Differential expression of MAGEA6 toggles autophagy to promote pancreatic cancer progression
Figures
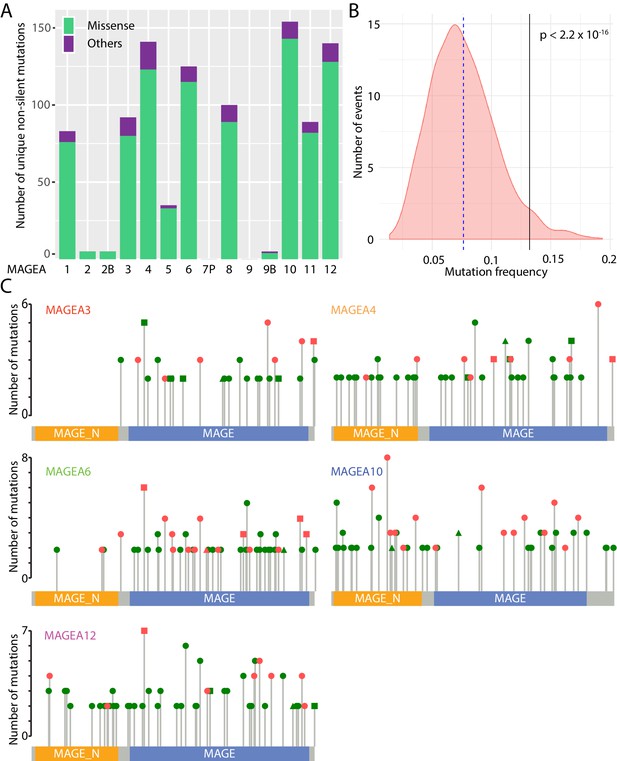
Mutational landscape of MAGEA genes in pan-cancer study.
(A) Histogram of missense mutations and others (nonsense, frameshift, in-frame indels, and splice site mutations) across the entire MAGEA family. (B) Mutation frequency analysis of MAGEA genes. Pink area represents the mutation frequency distribution of 1000 randomly selected genes with sizes similar to MAGEA genes. Blue dotted line and black line indicate the mean mutation frequency of the 1000 genes and the MAGEA gene respectively. P value was calculated by a one-sample Wilcoxon test. (C) Lollipop diagrams indicate the distribution and number of recurrent mutations in individual MAGEA genes. Reported amino acid changes: missense mutations (dot), others (triangle), and both (square) are marked in pink for those selected for immunoblot analysis in Figure 2.
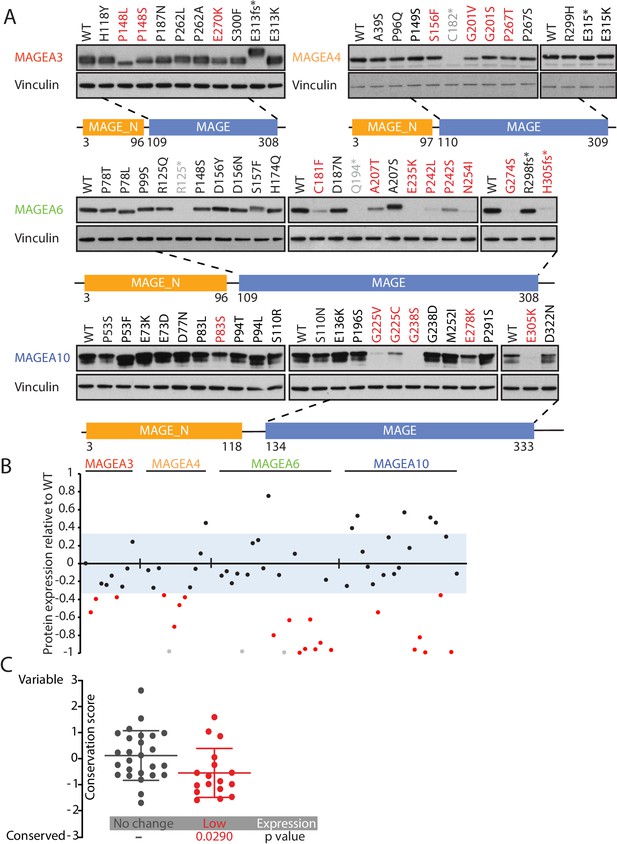
Cancer-specific mutations of MAGEA genes reduce their protein expression.
(A) Immunoblot analysis of MAGEA3, A4, A6 and A10 variants expressed in HEK293T cells. Variants that are not recognized by the antibody (gray), variants expressed at levels 33% (three standard deviation determined by the expression deviation analysis in Figure 2—figure supplement 2) or less than those of the wild-type (WT) (red) are indicated. MAGE_N: N-terminal MAGE domain. (B) Densitometry analysis of protein expression of the MAGEA variants in (A). Each variant is represented by a dot, and the dots are shown in the same order and colors as in (A). The blue area represents three standard deviations, determined from Figure 2—figure supplement 2. (C) Conservation score analysis (mean ± standard deviation) of amino acids that show reduced protein expression and those that show unchanged protein expression when mutated. P value was calculated by two-tailed unpaired t-test (N = 26 for no change cohort, N = 17 for low-expression cohort).
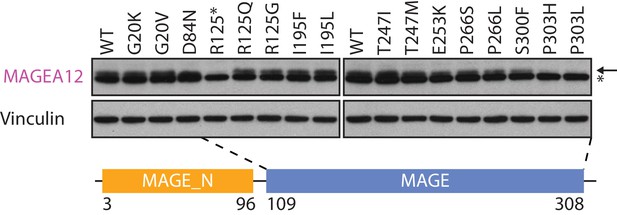
Immunoblot analysis of MAGEA12 variants expressed in HEK293T cells.
MAGEA12 and the non-specific band recognized by MAGEA12 antibody are indicated as ← and *, respectively.
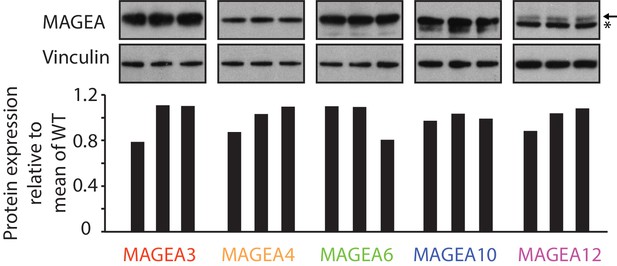
Immunoblot (top) and densitometry plot (bottom) of the expression deviation analysis.
Standard deviation of all 15 samples = 0.1118.
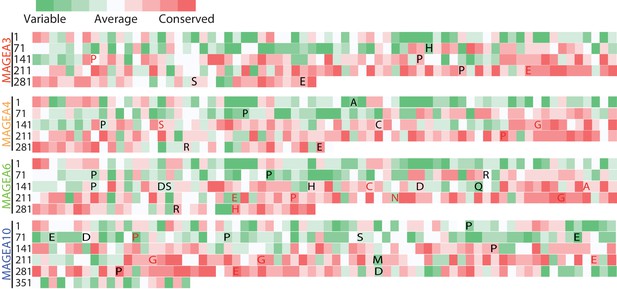
Evolutionary conservation study of MAGEAs.
Wild-type amino acids of recurrent mutations are shown and colored as in Figure 2A.
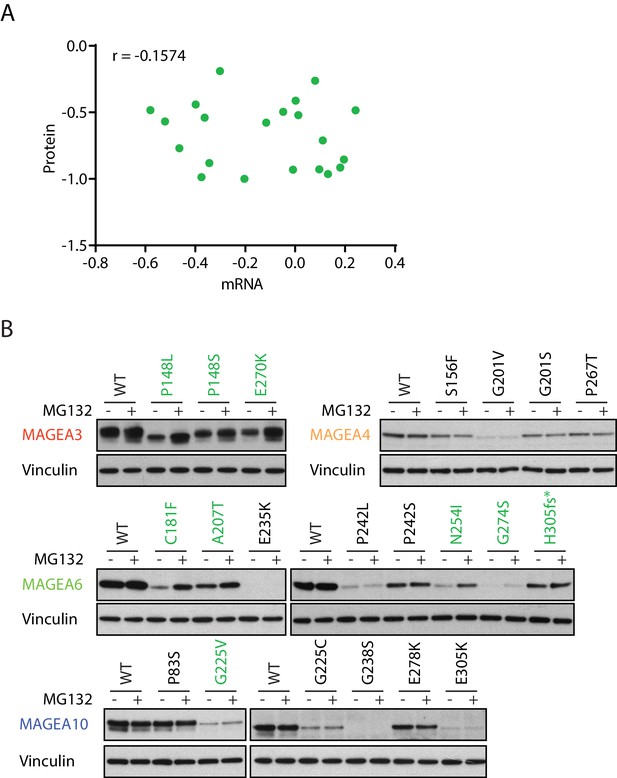
MAGEA variants are degraded through the ubiquitin proteasome pathway.
(A) Correlation study of protein vs. mRNA expression of MAGEA variants. MAGEA variant protein and mRNA expression levels shown in Figure 3B and Figure 3—figure supplement 1, respectively, were analyzed after normalized to their corresponding wild-type. Pearson correlation coefficient r (N = 21) was calculated using GraphPad. (B) Immunoblot of MAGEA variants expressed in HEK293T cells with or without MG132 treatment for 9 hr. Variants that showed increased protein levels under MG132 treatment are in green.
qRT-PCR analysis of the MAGEA variants in Figure 3B.
qRT-PCR primer RT#3 was used, and MAGEA expression is normalized to that of parental HEK293T cells. *P value < 0.05; two-tailed unpaired t-test.
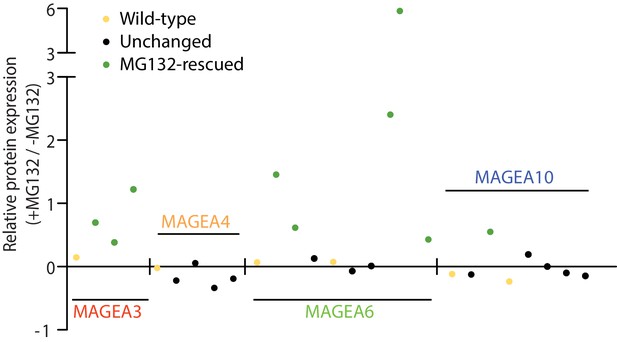
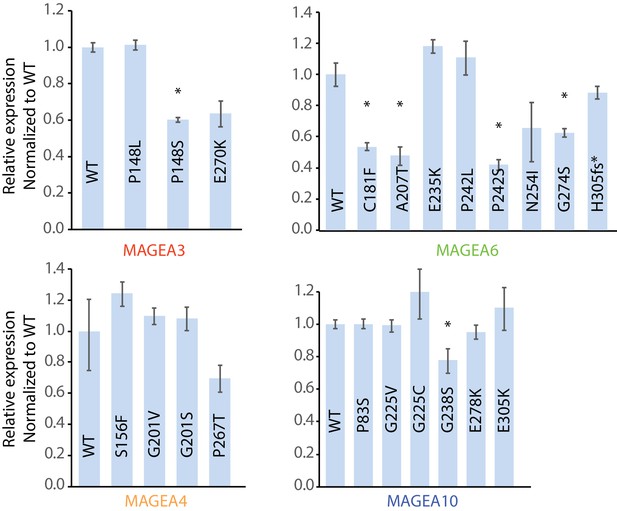
Densitometry analysis of protein expression of the MAGEA variants (+MG132/ -MG132) in Figure 3B.
Each dot represents a variant.
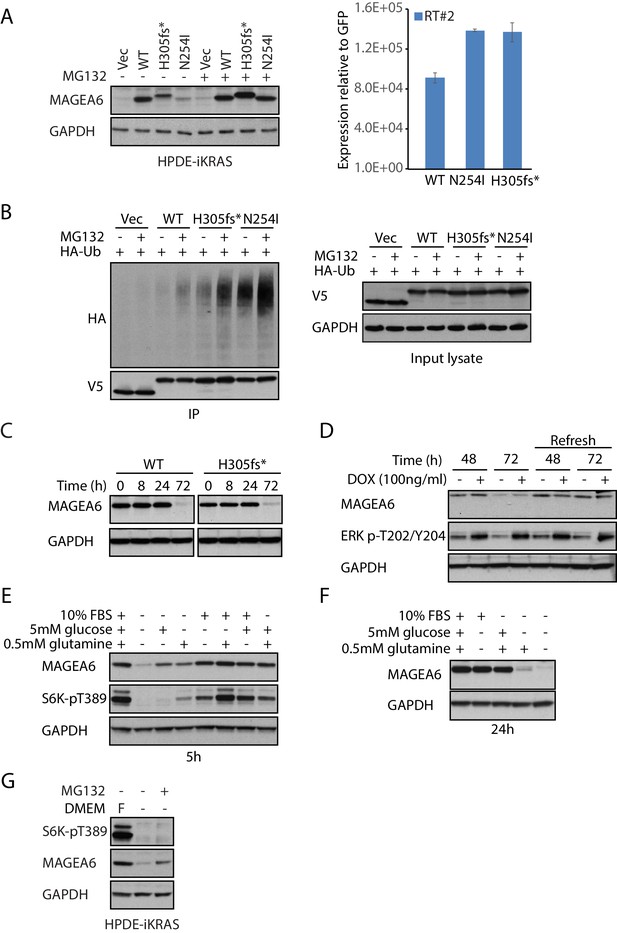
Cancer-specific mutations and glucose/glutamine depletion stimulate proteasome-dependent MAGEA6 degradation.
(A) Immunoblots (left) and qRT-PCR (right) analysis of HPDE-iKRAS cells expressing GFP (Vec) and MAGEA6 variants with or without MG132. WT: wild-type, GAPDH: glyceraldehyde 3-phosphate dehydrogenase. (B) V5 pull-down assays of HEK293T lysate expressing V5-tagged GFP/MAGEA6 variants with or without MG132 treatment for 6 hr. Immunoblots of the pulled-down samples (left) and total lysate (right) are shown. HA: hemagglutinin, Ub: ubiquitin. (C) Immunoblot analysis of MAGEA6 in the transduced HPDE-iKRAS cells cultured in KSFM for the indicated times and (D) in the presence or absence of doxycycline at the indicated time points. KSFM was refreshed every 24 hr in the last four lanes throughout the experiment. (E) Immunoblot analysis of HPDE-iKRAS cells cultured in DMEM– supplemented with PBS, FBS, glutamine, or glucose, as indicated, for 5 hr and (F) 24 hr. (G) Immunoblot analysis of HPDE-iKRAS cells cultured in DMEM– or in FBS-, glucose-, and glucose-supplemented DMEM (F) with or without MG132 treatment for 6 hr.
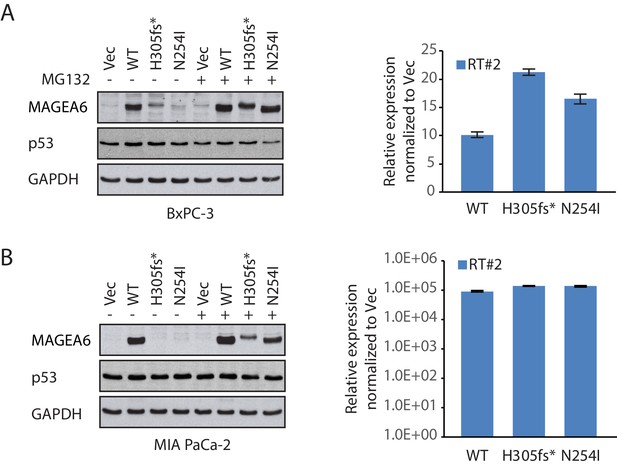
Immunoblot and qRT-PCR analysis of MAGEA6 in PDAC cell models in response to MG132.
Immunoblot (left) and qRT-PCR (right) analysis of MAGEA6 in BxPC-3 (A) and MIA PaCa-2 (B) in the presence or absence of MG132.
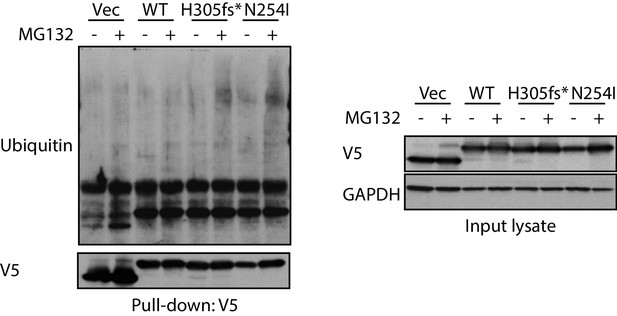
V5 pull-down assays to examine endogenous polyubiquitination signal on MAGEA6 variants with or without MG132 treatment.
Immunoblots of V5-tag pulled-down samples (left) and total lysate (right) are shown.
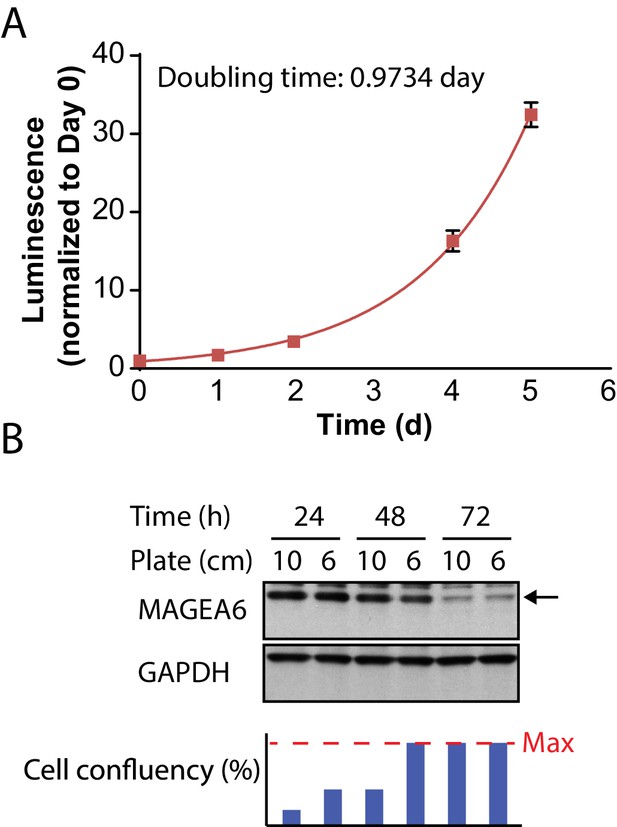
MAGEA6 protein expression is independent of cell confluency.
(A) Cell proliferation analysis of HPDE-iKRAS cells (mean ± standard deviation of replicates, N = 3). (B) Immunoblot study of the transduced HPDE-iKRAS cells with the estimated cell confluency (bottom).
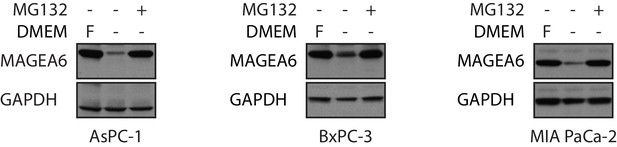
MAGEA6 protein expression is regulated by nutrient levels in culture media.
Immunoblot analysis of MAGEA6 in transduced PDAC lines AsPC-1 (left), BxPC-3 (middle), and MIA PaCa-2 (right) in nutrient-depleted DMEM (-) with or without MG132. Fully nutrient-supplemented DMEM (f) served as a control.
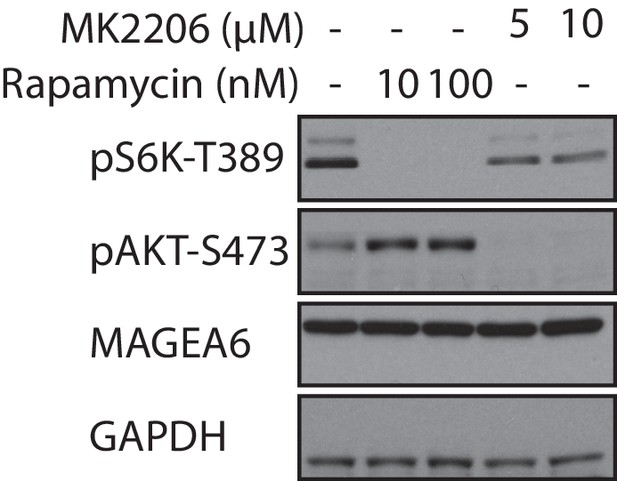
MAGEA6 protein expression is not regulated by AKT and mTORC1 kinases.
HPDE-iKRAS cells expressing wild-type MAGEA6 were treated with AKT and mTORC1 inhibitors (MK2206 and rapamycin, respectively) as indicated. Immunoblots of AKT and S6K phosphorylation status confirmed the efficacy of these inhibitors.
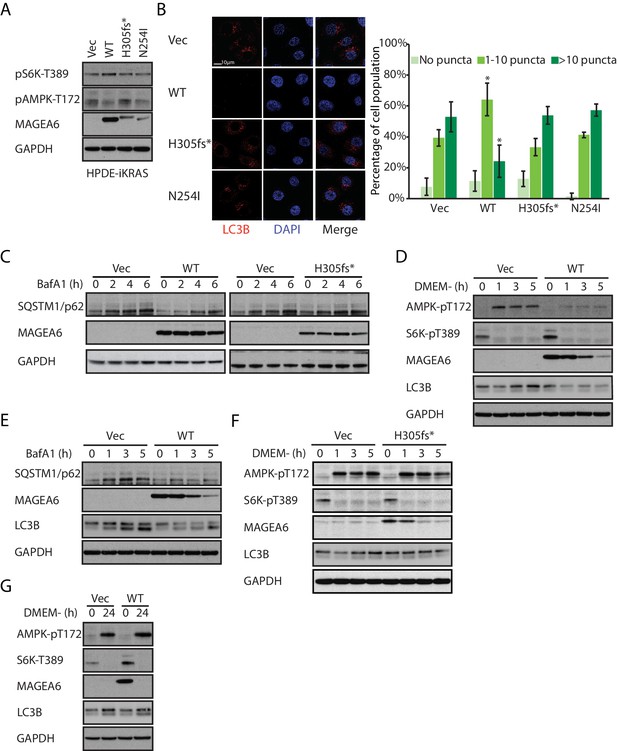
Overexpression of wild-type MAGEA6, but not mutant MAGEA6, suppresses autophagy in PDAC cell lines.
(A) Immunoblot analysis of autophagy signaling in HPDE-iKRAS cells expressing GFP (Vec) and MAGEA6 variants. (B) Immunofluorescence staining of LC3B puncta in the transduced HPDE-iKRAS cells. Representative photos (left) and statistical analysis (mean ± standard deviation of counted cells, N=~100 per cohort) are shown. *p=0.002; two-tailed unpaired t-test. (C) Immunoblot analysis of autophagy substrate SQSTM1/p62 in the transduced HPDE-iKRAS cells treated with BafA1 for the indicated time points., Immunoblot analysis of (D) autophagy signaling and (E) SQSTM1/p62 accumulation in wild-type MAGEA6 expressing and (F) autophagy signaling in MAGEA6H305fs* expressing HPDE-iKRAS cells under nutrient-deficient conditions. (G) Immunoblot analysis of autophagy signaling in wild-type MAGEA6 expressing cells under prolonged nutrient-deficient conditions.
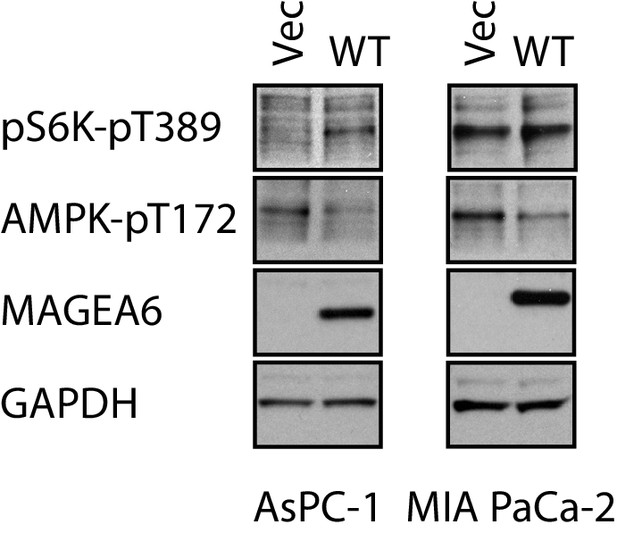
Immunoblot analysis of autophagy signaling of transduced AsPC-1 and MIA PaCa-2.
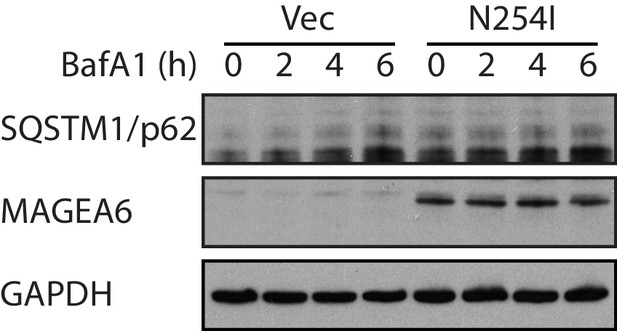
Immunoblot analysis of the accumulation of autophagy substrate SQSTM/p62 in the transduced HPDE-iKRAS cells under BafA1 for the indicated time points.
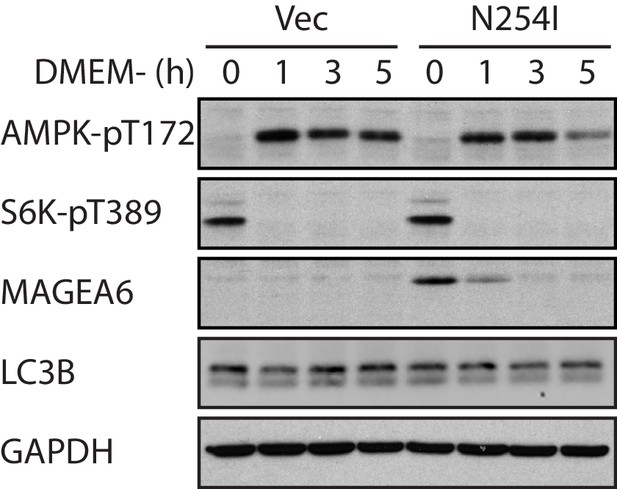
Immunoblot analysis of autophagy activity in transduced HPDE-iKRAS cells under nutrient-depleted conditions as indicated.
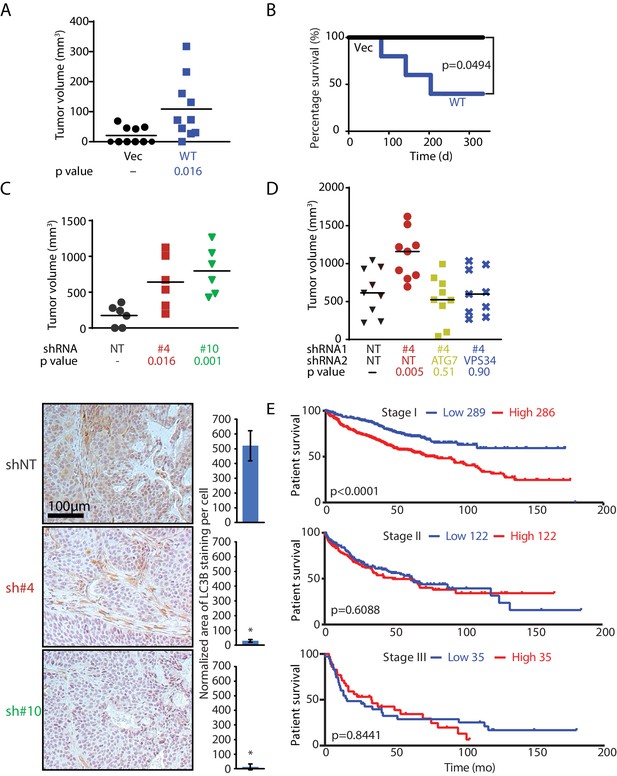
MAGEA6 mutations and expression variation manipulate autophagy to promote tumor progression at different stages.
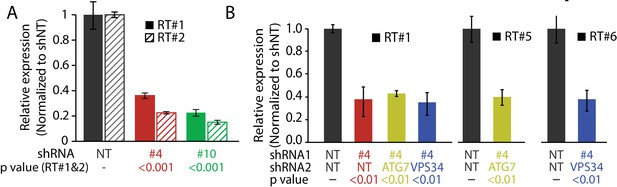
(A) Tumor volume plot (50 d after injection) and Kaplan-Meier survival plot (B) of xenograft assay using HPDE-iKRAS cells expressing GFP (Vec) or MAGEA6 (five mice, two injections each, N = 10). P value was calculated by two-tailed unpaired t-test in (A) and log-rank test in (B). (C) (Top) Tumor volume plot (100 d after injection) of xenograft assay using BxPC-3 cells transduced with MAGEA6 shRNAs (#4 and #6) and non-targeting shRNA (NT). P value was calculated by two-tailed unpaired t-test (three mice, two injections each, N = 6). (Bottom) Representative photos and quantification of LC3B immunohistochemical staining in the xenograft tumor samples. P value was calculated by two-tailed unpaired t-test (>150 cells analyzed per cohort). (D) Tumor volume plot (100 d after injection) of xenograft assay using BxPC-3 cells transduced with MAGEA6 (#4), ATG7, VPS34 shRNAs and non-targeting shRNA (NT). P value was calculated by two-tailed unpaired t-test (nine mice, one injection each, N = 9). (E) Patient survival analysis of low and high MAGEA6 expression in stage I (top), stage II (middle), and stage III (bottom) lung cancer. Number of patients analyzed per cohort is shown (Győrffy et al., 2013). P value was calculated by log-rank test.
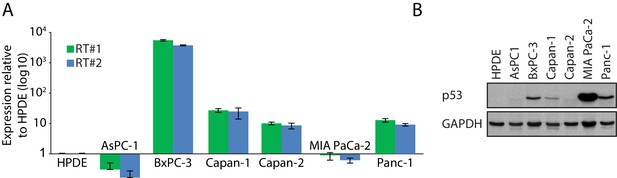
MAGEA6 expression analysis in PDAC cell line panel.
qRT-PCR of endogenous MAGEA6 expression (A) and immunoblot analysis of endogenous p53 (B) in HPDE and six other PDAC cell lines.
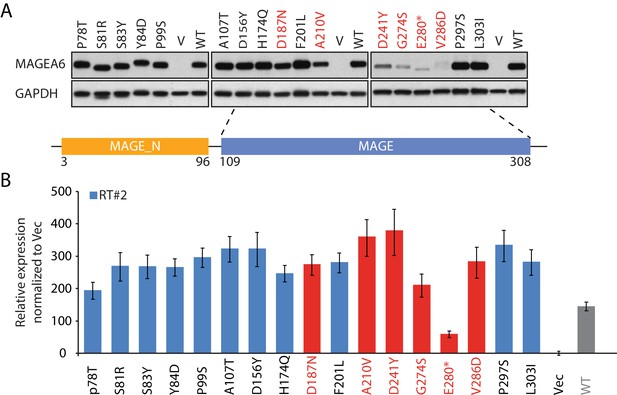
Gene and protein expression analysis of MAGEA6 variants identified from ICGC lung cancer patients.
Immunoblot (A) and qRT-PCR analysis (B) of MAGEA6 variants identified from the ICGC lung cancer database. MAGEA6 variant expression levels are normalized to those of HEK293T cells transfected with GFP (Vec). MAGEA6 variants with low protein expression are in red.
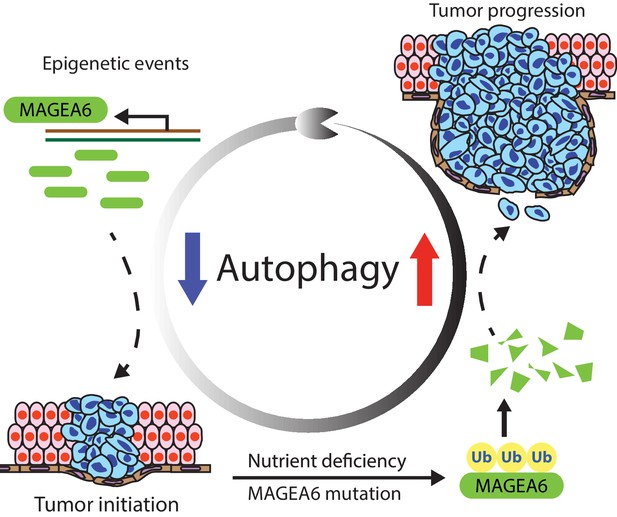
MAGEA6 drives PDAC via manipulation of autophagy.
In the proposed model, MAGEA6 expression is activated via epigenetic regulation at an early disease stage to suppress autophagy and promote tumor initiation. MAGEA6 mutations and metabolic stress during subsequent tumor development lead to MAGEA6 degradation, reactivation of autophagy, and thus better tumor survival at the late disease stages.
Additional files
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/48963/elife-48963-supp1-v2.docx
-
Supplementary file 2
List of clinical trials of cancer immunotherapy targeting MAGEA antigens.
Clinical trials of MAGEA-targeted immunotherapy reported from clinicaltrials.gov.
- https://cdn.elifesciences.org/articles/48963/elife-48963-supp2-v2.xlsx
-
Supplementary file 3
MAGEA variants reported in skin, lung and pancreas cancer patients.
Recurrent mutations of MAGEA3, A4, A6, A10 and A12 genes identified from skin, lung and pancreas cancer patients in ICGC, TCGA and COSMIC databases.
- https://cdn.elifesciences.org/articles/48963/elife-48963-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/48963/elife-48963-transrepform-v2.docx