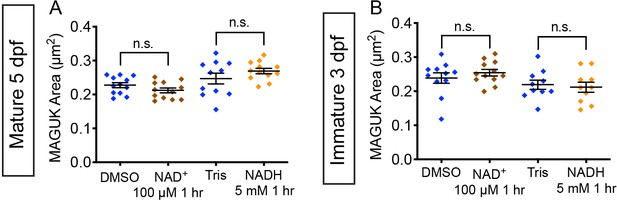
NAD+ and NADH treatment do not impact postsynapse size.
(A-B) Quantification of postsynapse size assayed by MAGUK immunolabel in mature (A) and immature (B) neuromasts indicate the treatment with 100 μM NAD+ and 5 mM NADH do not significantly alter postsynapse size compared to controls, n ≥ 9 neuromasts per treatment. Error bars represent SEM. A Welch’s unequal variance t-test was used for comparisons.
-
Figure 7—figure supplement 1—source data 1
Summary of MAGUK area after NAD+ and NADH treatment.
- https://cdn.elifesciences.org/articles/48914/elife-48914-fig7-figsupp1-data1-v2.xlsx
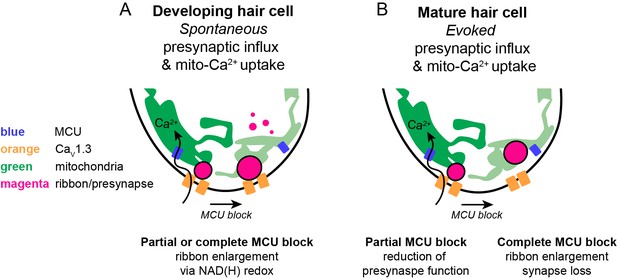
Schematic model of mito-Ca2+ in developing and mature hair cells.
(A) In developing hair cells, spontaneous presynaptic-Ca2+ influx is linked to mito-Ca2+ uptake. Together these Ca2+ signals function to regulate ribbon size during ribbon formation. When the CaV1.3 or MCU channels are blocked, ribbon formation is increased leading to larger ribbons. These Ca2+ signals regulate ribbon formation via NAD(H) redox. MCU block lowers mito-Ca2+, increases the NAD+/NADH ratio and promotes ribbon formation. (B) In mature hair cells, evoked presynaptic-Ca2+ influx is linked to mito-Ca2+ uptake. When the MCU is partially blocked there is a reduction in presynaptic-Ca2+ influx. When the MCU is completely blocked there are synaptopathic consequences; ribbons are enlarged and synapses are lost.
Videos
Airyscan image of MitoGCaMP3 and Rib a-tagRFP at the base of a single live hair cell.
Spontaneous ∆F GCaMP6sCAAX (left) and ∆F MitoRGECO1 (right) signals acquired at 3 dpf, 25 s per frame.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Danio rerio) | Tübingen | ZIRC | RRID: ZIRC_ZL57; ZFIN ID: ZDB-GENO-990623–3 | |
| Strain, strain background (Danio rerio) | TL | ZIRC | RRID: ZIRC_ZL86; ZFIN ID: ZDB-GENO-990623–2 | |
| Genetic reagent (Danio rerio) | Tg(myo6b:GCaMP6s-CAAX)idc1; GCaMP6sCAAX; 6sCAAX; GCaMP6s | (Jiang et al., 2017) | RRID: ZFIN_ZDB-ALT-170113-3; https://zfin.org/ZDB-ALT-170113-3 | Membrane-localized calcium biosensor |
| Genetic reagent (Danio rerio) | Tg(myo6b:RGECO1)vo10Tg; CytoRGECO1 | (Maeda et al., 2014) | RRID: ZFIN_ZDB-ALT-150114-2; https://zfin.org/ZDB-ALT-150114-2 | Cell-fill calcium biosensor |
| Genetic reagent (Danio rerio) | Tg(myo6b:GCaMP3)w78Tg; CytoGCaMP3 | (Esterberg et al., 2013) | RRID: ZFIN_ZDB-ALT-130514-1; https://zfin.org/ZDB-ALT-130514-1 | Cell-fill calcium biosensor |
| Genetic reagent (Danio rerio) | Tg(myo6b:mitoGCaMP3)w119Tg; MitoGCaMP3; Mito | (Esterberg et al., 2014) | RRID: ZFIN_ZDB- ALT-141008–1; https://zfin.org/ZDB-ALT-141008-1 | Mitochondria-localized calcium biosensor |
| Genetic reagent (Danio rerio) | Tg(myo6b:ribeye a-tagRFP)idc11Tg; Rib a-RFP; Rib a-tagRFP; Rib a | (Sheets, 2017) | RRID: ZFIN_ZDB- ALT-190102–4; https://zfin.org/ZDB-ALT-190102-4 | Ribbon-localized fluorescent protein |
| Genetic reagent (Danio rerio) | Tg(myo6b:mitoRGECO1)idc12Tg; MitoRGECO1 | This paper | RRID: ZFIN_ZDB-ALT-190102-5; https://zfin.org/ZDB-ALT-190102-5 | Mitochondria-localized calcium biosensor. See Materials and methods,‘Cloning and Transgenic Fish Production’ |
| Genetic reagent (Danio rerio) | Tg(myo6b:Rex-YFP)idc13Tg; Rex-YFP | This paper | RRID: ZFIN_ZDB- ALT-190102–6; https://zfin.org/ZDB-ALT-190102-6 | Cell-fill NAD+/NADH ratio biosensor. See Materials and methods, ‘Cloning and Transgenic Fish Production’ |
| Antibody | Ribbon label: Mouse anti-Ribeye b IgG2a; Ribeye b; Ribeye; Rib b | (Sheets et al., 2011) | N/A | 1:10,000 |
| Antibody | PSD label: Mouse anti-pan-MAGUK IgG1; MAGUK | NeuroMab | RRID: AB_10673115; K28/86, #75–029 | 1:500 |
| Antibody | Hair cell label: Rabbit anti-Myosin VIIa | Proteus | RRID: AB_10015251; #25–6790 | 1:1000 |
| Antibody | goat anti-mouse IgG2a, Alexa Fluor 488 | ThermoFisher Scientific | RRID: AB_2535771; #A-21131 | 1:1000 |
| Antibody | goat anti-rabbit IgG (H+L) Alexa Fluor 568 | ThermoFisher Scientific | RRID: AB_143157; #A-11011 | 1:1000 |
| Antibody | goat anti-mouse IgG1 Alexa Fluor 647 | ThermoFisher Scientific | RRID: AB_2535809; #A-21240 | 1:1000 |
| Recombinant DNA reagent | Plasmid: 5E-pmyo6b | (Kindt et al., 2012) | N/A | |
| Recombinant DNA reagent | Plasmid: 3E-polyA | (Kwan et al., 2007) | #302 | |
| Recombinant DNA reagent | Plasmid: pDestTol2CG2 | (Kwan et al., 2007) | #395 | |
| Recombinant DNA reagent | Plasmid: pC1-Rex-YFP | (Bilan et al., 2014) | Addgene #48247 | |
| Recombinant DNA reagent | Plasmid: pDONR221 | ThermoFischer | Cat #12536017 | |
| Recombinant DNA reagent | Plasmid: CMV-R-GECO1 | (Zhao et al., 2011) | Addgene #32444 | |
| Recombinant DNA reagent | Plasmid: pME-Rex-YFP | This paper | N/A | See Materials and methods, ‘Cloning and Transgenic Fish Production’ |
| Recombinant DNA reagent | Plasmid: pME-mitoRGECO1 | This paper | N/A | See Materials and methods, ‘Cloning and Transgenic Fish Production’ |
| Sequence-based reagent | RexYFP attB FWD | This paper | PCR primers | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGCCACCATGAAGGTCCCCGAAGCG; Made by Integrated DNA Technologies (IDT). |
| Sequence-based reagent | REX-YFP attB REV | This paper | PCR primers | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCACCCCATCATCTCTTCCCG |
| Sequence-based reagent | RGECO1 FWD1 | This paper | PCR primers | [ATGTCCGTCCTGACGCCGCTGCTGCTGCGGGGCTTGACAGGCTCGGCCCGGCGGCTCCCAGTGCCGCGCGCCAAGATCCATTCGTTGGGGGATCCA]-GTCGACTCTTCACGTCGTAAGTG; Made by IDT. |
| Sequence-based reagent | RGECO1 REV1 | This paper | PCR primers | CTACTTCGCTGTCATCATTTGTACAAACTC; Made by IDT. |
| Sequence-based reagent | RGECO1 attB FWD2 | This paper | PCR primers | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCACCATGTCCGTCCTGACGCCGC; Made by IDT. |
| Sequence-based reagent | RGECO1 attB REV2 | This paper | PCR primers | GGGGACCACTTTGTACAAGAAAGCTGGGTGCTACTTCGCTGTCATCATTTGTACAAACTC; Made by IDT. |
| Peptide, recombinant protein | α-bungarotoxin | Tocris | 2133 | |
| Chemical compound, drug | Isradipine; Israd | Sigma-Aldrich | I6658 | |
| Chemical compound, drug | BayK 8644; BayK | Sigma-Aldrich | B133 | |
| Chemical compound, drug | Ru360 | Millipore | 557440 | |
| Chemical compound, drug | NAD+ | Sigma-Aldrich | N1511 | |
| Chemical compound, drug | NADH | Cayman Chemical | 16038 | |
| Chemical compound, drug | Tricaine; MESAB | Sigma-Aldrich | A5040 | |
| Chemical compound, drug | Paraformaldehyde: EM | Electron Microscopy Sciences | 15710 | |
| Chemical compound, drug | Glutaraldehyde: EM | Electron Microscopy Sciences | 16210 | |
| Chemical compound, drug | Paraformaldehyde: IHC | ThermoFisher Scientific | 28906 | |
| Chemical compound, drug | TRO 19622; TRO | Cayman Chemical | 21264 | Vortex vigorously to dissolve in embryo media + 0.1% DMSO |
| Software, algorithm | Prism (v. 8) | Graphpad Software | RRID: SCR_002798; https://www.graphpad.com | |
| Software, algorithm | Adobe Illustrator | Adobe | RRID: SCR_014198; https://www.adobe.com | |
| Software, algorithm | FIJI is just ImageJ | NIH | RRID: SCR_003070; https://fiji.sc | |
| Software, algorithm | Zen | Zeiss | RRID: SCR_01367; https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html | |
| Software, algorithm | Prairie View | Bruker Corporation | RRID: SCR_017142; https://www.bruker.com/products/fluorescence-microscopes/ultima-multiphoton-microscopy/ultima-in-vitro/overview.html | |
| Software, algorithm | Python Programming Language | Python Software Foundation | RRID: SCR_008394; https://www.python.org/ |





