Identification of TMEM206 proteins as pore of PAORAC/ASOR acid-sensitive chloride channels
Figures
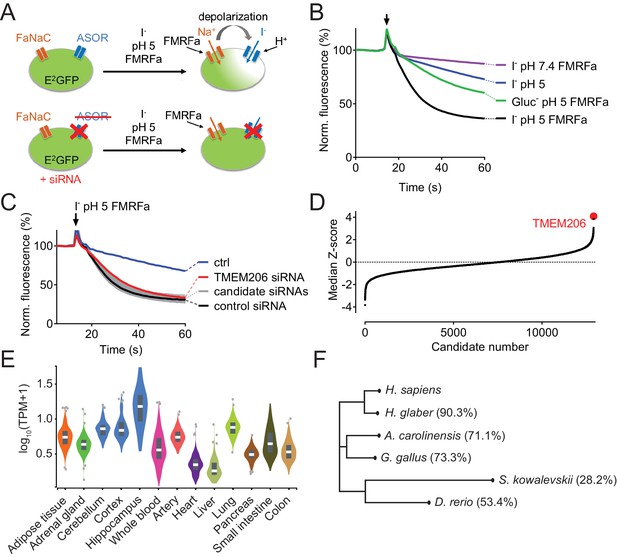
Identification of TMEM206 as ASOR component.
(A) Screening assay. Engineered HeLa cells inducibly expressing iodide-sensitive E2GFP and FRMFamide-gated Na+ channel FaNaC were acutely exposed to an acidic solution (pH 5) containing 20 μM FMRFamide and 100 mM I-. Iodide influx through ASOR is stimulated both by acidic pH and the depolarization caused by FaNaC-mediated Na+-influx and induces quenching of E2GFP fluorescence. Fluorescence quenching is reduced by ASOR knock-down. (B) Assay verification under the conditions used for screening. Addition of I- and FRMFamide at pH 5 (arrow) induces rapid quenching of fluorescence. Less rapid quenching upon omission of either I-, FMRFamide or acidic pH suggests that it is caused by I- influx through ASOR, as further supported by inhibition by PS (pregnenolone sulfate) and DIDS (4,4'-diisothiocyano-2,2'-stilbenedisulfonic acid) (Figure 1—figure supplement 1). (C) Fluorescence curves from a 384-well plate treated with siRNA against 280 genes, including TMEM206. Fluorescence quenching is specifically slowed by siRNA against TMEM206. (D) Distribution of median Z-scores (mean of 3 replicates) from filtered hits (see Materials and methods). TMEM206 was the top hit. (E) Tissue expression of TMEM206 extracted from the GTEx database (https://gtexportal.org/home/; TPM, transcripts per million). (F) Dendrogram depicting similarity between TMEM206 orthologs from human (Homo sapiens), African naked mole-rat (Heterocephalus glaber), chicken (Gallus gallus), green anole lizard (Anolis carolinensis), zebrafish (Danio rerio) and from the hemichordate acorn worm (Saccoglossus kowalewski). Amino-acid sequence identity to human TMEM206 given in brackets. Dendrogram based on a Clustal Omega protein alignment fed into the Simple Phylogeny tool at EMBL-EBI (https://www.ebi.ac.uk/services).
-
Figure 1—source data 1
Raw data for Figure 1B-E.
- https://doi.org/10.7554/eLife.49187.004
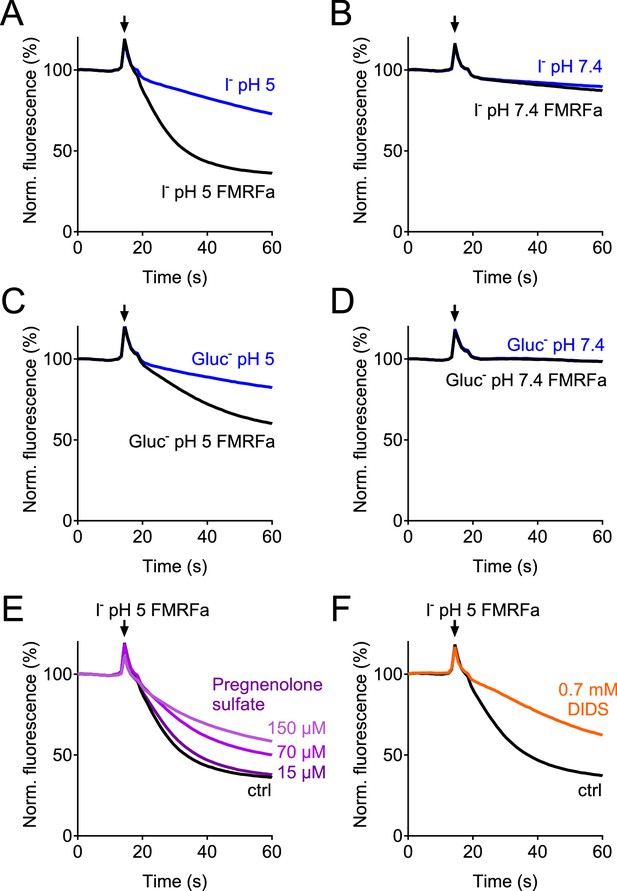
Verification of the assay for the genome-wide siRNA screen.
(A–D) Controls in which E2GFP-2A-FaNaC-expressing cells were subjected to solutions containing I- (A, B) or gluconate (C, D) at pH 5 (A, C) or pH 7.4 (B, D). Only depolarization induced by activation of FaNaC by FMRFamide in the presence of I- and acidic pH (A) produced rapid quenching of E2GFP fluorescence. (E–F) ASOR inhibitors pregnenolone sulfate (PS, (E) and DIDS (F) inhibited fluorescence quenching. PS was added acutely together with the acidic solution. Cells were preincubated with DIDS, which induced a large increase in the absolute signal due to its intrinsic fluorescence.
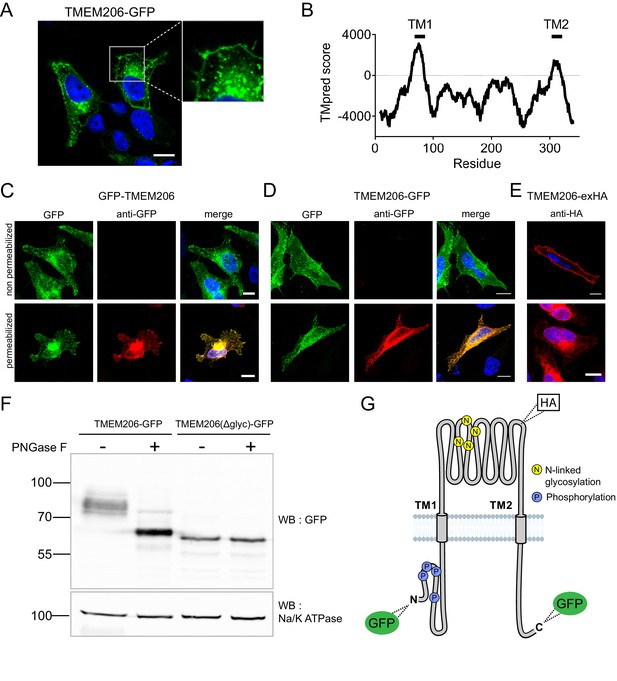
Subcellular localization and transmembrane topology of TMEM206.
(A) Subcellular localization of TMEM206 (fused to GFP at the C-terminus) in transfected HeLa cells. A similar localization was observed when the tag was attached to the N-terminus. Scale bar: 10 μm (B) Hydropathy analysis of TMEM206 using the TMpred server (https://embnet.vital-it.ch/software/TMPRED_form.html) suggests the presence of two transmembrane domains. (C, D) Detection of GFP by its fluorescence (green) or immunocytochemistry (red) in cells transfected with GFP-TMEM206 (C) or TMEM206-GFP (D), without (top panels) or with (lower panels) plasma membrane permeabilization. Scale bars: 10 μm. (E) A HA-epitope inserted after residue 271 between TM1 and TM2 (G) in the TMEM206-exHA mutant was detected in non-permeabilized cells. (F) Western blot of membranes from HEK cells transfected with TMEM206-GFP or TMEM206(Δglyc)-GFP in which all four predicted N-linked glycosylation sites between TM1 and TM2 were disrupted by mutagenesis. Note the lower molecular weight of TMEM206(Δglyc)-GFP. Deglycosylation of membrane proteins by PNGaseF reduced the molecular weight of the WT, but not the TMEM206(Δglyc)-GFP protein. (G) Schematic topology of TMEM206. Predicted glycosylation sites (N), and phosphorylated sites (P) identified in mass spectrometry (https://www.phosphosite.org/) are indicated, as well as the location of added epitopes.
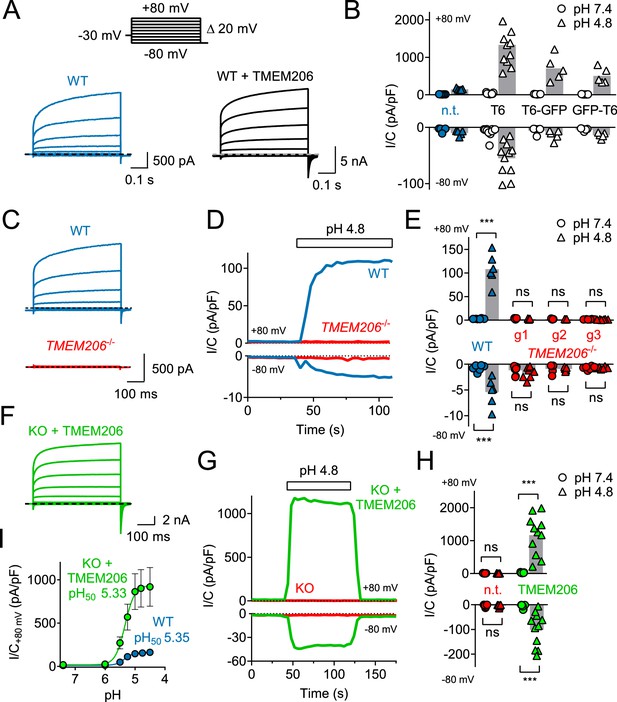
TMEM206 mediates ICl,H ASOR currents.
(A) Voltage-clamp traces of non-transfected (n.t.) and TMEM206-GFP transfected HEK cells at pHo = 4.8, using the protocol shown at the top. (B) Current densities (I/C, at indicated pHo) at +80 and −80 mV for non-transfected (n.t.) HEK cells and cells transfected with human TMEM206 (T6) fused N- or C-terminally to GFP, or co-expressing untagged TMEM206 with GFP from a separate pEGFP-N1 vector. Transfection increased current levels by 5- to 15-fold (bars, mean). (C) CRISPR-Cas9 mediated genomic disruption of TMEM206 in HEK cells (by guide RNA g1) abolished native acid-activated ASOR currents (ICl,H) as determined at pHo 4.8. Clamp protocol as in (A). (D) Outward and (smaller) inward ICl,H currents rapidly activate in WT, but not TMEM206−/− HEK cells when pHo is lowered from 7.4 to 4.8. (E) Native ICl,H of HEK cells is abolished in three independent TMEM206−/− cell lines generated with different guide RNAs (g1 – g3). (bars, mean; ***, p<0.001, one-way ANOVA with Bonferroni correction) (F) Voltage-clamp traces of TMEM206−/− HEK cells transfected with human TMEM206 reveal large ICl,H. Clamp protocol as in (A). (G) Rapid activation and deactivation of ICl,H in TMEM206-transfected TMEM206−/− HEK cells when pHo is changed between 7.4 and 4.8. Currents monitored using a ramp protocol. (H) ICl,H densities at indicated voltages and pHo of non-transfected (n.t.) or TMEM206-transfected TMEM206−/− HEK cells (bars, mean; ***, p<0.001, one-way ANOVA with Bonferroni correction). (I) pHo-dependence of ICl,H (at +80 mV) from native HEK cells and TMEM206-transfected TMEM206−/− HEK cells. pH50, pHo at which current is half maximal.
-
Figure 3—source data 1
Raw data for Figure 3.
- https://doi.org/10.7554/eLife.49187.008
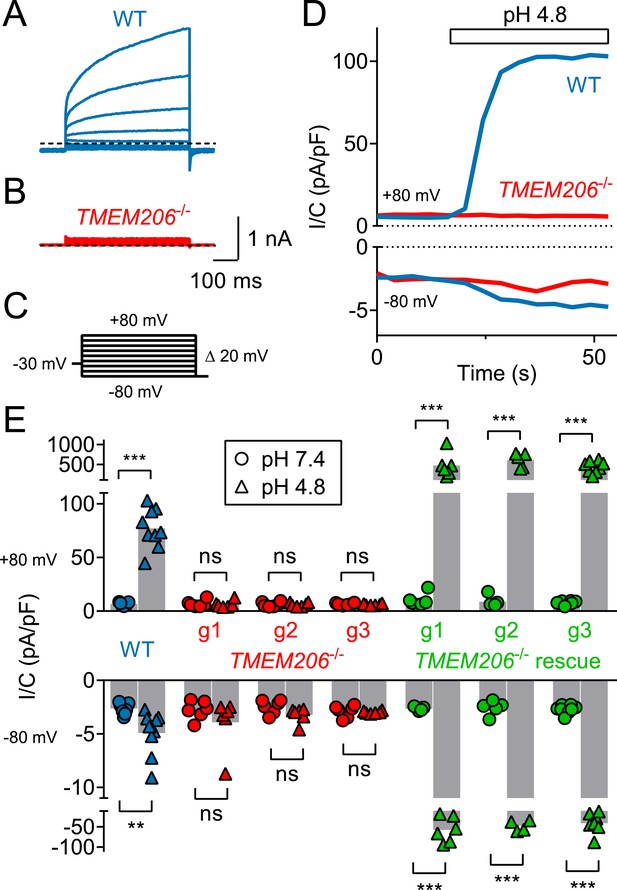
TMEM206 mediates ICl,H ASOR currents in HeLa cells.
(A–C) Voltage-clamp traces of wild type (WT, (A) and TMEM206−/− HeLa cells (B) at pHo = 4.8, using the protocol shown in (C). (D) Outward and (smaller) inward ICl,H currents rapidly activate in WT, but not TMEM206−/− HeLa cells when pHo is lowered from 7.4 to 4.8. (E) Current densities (at pHo 7.4 and pHo 4.8) at +80 and −80 mV for non-transfected WT HeLa cell, three independent TMEM206−/− clones, and TMEM206-transfected TMEM206−/− HeLa cells (rescue). Rescue current densities exceeded those in WT cells by ~10 fold (g1–g3: guide RNAs used for CRISPR-Cas9-mediated gene disruption, see Table 1). Bars, mean; **, p<0.05; ***, p<0.001; one-way ANOVA with Bonferroni correction.
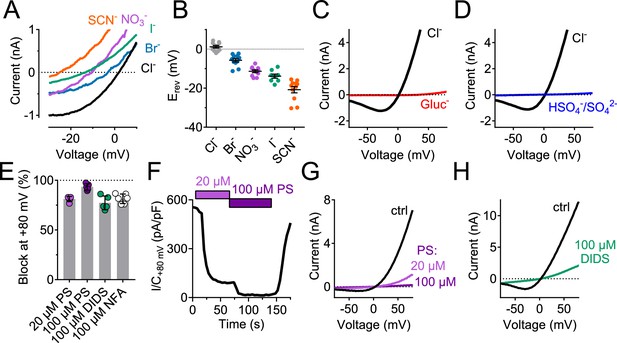
Ion selectivity and inhibitor sensitivity of TMEM206/ASOR channels.
(A) Example traces of ICl,H (elicited by voltage ramps) from TMEM206-transfected TMEM206−/− HEK cells upon equimolar replacement of extracellular NaCl (150 mM) by the sodium salts of the indicated anions. The intracellular solution contained 150 mM CsCl. The voltage at which I = 0 defines the reversal potential. (B) Reversal potentials Erev. (C) Upon replacement of extracellular Cl- by gluconate, or (D) by HSO4-/SO42-, no currents discernible from background were detected (at pHo 4.8). (E) Mean inhibition (%) of ICl,H from TMEM206-transfected TMEM206−/− HEK cells (measured at +80 mV, pHo 4.8) by various inhibitors of ASOR. PS, pregnenolone sulfate; DIDS, 4,4'-diisothiocyano-2,2'-stilbenedisulfonic acid; NFA, niflumic acid. (F) Fast and reversible block of ICl,H by PS. (G) Example I/V curves of ICl,H from TMEM206-transfected TMEM206−/− HEK cells exposed to different PS concentrations. (H) Same for block by DIDS.
-
Figure 4—source data 1
Raw data for Figure 4.
- https://doi.org/10.7554/eLife.49187.011
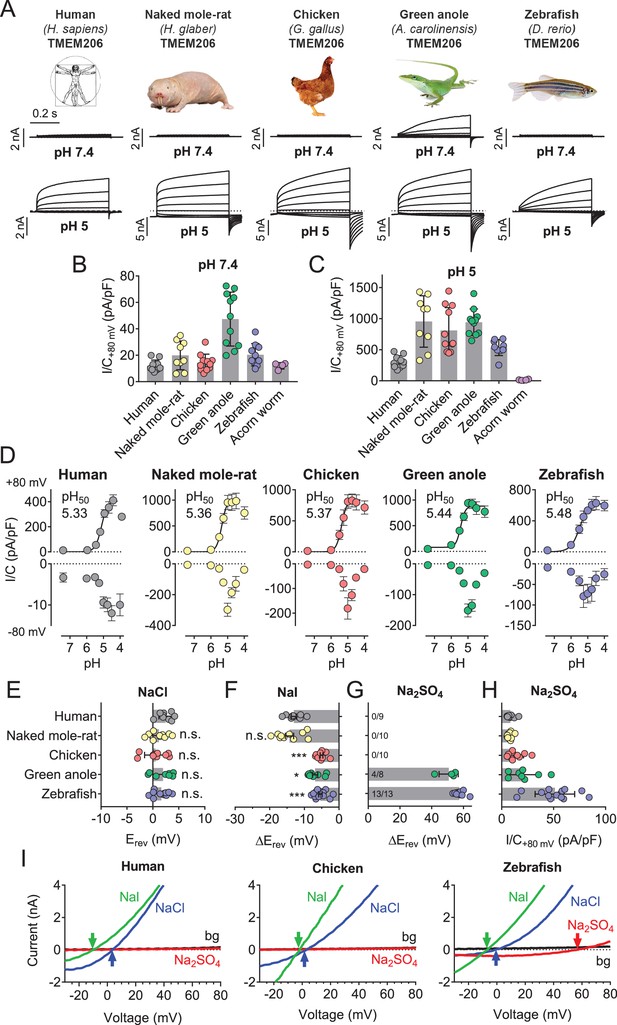
Properties of TMEM206 orthologs.
(A) Typical voltage clamp-traces of indicated TMEM206 orthologs transiently expressed in TMEM206−/− HEK cells at pHo 7.4 (top traces) or pHo 5.0 (bottom traces). Clamp protocol as in Figure 3A. (B) Mean current amplitudes of tested orthologs at pHo7.4 and (C) pHo 5.0. The ortholog from acorn worm failed to give currents because of its retention in the endoplasmic reticulum (Figure 5—figure supplement 1F). (D) pHo-dependence of currents from various orthologs at +80 mV (top) and –80 mV (bottom) as determined from voltage ramps. (E–G) Reversal potentials Erev of indicated orthologs with external NaCl (E), Nal (F) and Na2SO4 (G). Significant currents with Na2SO4 could be measured only for green anole and zebrafish. ΔErev, difference to Erev for NaCl. All currents measured at pHo 5.25. (H) Current densities (at +80 mV, pHo 5.25) with external Na2SO4 (I) Example traces showing determination of Erev (indicated by arrows) for human, chicken and zebrafish orthologs. bg, background current at pHo 7.4 (n = 8–13 cells; *, p<0.033; ***, p<0.001; Kruskal-Wallis test, Dunn’s multiple comparison correction; error bars, SD (B,C) or SEM (D–H)).
-
Figure 5—source data 1
Raw data for Figure 5.
- https://doi.org/10.7554/eLife.49187.014
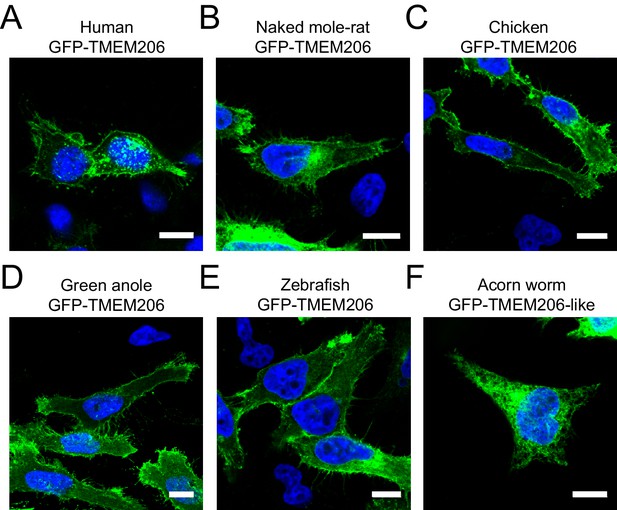
Subcellular localization of different GFP-tagged TMEM206 orthologs after transfection into HeLa TMEM206−/− cells.
All proteins were fused with GFP at the N-terminus to allow their detection by fluorescence. Scale bars: 10 μm.
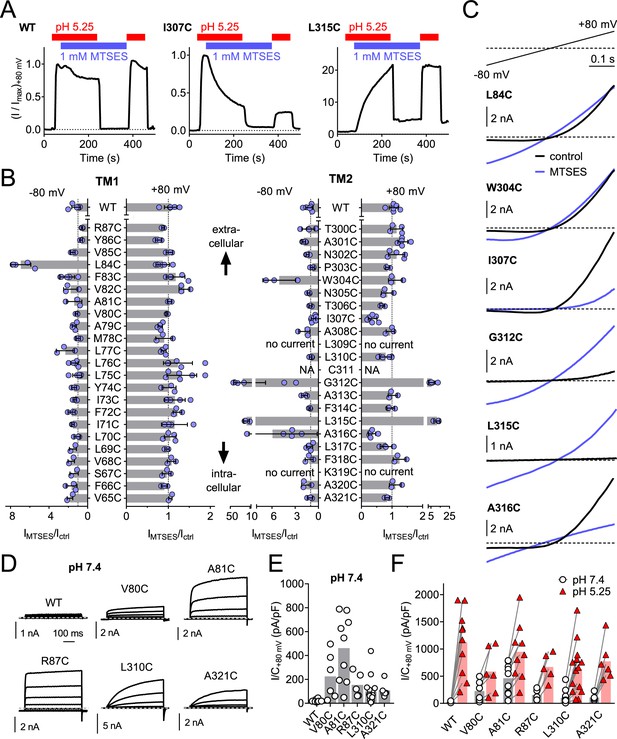
Substituted cysteine accessibility scan of TMEM206 transmembrane domains.
(A) Effect of MTSES on currents from WT and mutant TMEM206-transfected TMEM206−/− cells. MTSES had no effect on WT TMEM206, but acutely and irreversibly decreased or increased ICl,H of I307C and L315C mutants, respectively. Note that acidic pHo does not interfere with MTSES reactivity. The protocol was designed to be able to detect potential effects of MTSES at pH 7.4. (B) Ratio of current before (Ictrl) and after (IMTSES) exposure to MTSES, at +80 and −80 mV. Mutants are grouped by location within the two TMDs of TMEM206. For current amplitudes of non-modified cysteine mutants, see Figure 6—figure supplement 1. (C) Current traces of selected mutants elicited by voltage ramps (top) before and after MTSES-application. (D–F) Cysteine mutants showing currents at pHo 7.4. (D) Voltage-clamp traces of WT TMEM206 and indicated mutants at pH 7.4. Clamp protocol as in Figure 3A. (E) Mean current densities at pH 7.4. (F) All mutants still responded to low pHo.
-
Figure 6—source data 1
Raw data for Figure 6.
- https://doi.org/10.7554/eLife.49187.018
Acid-induced current densities of cysteine mutants.
(A, B) Maximal current densities (I/C) at +80 mV and pH 5.25 for all cysteine mutants in transmembrane domains 1 (TM1) and 2 (TM2) are shown. Bars, mean.
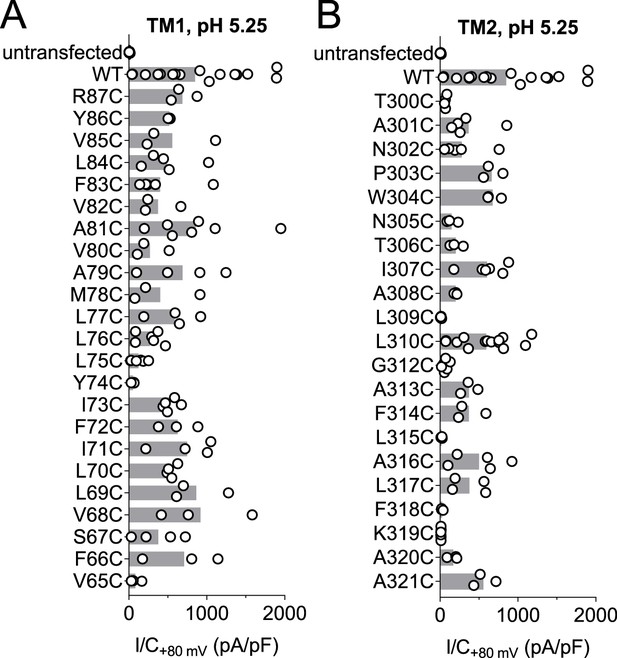
Localization of studied TMEM206 residues.
(A) Visualization of the human TMEM206 protein (using software provided by http://wlab.ethz.ch/protter) and helical wheel diagrams of TM1 and TM2. Selected residues are color-coded as explained. In helical wheels, (N) and (C) label the N- and C-terminal end of the displayed polypeptide segments. (B) Alignment of protein sequences encoding TM1 and TM2 from the TMEM206 orthologs studied in this work. Residues color-coded as in (A). With the exception of R87, all residues found to have interesting functional effects on TMEM206/ASOR currents are conserved.
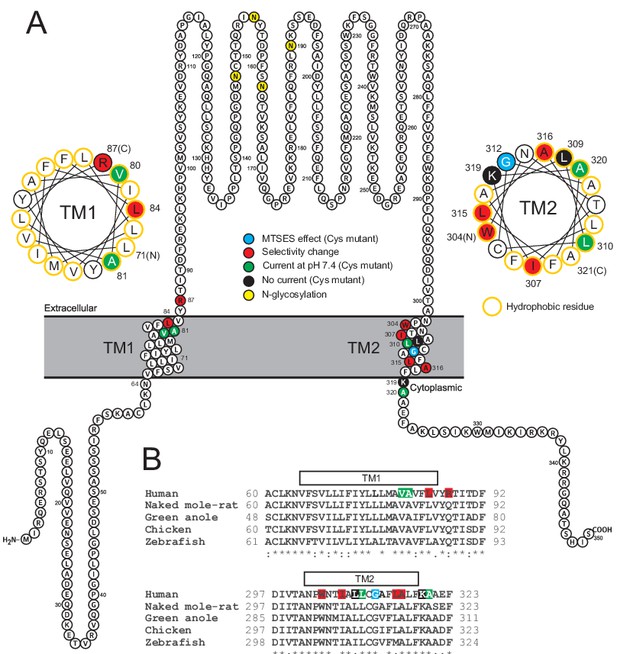
Ion selectivity changes of mutants in transmembrane domains.
(A–D) Current traces of WT (A), L84C (B), I307W (C) and L315D (D) TMEM206 expressed in TMEM206−/− HEK cells measured with indicated extracellular solutions at pH 5.25. Reversal potentials Erev indicated by arrows. With WT and L315D TMEM206 currents in Na2SO4 were too small for Erev determination. Currents were elicited by voltage ramps as in Figure 6C. (E) Reversal potential Erev with extracellular NaCl (***, p<0.001 vs. WT; one-way ANOVA, Bonferroni correction). (F, G) Shift of reversal potentials (ΔErev) with extracellular NaI (F) or Na2SO4 (G) relative to that measured with NaCl (NA, not applicable (currents not significantly above background or Erev not stable over time (I307C)). (H) Current densities at +80 mV with extracellular Na2SO4 at pH 5.25. *, p<0.033 and ***, p<0.001 vs. WT; one-way ANOVA, Bonferroni correction. No statistical analysis was performed on the data in (G).
-
Figure 7—source data 1
Raw data for Figure 7.
- https://doi.org/10.7554/eLife.49187.021
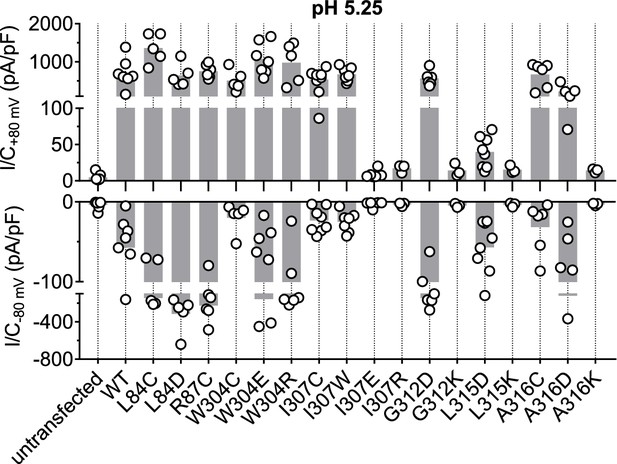
Acid-induced current densities of TMEM206 mutants.
Maximal current densities (I/C) at +80 mV and −80 mV and pH 5.25 for the indicated mutants. Note segmented y axis. Bars, mean.
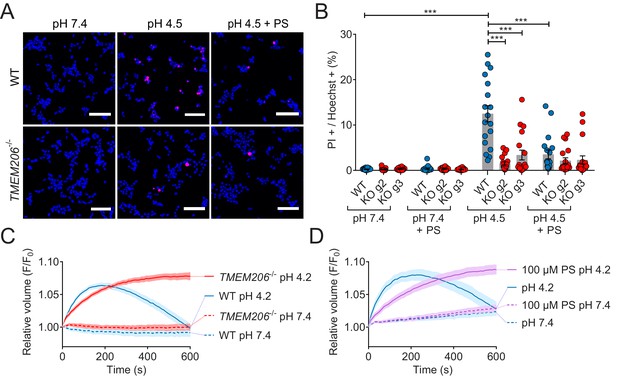
Role of TMEM206/ASOR channels in acid-induced cell death and volume regulation.
(A) Representative pictures of WT and TMEM206−/− HEK293 cells (clone 2E5) double stained with propidium iodide (PI, red) and Hoechst 33342 (blue) after incubation for 2 hr with control (pH 7.4) or acidic (pH 4.5) solution, also containing the potent inhibitor of TMEM206 pregnenolone sulfate (PS) at 100 µM or not. In this assay, dead cells are stained with PI, membrane impermeant, whereas Hoechst 33342, which is permeant, stains nuclei of all cells. Scale bars: 100 µm (B) Quantification of PI positive cells over the total number of cells, determined by Hoechst staining, in WT and two independent TMEM206−/− HEK clones generated with different guide RNAs (g2, g3). For quantification, 10x objective microscopic fields were randomly chosen. Bars represent mean ± SEM. Data were acquired in three independent experiments and analyzed by using one-way ANOVA with post hoc multiple comparisons using the Tukey’s test: ***p<0.001 (C,D) Influence of TMEM206 ablation (C) or block by 100 µM pregnenolone sulfate (PS) (D) on acid-induced cell volume changes of HEK293 cells as monitored by calcein fluorescence. Mean of eight measurements; error range, SEM. Similar results were obtained in three experiments, and similar effects were observed with HeLa cells (Figure 8—figure supplement 1).
-
Figure 8—source data 1
Raw data for Figure 8.
- https://doi.org/10.7554/eLife.49187.024
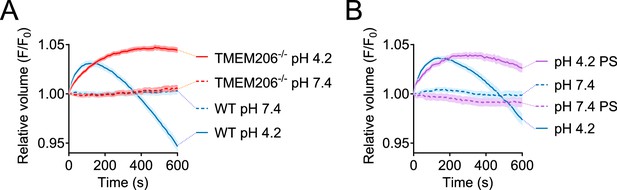
Role of TMEM206/ASOR ablation or block on acid-induced cell volume changes in HeLa cells.
Influence of TMEM206 ablation (A) or block by 100 µM pregnenolone sulfate (PS) (B) on acid-induced cell volume changes of HeLa cells as monitored by calcein fluorescence. Mean of eight measurements; error range, SEM.
Tables
TMEM206 Knockout Cell Lines.
https://doi.org/10.7554/eLife.49187.009| Cell line | Clone | sgRNA sequence | Genetic modification | Protein modification | Figures |
|---|---|---|---|---|---|
| HEK293 TMEM206−/− | 1D4 | CAGCTGTAAGCACCATTACG | a1: duplication of a395 a2: Δa395 | 1: Y132stop between TMD1 and TMD2 2: Y132S-fs between TMD1 and TMD2 | 3E |
| 2E5 | GGACCGAGAAGACGTTCTTC | Homozygous Δ2nt (g189-a190) | N64R-fs before TMD1 | 3 C-H; 4; 5; 6; 6-S1; 7; 7-S1; 8; | |
| 3D9 | AAGGAGACGGTCAGAGTCCA | Homozygous duplication of t110 | Q38P-fs before TMD1 | 3E; 8A-B | |
| HeLa TMEM206−/− | 1G1 | CAGCTGTAAGCACCATTACG | duplication of a395 | Y132stop between TMD1 and TMD2 | 3-S1 |
| 2C6 | GGACCGAGAAGACGTTCTTC | a1: Δ11nt (g178-a188) a2: duplication of g189 | 1: A60E-fs 2: N64E-fs before TMD1 | 3-S1 | |
| 3F8 | AAGGAGACGGTCAGAGTCCA | duplication of t110 | Q38P-fs before TMD1 | 2A,C,D; 8C,D; 3-S1 |
-
a = allele; fs = frameshift; nt = nucleotide; TMD = transmembrane domain; Δ deletion.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 WT | Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany | ACC No. 305; RRID: CVCL_0045 | |
| Cell line (Homo sapiens) | HEK293 TMEM206-/- | this paper | generated from WT HEK293, different clones (Table 1) | |
| Cell line (Homo sapiens) | HeLa WT | Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany | ACC No. 57; RRID: CVCL_0030 | |
| Cell line (Homo sapiens) | HeLa TREx | Thermo Fisher | Cat. No. R71407; RRID: CVCL_D587 | |
| Cell line (Homo sapiens) | HeLa-E2GFP-2A-FaNaC | this paper | see below for FaNaC/E2GFP | generated from HeLa TREx, clone E2F-5 |
| Cell line (Homo sapiens) | HeLa TMEM206-/- | this paper | generated from HeLa-E2GFP-2A-FaNaC, different clones (Table 1) | |
| Transfected construct (S. pyogenes) | pSpCas9(BB)−2A-GFP (PX458) | PMID: 24157548 | RRID: Addgene_48138 | |
| Gene (Homo sapiens) | Human TMEM206 | this paper | RefSeq: NM_018252.2; UniProtKB: Q9H813 | Cloned from human brain cDNA library (Clontech) |
| Gene (Heterocephalus glaber) | Naked mole-rat TMEM206 | this paper | RefSeq: XM_004853543.3; UniProtKB: A0A0N8ETT8 | human codon-optimized, synthesized by Life Technologies |
| Gene (Gallus gallus) | Chicken TMEM206 | this paper | RefSeq: XM_419431.6; UniProtKB: A0A1D5PSZ0 | human codon-optimized, synthesized by Life Technologies |
| Gene (Anolis carolinensis) | Green anole TMEM206 | this paper | RefSeq: XM_003216011.3; UniProtKB: G1KFB8 | human codon-optimized, synthesized by Life Technologies |
| Gene (Danio rerio) | Zebrafish TMEM206 | this paper | RefSeq: NM_001291762.1; UniProtKB: X1WG42 | human codon-optimized, synthesized by Life Technologies |
| Gene (Saccoglossus kowalevskii) | Acorn worm TMEM206-like | this paper | RefSeq: XM_006811230.1 | human codon-optimized, synthesized by Life Technologies |
| Gene (Cornu aspersum) | FaNaC | PMID: 7501021; PMID: 16034422 | GenBank: X92113.1; UniProtKB: Q25011 | |
| Gene (Aequorea victoria) | E2GFP (GFP S65T/T203Y) | PMID: 17434942, PMID: 20581829 | subcloned from ClopHensor (Addgene #25938) | |
| Antibody | anti-GFP (chicken polyclonal) | Aves Lab | Cat. No. GFP-1020; RRID: AB_10000240 | (1/1,000) |
| Antibody | anti-HA (rabbit monoclonal) | Cell Signaling | Cat. No. 3724;RRID: AB_1549585 | (1/1,000) |
| Antibody | anti-GFP (rabbit polyclonal) | Thermo Fisher | Cat. No. A-11122; RRID: AB_221569 | (1/1,000) |
| Antibody | anti-Na/K-ATPase (mouse monoclonal) | Millipore | Cat. No. 05–369; RRID: AB_309699 | (1/10,000) |
| Antibody | anti-rabbit AlexaFluor 633 (goat polyclonal) | Invitrogen | Cat. No. A21071; RRID: AB_2535732 | (1/2,000) |
| Antibody | anti-chicken HRP (rabbit polyclonal) | Sigma | Cat. No. A-9792; RRID: AB_258473 | (1/10,000) |
| Antibody | anti-mouse HRP Affinipure (goat polyclonal) | Jackson ImmunoResearch | Cat. No. 115-035-003; RRID: AB_10015289 | (1/10,000) |
| Sequence-based reagent | Dharmacon ON-TARGETplus human siRNA library | Horizon Discovery | Cat. No. G-105005-E2-025 | arrayed in 384-well plates |
| Chemical compound, drug | FMRFamide (H-Phe-Met-Arg-Phe-NH₂) | Bachem | Cat. No. 4001566 | 5 mM stock in ddH2O, stored at −20°C for up to 6 months |
| Chemical compound, drug | MTSES (2-Sulfonatoethyl methanethiosulfonate) | Biotium | Cat. No. 91020 | 250 mM stock in ddH2O, freshly prepared, kept on ice for up to 4 hr |
| Chemical compound, drug | Pregnenolone sulfate (PS) | Sigma | Cat. No. P162 | 50 mM stock in DMSO (stored at −20°C for up to 6 months) for electrophysiology experiments or freshly diluted in PBS for cell death analysis |
| Chemical compound, drug | 4,4′-Diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS) | Sigma | Cat. No. D3514 | 100 mM stock in DMSO, stored at −20°C for up to 6 months |
| Chemical compound, drug | Niflumic acid (NFA) | Sigma | Cat. No. N0630 | 300 mM stock in DMSO, stored at −20°C for up to 6 months |
| Chemical compound, drug | Hoechst 33342 | ImmunoChemistry Technologies | Cat. No. ICT-639 | |
| Chemical compound, drug | Propidium iodide | Thermo Fisher | Cat. No. P3566 | |
| Chemical compound, drug | Calcein AM | Thermo Fisher | Cat. No. 65-0853-39 | 10 mM stock in DMSO, stored at −20°C for up to 6 months |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49187.025





